A Cross-Tissue Investigation of Molecular Targets and Physiological Functions of Nsun6 Using Knockout Mice
Abstract
:1. Introduction
2. Results
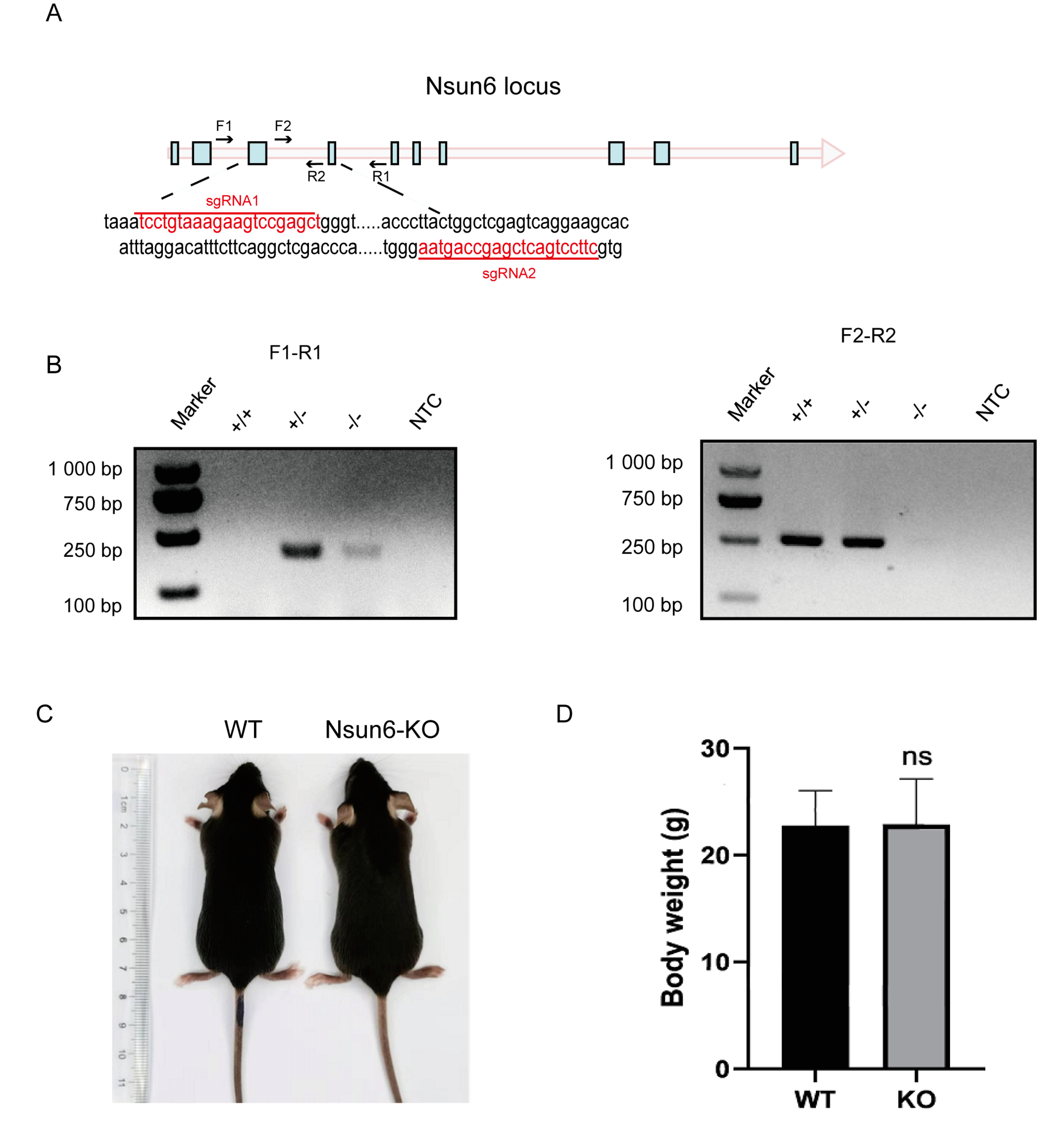
2.1. Nsun6 Gene Is Not Essential for the Development and Breeding of Mice
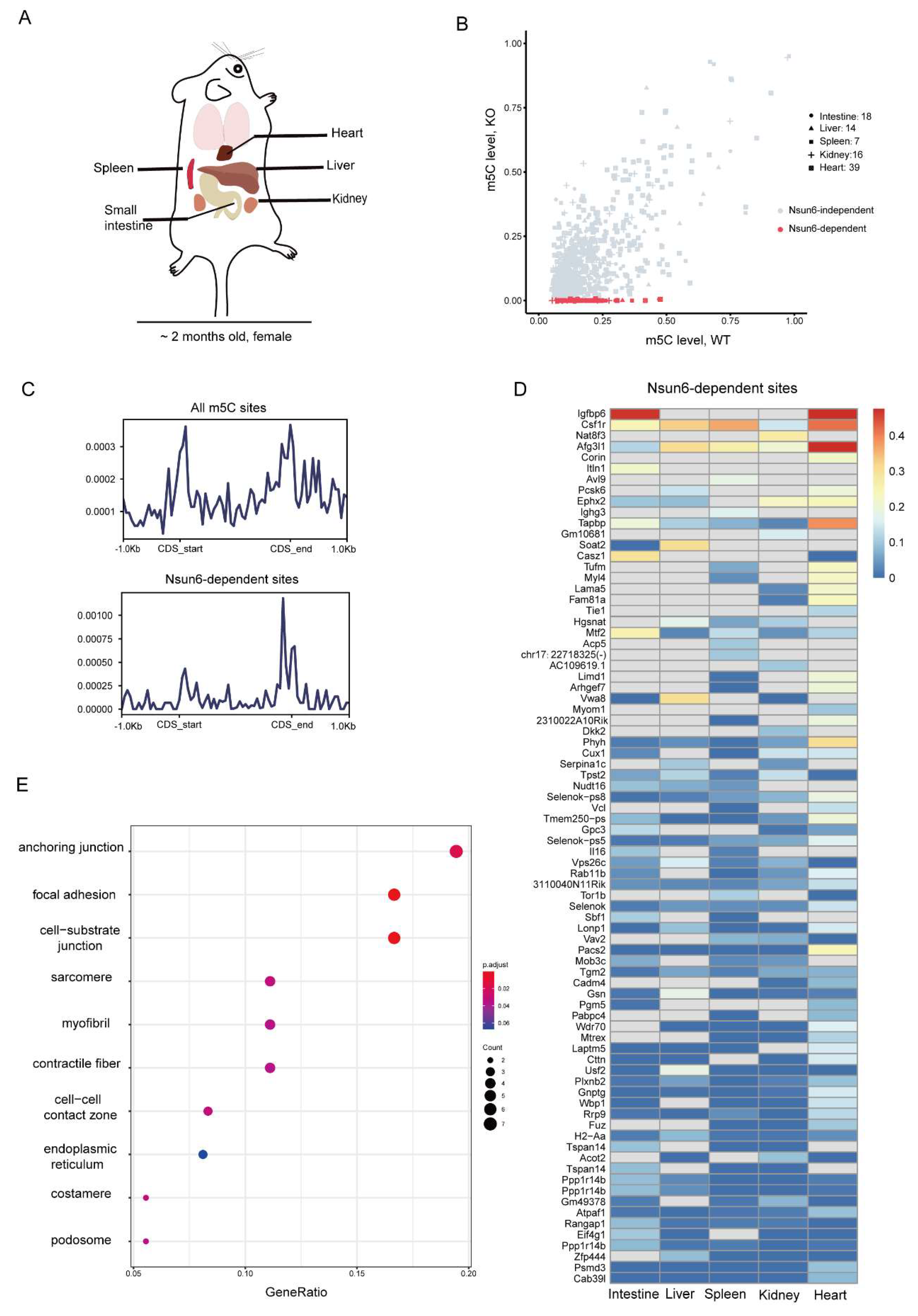
2.2. Identification of High-Confident Nsun6-Dependent m5C Sites across Tissues
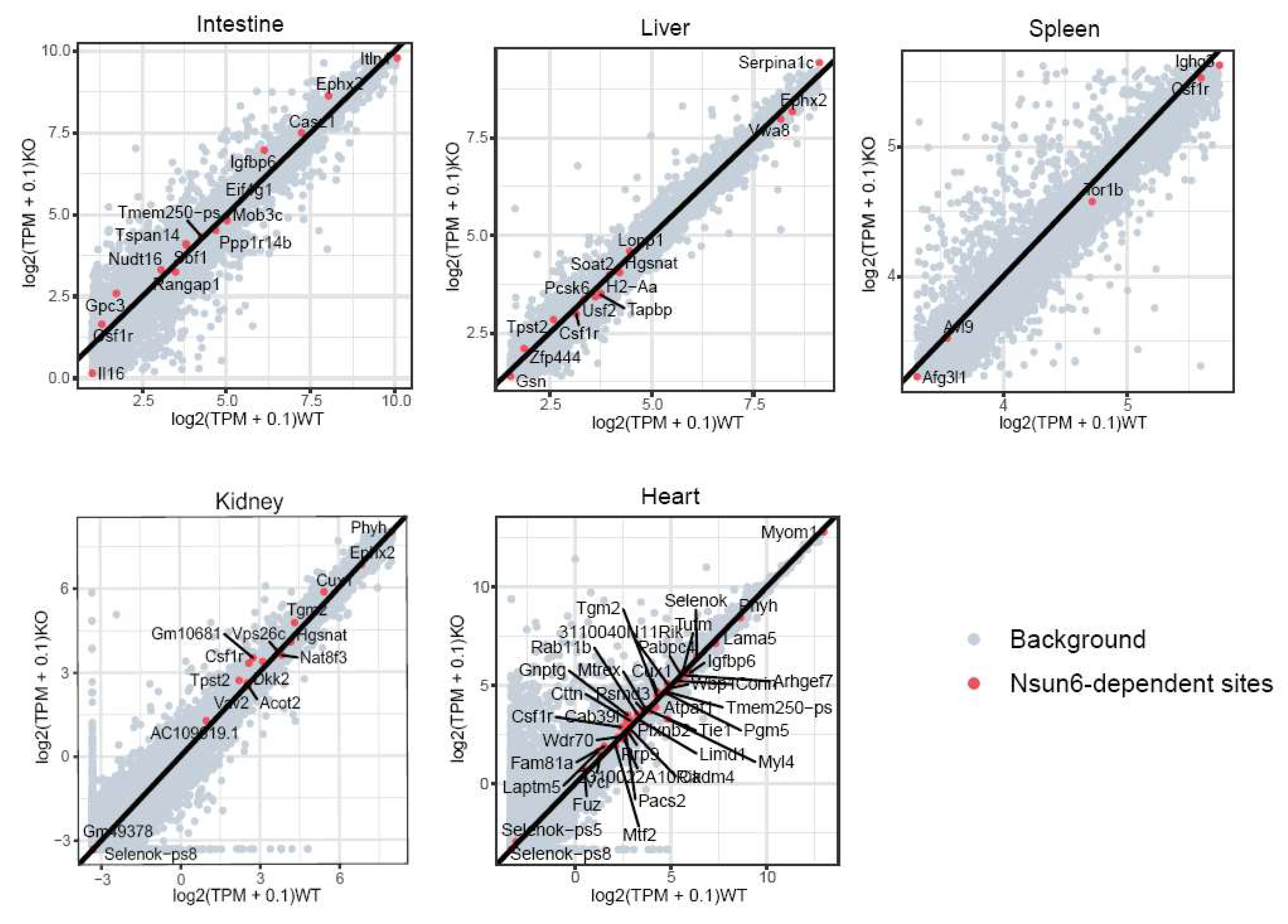
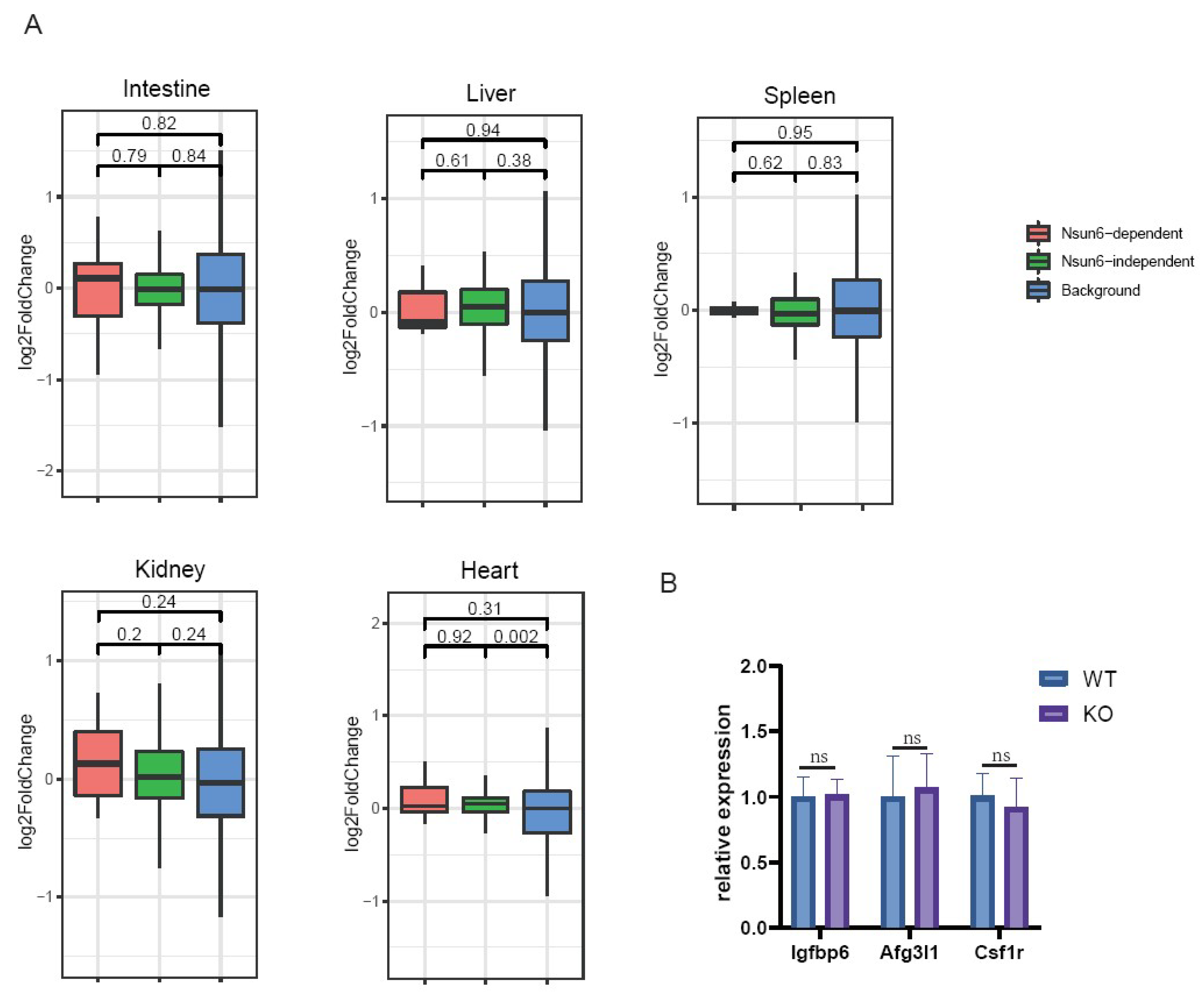
2.3. Nsun6 Deficiency Induces Limited Transcriptome Changes across Tissues
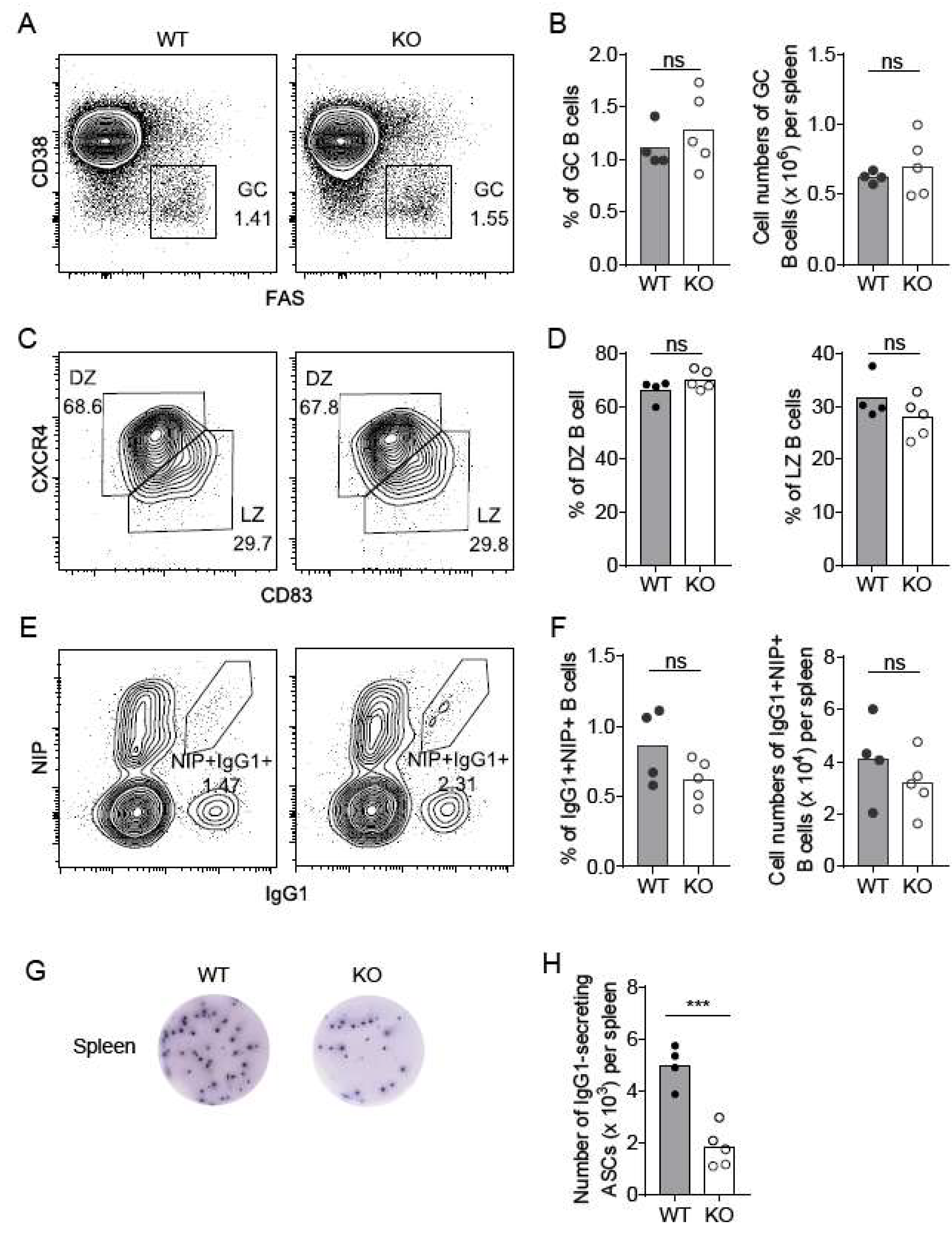
2.4. Nsun6-KO Impairs the Formation of Antibody-Secreting Plasma Cells in the Spleen
2.5. Nsun6-Mediated m5C Modification Does Not Affect RNA Stability across Tissues
3. Discussion
4. Methods
4.1. Generation of the Nsun6-Knockout Mouse Strain
4.2. Total RNA Extraction
4.3. mRNA Capture
4.4. mRNA Library Construction
4.5. mRNA Bisulfite Sequencing
4.6. Western Blot
4.7. Detection of the m5C Modification
4.8. Nsun6-Dependent Site Identification
