Kinase Activity of PAR1b, Which Mediates Nuclear Translocation of the BRCA1 Tumor Suppressor, Is Potentiated by Nucleic Acid-Mediated PAR1b Multimerization
Abstract
:1. Introduction
2. Results
2.1. Nucleic Acids Mediate Multimerization of PAR1b
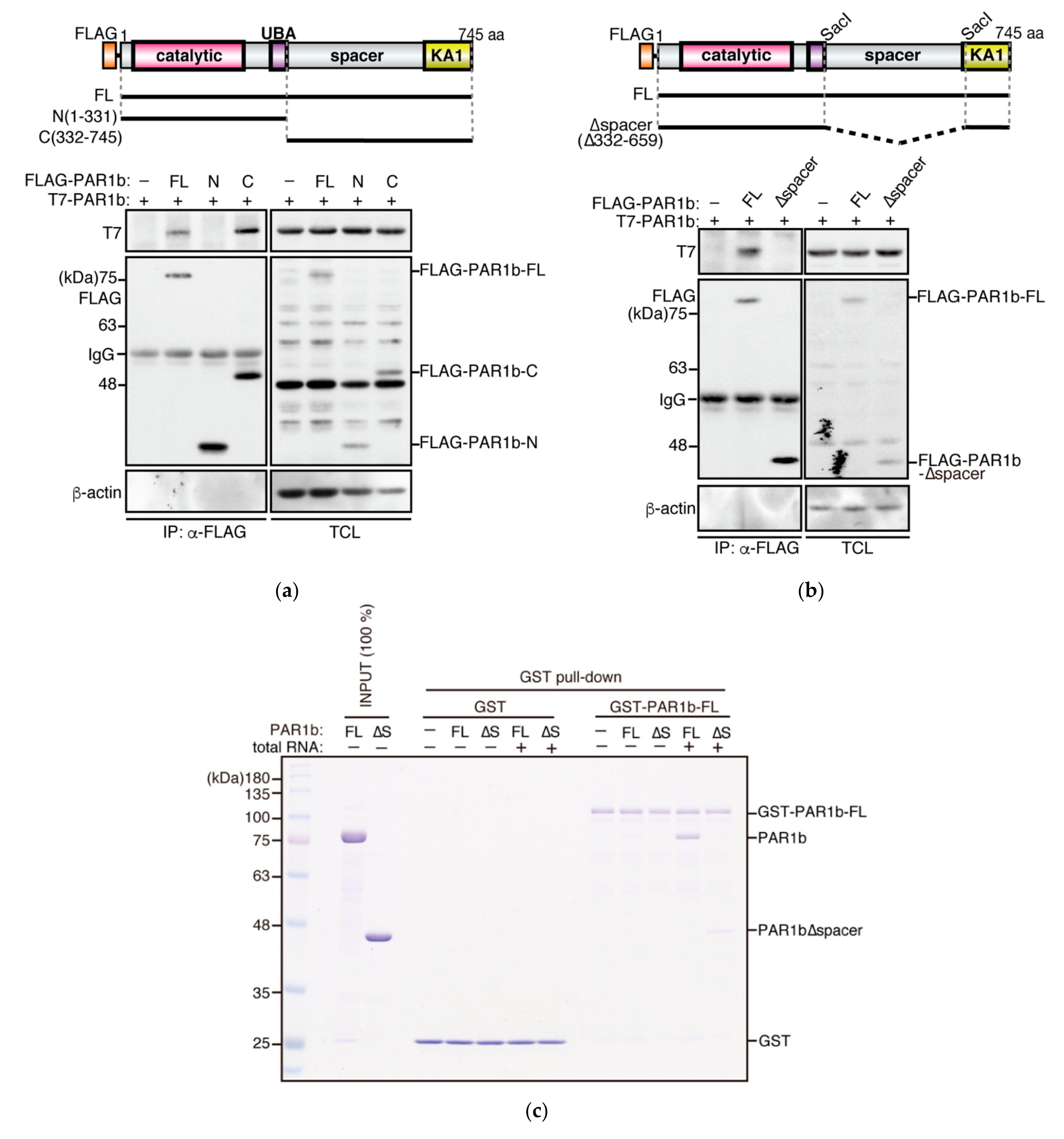
2.2. PAR1b Multimerizes through the Spacer Region
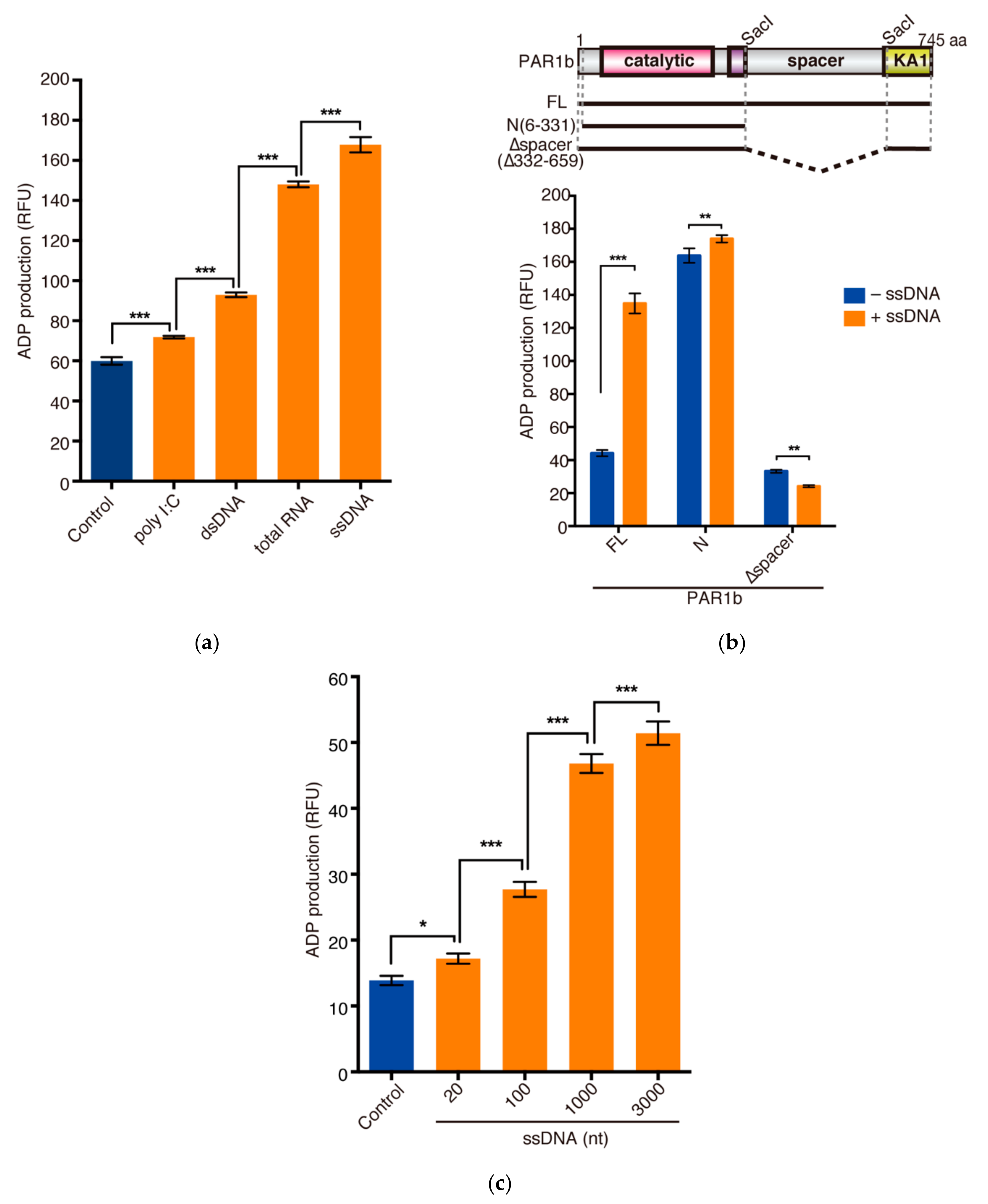
2.3. Nucleic Acids Potentiate the Kinase Activity of PAR1b through the Spacer Region
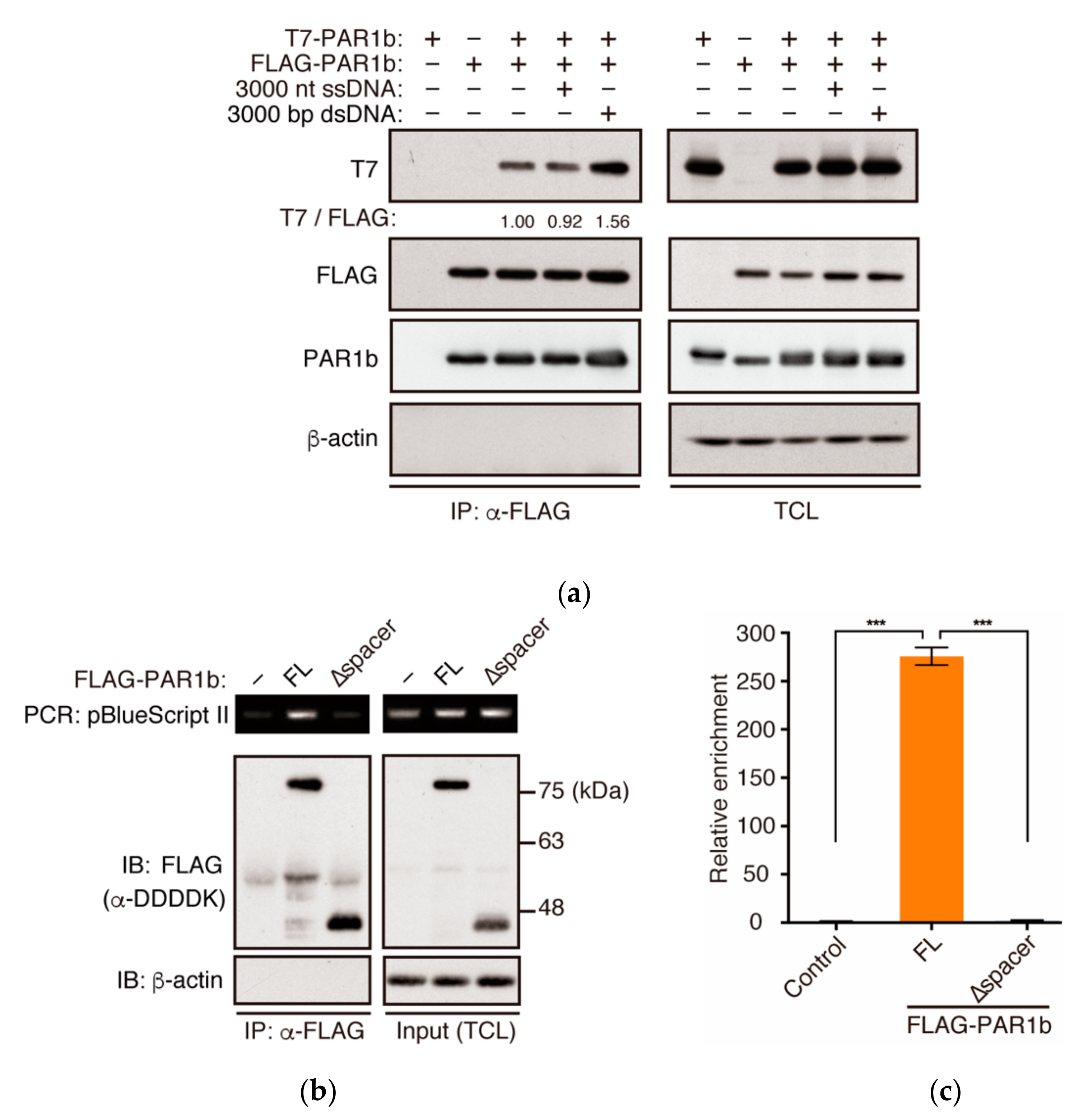
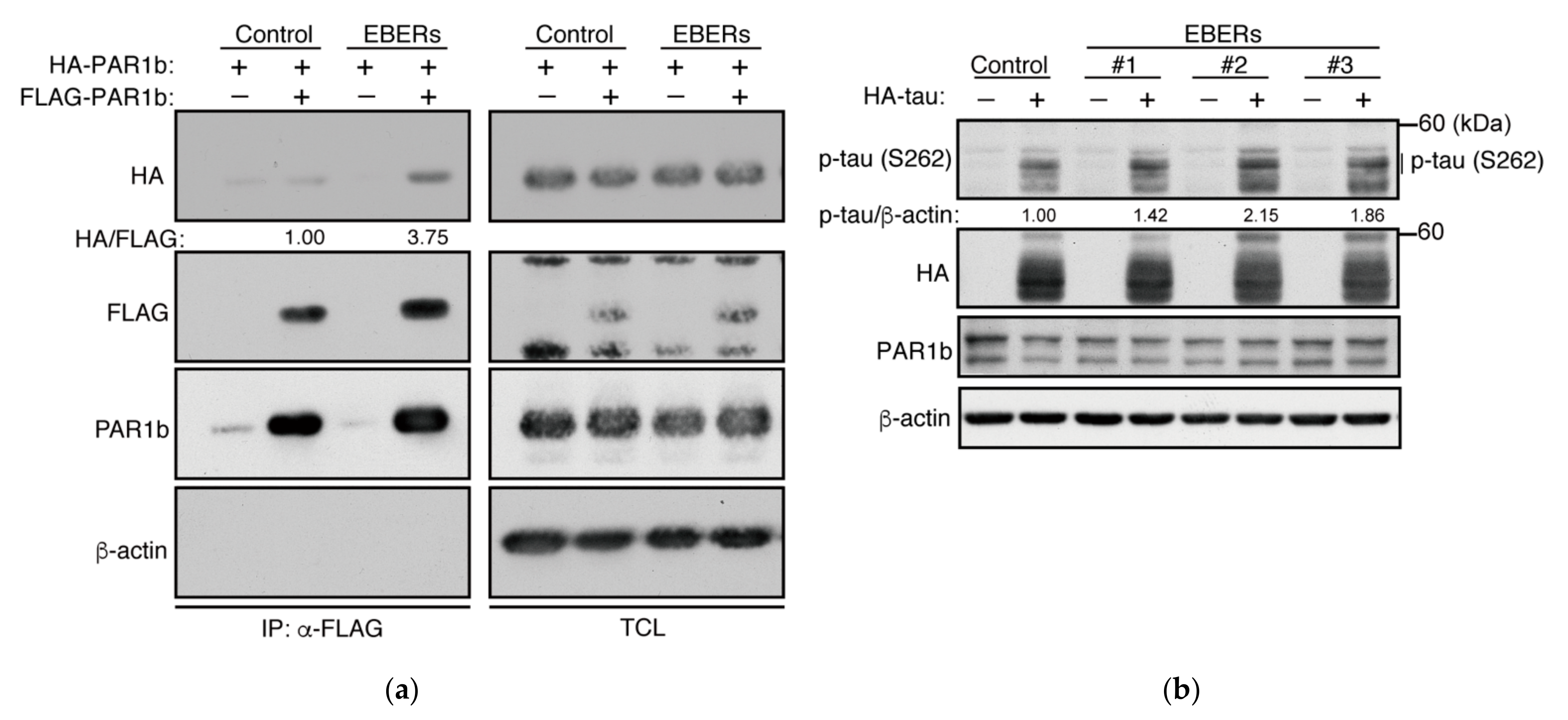
2.4. PAR1b Can Multimerize in a DNA-Dependent Manner in Cells
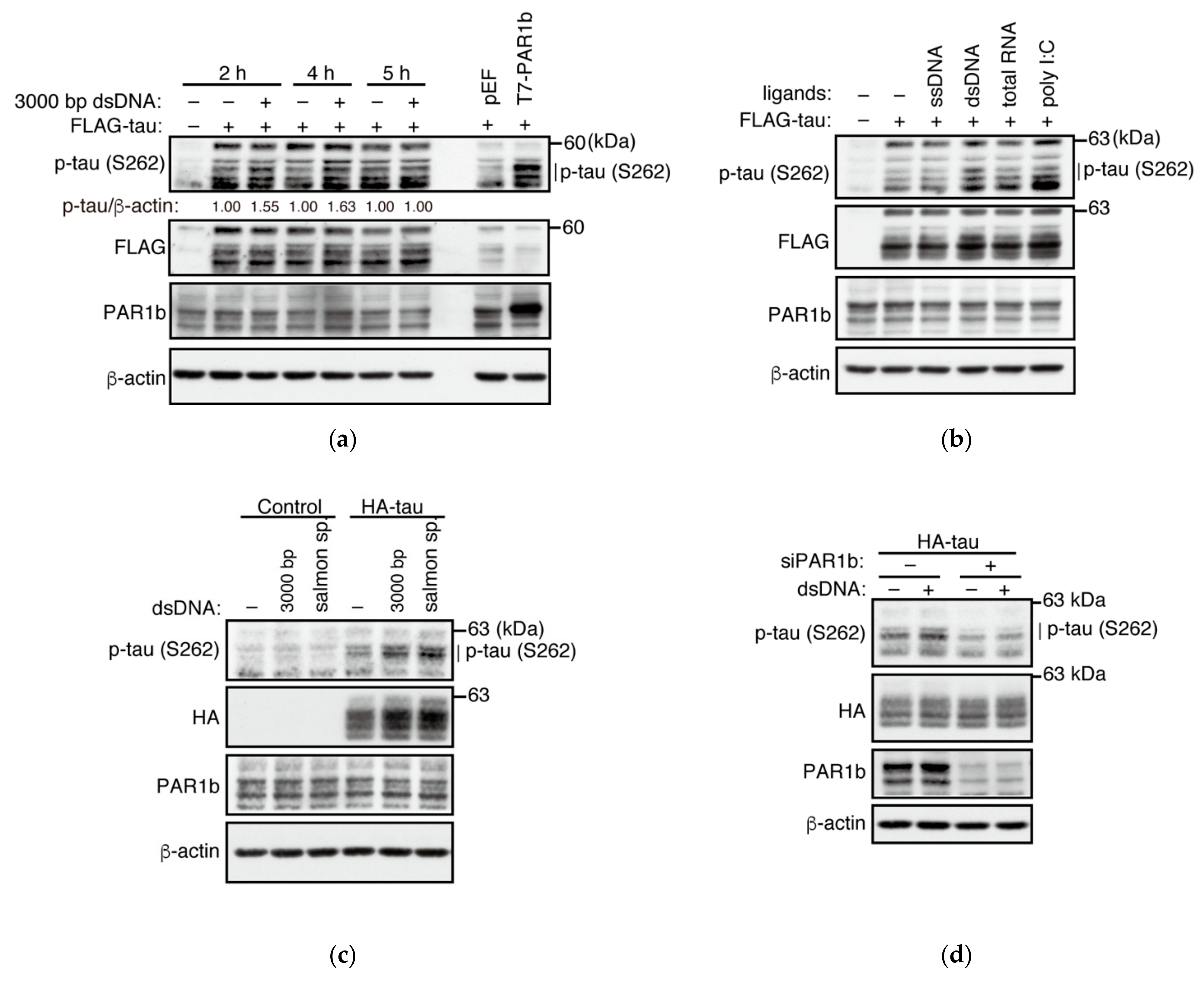
2.5. dsDNA Potentiates the Kinase Activity of PAR1b
2.6. The Effect of Epstein–Barr Virus EBERs on PAR1b
3. Discussion
4. Materials and Methods
4.1. Bacterial Expression Vectors
4.2. Antibodies
4.3. Protein Expression and Purification
4.4. GST Pull-Down Assay
4.5. Nucleic Acid Ligands
4.6. In Vitro Kinase Assay
4.7. Mammalian Expression Vectors
4.8. Cell Culture and Transfection
4.9. Generation of Stably Transfected Cell Lines
4.10. Immunoprecipitation and Immunoblotting
4.11. DNA-Immunoprecipitation (DNA-IP)
4.12. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marx, A.; Nugoor, C.; Panneerselvam, S.; Mandelkow, E. Structure and Function of Polarity-Inducing Kinase Family MARK/Par-1 within the Branch of AMPK/Snf1-Related Kinases. FASEB J. 2010, 24, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Ooki, T.; Murata-Kamiya, N.; Komura, D.; Tahmina, K.; Wu, W.; Takahashi-Kanemitsu, A.; Knight, C.T.; Kunita, A.; Suzuki, N.; et al. Helicobacter pylori CagA Elicits BRCAness to Induce Genome Instability That May Underlie Bacterial Gastric Carcinogenesis. Cell Host Microbe 2021, 29, 941–958. [Google Scholar] [CrossRef] [PubMed]
- Böhm, H.; Brinkmann, V.; Drab, M.; Henske, A.; Kurzchalia, T.V. Mammalian Homologues of C. elegans PAR-1 Are Asymmetrically Localized in Epithelial Cells and May Influence Their Polarity. Curr. Biol. 1997, 7, 603–606. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, A.; Hirata, M.; Kamimura, K.; Maniwa, R.; Yamanaka, T.; Mizuno, K.; Kishikawa, M.; Hirose, H.; Amano, Y.; Izumi, N.; et al. aPKC Acts Upstream of PAR-1b in Both the Establishment and Maintenance of Mammalian Epithelial Polarity. Curr. Biol. 2004, 14, 1425–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drewes, G.; Trinczek, B.; Illenberger, S.; Biernat, J.; Schmitt-Ulms, G.; Meyer, H.E.; Mandelkow, E.