4.9. Mice Immunization
4.10. Flow Cytometry
4.11. ELISPOT Assay
4.12. RNA-seq Analysis
4.13. t-SNE Visualization
4.14. GO Enrichment Analysis
4.15. GSEA Analysis
4.16. Motif Analysis of m5C Sites
4.17. Data Availability
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.S.; Yang, W.L.; Zhao, Y.L.; Yang, Y.G. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA 2021, 12, e1639. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1alpha. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heissenberger, C.; Rollins, J.A.; Krammer, T.L.; Nagelreiter, F.; Stocker, I.; Wacheul, L.; Shpylovyi, A.; Tav, K.; Snow, S.; Grillari, J.; et al. The ribosomal RNA m(5)C methyltransferase NSUN-1 modulates healthspan and oogenesis in Caenorhabditis elegans. Elife 2020, 9, e56205. [Google Scholar] [CrossRef]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef] [Green Version]
- Van Haute, L.; Dietmann, S.; Kremer, L.; Hussain, S.; Pearce, S.F.; Powell, C.A.; Rorbach, J.; Lantaff, R.; Blanco, S.; Sauer, S.; et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016, 7, 12039. [Google Scholar] [CrossRef] [Green Version]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Ontiveros, R.J.; Owens, M.C.; Liu, M.Y.; Ghanty, U.; Kohli, R.M.; Liu, K.F. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J. Biol. Chem. 2021, 296, 100087. [Google Scholar] [CrossRef]
- Kawarada, L.; Suzuki, T.; Ohira, T.; Hirata, S.; Miyauchi, K.; Suzuki, T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017, 45, 7401–7415. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, L.; Han, X.; Yang, W.L.; Zhang, M.; Ma, H.L.; Sun, B.F.; Li, A.; Xia, J.; Chen, J.; et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol. Cell 2019, 75, 1188–1202.e1111. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Bandiera, R.; Popis, M.; Hussain, S.; Lombard, P.; Aleksic, J.; Sajini, A.; Tanna, H.; Cortes-Garrido, R.; Gkatza, N.; et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016, 534, 335–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco, S.; Kurowski, A.; Nichols, J.; Watt, F.M.; Benitah, S.A.; Frye, M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011, 