-M.; Mandelkow, E. Microtubule-Associated Protein/Microtubule Affinity-Regulating Kinase (P110mark): A Novel Protein Kinase That Regulates Tau-Microtubule Interactions and Dynamic Instability by Phosphorylation at the Alzheimer-Specific Site Serine 262. J. Biol. Chem. 1995, 270, 7679–7688. [Google Scholar] [CrossRef] [Green Version]
- Drewes, G.; Ebneth, A.; Preuss, U.; Mandelkow, E.-M.; Mandelkow, E. MARK, a Novel Family of Protein Kinases That Phosphorylate Microtubule-Associated Proteins and Trigger Microtubule Disruption. Cell 1997, 89, 297–308. [Google Scholar] [CrossRef]
- Wang, Y.; Mandelkow, E. Tau in Physiology and Pathology. Nat. Rev. Neurosci. 2016, 17, 22–35. [Google Scholar] [CrossRef]
- Timm, T.; Li, X.-Y.; Biernat, J.; Jiao, J.; Mandelkow, E.; Vandekerckhove, J.; Mandelkow, E.-M. MARKK, a Ste20-like Kinase, Activates the Polarity-inducing Kinase MARK/PAR-1. EMBO J. 2003, 22, 5090–5101. [Google Scholar] [CrossRef] [Green Version]
- Lizcano, J.M.; Göransson, O.; Toth, R.; Deak, M.; Morrice, N.A.; Boudeau, J.; Hawley, S.A.; Udd, L.; Mäkelä, T.P.; Hardie, D.G.; et al. LKB1 Is a Master Kinase That Activates 13 Kinases of the AMPK Subfamily, Including MARK/PAR-1. EMBO J. 2004, 23, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Timm, T.; Balusamy, K.; Li, X.; Biernat, J.; Mandelkow, E.; Mandelkow, E.-M. Glycogen Synthase Kinase (GSK) 3β Directly Phosphorylates Serine 212 in the Regulatory Loop and Inhibits Microtubule Affinity-Regulating Kinase (MARK) 2. J. Biol. Chem. 2008, 283, 18873–18882. [Google Scholar] [CrossRef] [Green Version]
- Matenia, D.; Griesshaber, B.; Li, X.; Thiessen, A.; Johne, C.; Jiao, J.; Mandelkow, E.; Mandelkow, E.-M. PAK5 Kinase Is an Inhibitor of MARK/Par-1, Which Leads to Stable Microtubules and Dynamic Actin. Mol. Biol. Cell 2005, 16, 4410–4422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nešić, D.; Miller, M.C.; Quinkert, Z.T.; Stein, M.; Chait, B.T.; Stebbins, C.E. Helicobacter pylori CagA Inhibits PAR1-MARK Family Kinases by Mimicking Host Substrates. Nat. Struct. Mol. Biol. 2010, 17, 130–132. [Google Scholar] [CrossRef] [PubMed]