7, e1002403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, S.; Tuorto, F.; Menon, S.; Blanco, S.; Cox, C.; Flores, J.V.; Watt, S.; Kudo, N.R.; Lyko, F.; Frye, M. The Mouse Cytosine-5 RNA Methyltransferase NSun2 Is a Component of the Chromatoid Body and Required for Testis Differentiation. Mol. Cell. Biol. 2013, 33, 1561–1570. [Google Scholar] [CrossRef] [Green Version]
- Haag, S.; Warda, A.S.; Kretschmer, J.; Gunnigmann, M.A.; Hobartner, C.; Bohnsack, M.T. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 2015, 21, 1532–1543. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.J.; Long, T.; Li, J.; Li, H.; Wang, E.D. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017, 45, 6684–6697. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, H.; Long, T.; Dong, H.; Wang, E.D.; Liu, R.J. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019, 47, 2041–2055. [Google Scholar] [CrossRef]
- Fang, L.; Wang, W.; Li, G.; Zhang, L.; Li, J.; Gan, D.; Yang, J.; Tang, Y.; Ding, Z.; Zhang, M.; et al. CIGAR-seq, a CRISPR/Cas-based method for unbiased screening of novel mRNA modification regulators. Mol. Syst. Biol. 2020, 16, e10025. [Google Scholar] [CrossRef]
- Liu, J.; Huang, T.; Zhang, Y.; Zhao, T.; Zhao, X.; Chen, W.; Zhang, R. Sequence- and structure-selective mRNA m(5)C methylation by NSUN6 in animals. Natl. Sci. Rev. 2021, 8, nwaa273. [Google Scholar] [CrossRef]
- Selmi, T.; Hussain, S.; Dietmann, S.; Heiss, M.; Borland, K.; Flad, S.; Carter, J.M.; Dennison, R.; Huang, Y.L.; Kellner, S.; et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021, 49, 1006–1022. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Li, B.; Luo, Y.X.; Lin, Q.; Liu, S.R.; Zhang, X.Q.; Zhou, H.; Yang, J.H.; Qu, L.H. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018, 22, 286–298. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wang, S.; Xing, Z.; Lin, A.; Liang, K.; Song, J.; Hu, Q.; Yao, J.; Chen, Z.; Park, P.K.; et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat. Cell Biol. 2017, 19, 106–119. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.; Chen, W.; Liu, J.; Gu, N.; Zhang, R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 2019, 26, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.M.; Williams, A.; Eisenbarth, S.C. Structure and function of the immune system in the spleen. Sci. Immunol. 2019, 4, eaau6085. [Google Scholar] [CrossRef] [PubMed]
- Suan, D.; Sundling, C.; Brink, R. Plasma cell and memory B cell differentiation from the germinal center. Curr. Opin. Immunol. 2017, 45, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, A.; Sun, B.F.; Yang, Y.; Han, Y.N.; Yuan, X.; Chen, R.X.; Wei, W.S.; Liu, Y.C.; Gao, C.C.; et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Schumann, U.; Zhang, H.N.; Sibbritt, T.; Pan, A.; Horvath, A.; Gross, S.; Clark, S.J.; Yang, L.; Preiss, T. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 2020, 18, 40. [Google Scholar] [CrossRef] [Green Version]
- Wagih, O. ggseqlogo: A versatile R package for drawing sequence logos. Bioinformatics 2017, 33, 3645–3647. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Huang, H.; Jiang, H.; Tian, C.; Tang, Y.; Gan, D.; Wen, X.; Song, Z.; He, Y.; Ou, X.; et al. A Cross-Tissue Investigation of Molecular Targets and Physiological Functions of Nsun6 Using Knockout Mice. Int. J. Mol. Sci. 2022, 23, 6584. https://doi.org/10.3390/ijms23126584
Wang W, Huang H, Jiang H, Tian C, Tang Y, Gan D, Wen X, Song Z, He Y, Ou X, et al. A Cross-Tissue Investigation of Molecular Targets and Physiological Functions of Nsun6 Using Knockout Mice. International Journal of Molecular Sciences. 2022; 23(12):6584. https://doi.org/10.3390/ijms23126584
Chicago/Turabian StyleWang, Wen, Hengjun Huang, Hao Jiang, Chi Tian, Yisen Tang, Diwen Gan, Xiaozhen Wen, Zhenyu Song, Yuhao He, Xijun Ou, and et al. 2022. "A Cross-Tissue Investigation of Molecular Targets and Physiological Functions of Nsun6 Using Knockout Mice" International Journal of Molecular Sciences 23, no. 12: 6584. https://doi.org/10.3390/ijms23126584