- Saadat, I.; Higashi, H.; Obuse, C.; Umeda, M.; Murata-Kamiya, N.; Saito, Y.; Lu, H.; Ohnishi, N.; Azuma, T.; Suzuki, A.; et al. Helicobacter pylori CagA Targets PAR1/MARK Kinase to Disrupt Epithelial Cell Polarity. Nature 2007, 447, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Emptage, R.P.; Lemmon, M.A.; Ferguson, K.M. Molecular Determinants of KA1 Domain-Mediated Autoinhibition and Phospholipid Activation of MARK1 Kinase. Biochem. J. 2017, 474, 385–398. [Google Scholar] [CrossRef]
- Emptage, R.P.; Lemmon, M.A.; Ferguson, K.M.; Marmorstein, R. Structural Basis for MARK1 Kinase Autoinhibition by Its KA1 Domain. Structure 2018, 26, 1137–1143.e3. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.-R.; Tsai, P.-I.; Chen, G.-C.; Chou, H.-J.; Huang, Y.-P.; Chen, Y.-H.; Lin, M.-Y.; Kimchi, A.; Chien, C.-T.; Chen, R.-H. DAPK Activates MARK1/2 to Regulate Microtubule Assembly, Neuronal Differentiation and Tau Toxicity. Cell Death Differ. 2011, 18, 1507–1520. [Google Scholar] [CrossRef]
- Lebleu, B.; Sen, G.C.; Shaila, S.; Cabrer, B.; Lengyel, P. Interferon, Double-Stranded RNA, and Protein Phosphorylation. Proc. Natl. Acad. Sci. USA 1976, 73, 3107–3111. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.I.; Hunt, T.; Jackson, R.J.; Anderson, C.W. Double-Stranded DNA Induces the Phosphorylation of Several Proteins Including the 90 000 Mol. Wt. Heat-Shock Protein in Animal Cell Extracts. EMBO J. 1985, 4, 139–145. [Google Scholar] [CrossRef]
- Nishikawa, H.; Hayashi, T.; Arisaka, F.; Senda, T.; Hatakeyama, M. Impact of Structural Polymorphism for the Helicobacter pylori CagA Oncoprotein on Binding to Polarity-Regulating Kinase PAR1b. Sci. Rep. 2016, 6, 30031. [Google Scholar] [CrossRef]
- Hubaux, R.; Thu, K.L.; Vucic, E.A.; Pikor, L.A.; Kung, S.H.Y.; Martinez, V.D.; Mosslemi, M.; Becker-Santos, D.D.; Gazdar, A.F.; Lam, S.; et al. Microtubule Affinity-Regulating Kinase 2 Is Associated with DNA Damage Response and Cisplatin Resistance in Non-Small Cell Lung Cancer. Int. J. Cancer 2015, 137, 2072–2082. [Google Scholar] [CrossRef]
- Nagase, L.; Murata-Kamiya, N.; Hatakeyama, M. Potentiation of Helicobacter Pylori CagA Protein Virulence through Homodimerization. J. Biol. Chem. 2011, 286, 33622–33631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marx, A.; Nugoor, C.; Müller, J.; Panneerselvam, S.; Timm, T.; Bilang, M.; Mylonas, E.; Svergun, D.I.; Mandelkow, E.-M.; Mandelkow, E. Structural Variations in the Catalytic and Ubiquitin-Associated Domains of Microtubule-Associated Protein/Microtubule Affinity Regulating Kinase (MARK) 1 and MARK2. J. Biol. Chem. 2006, 281, 27586–27599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goedert, M.; Jakes, R.; Spillantini, M.G.; Hasegawa, M.; Smith, M.J.; Crowther, R.A. Assembly of Microtubule-Associated Protein Tau into Alzheimer-like Filaments Induced by Sulphated Glycosaminoglycans. Nature 1996, 383, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Kampers, T.; Friedhoff, P.; Biernat, J.; Mandelkow, E.-M.; Mandelkow, E. RNA Stimulates Aggregation of Microtubule-Associated Protein Tau into Alzheimer-like Paired Helical Filaments. FEBS Lett. 1996, 399, 344–349. [Google Scholar] [CrossRef]
- Miller, K.N.; Victorelli, S.G.; Salmonowicz, H.; Dasgupta, N.; Liu, T.; Passos, J.F.; Adams, P.D. Cytoplasmic DNA: Sources, Sensing, and Role in Aging and Disease. Cell 2021, 184, 5506–5526. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A Meta-Predictor of Intrinsically Disordered Amino Acids. Biochim. Biophys. Acta BBA Proteins Proteom. 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [Green Version]
- Preitner, N.; Quan, J.; Nowakowski, D.W.; Hancock, M.L.; Shi, J.; Tcherkezian, J.; Young-Pearse, T.L.; Flanagan, J.G. APC Is an RNA-Binding Protein, and Its Interactome Provides a Link to Neural Development and Microtubule Assembly. Cell 2014, 158, 368–382. [Google Scholar] [CrossRef] [Green Version]
- Tochio, N.; Koshiba, S.; Kobayashi, N.; Inoue, M.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Tomo, Y.; Motoda, Y.; et al. Solution Structure of the Kinase-Associated Domain 1 of Mouse Microtubule-Associated Protein/Microtubule Affinity-Regulating Kinase 3. Protein Sci. 2006, 15, 2534–2543. [Google Scholar] [CrossRef]
- Luecke, S.; Holleufer, A.; Christensen, M.H.; Jønsson, K.L.; Boni, G.A.; Sørensen, L.K.; Johannsen, M.; Jakobsen, M.R.; Hartmann, R.; Paludan, S.R. cGAS Is Activated by DNA in a Length-Dependent Manner. EMBO Rep. 2017, 18, 1707–1715. [Google Scholar] [CrossRef]
- Morrone, S.R.; Wang, T.; Constantoulakis, L.M.; Hooy, R.M.; Delannoy, M.J.; Sohn, J. Cooperative Assembly of IFI16 Filaments on dsDNA Provides Insights into Host Defense Strategy. Proc. Natl. Acad. Sci. USA 2014, 111, E62–E71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peisley, A.; Lin, C.; Wu, B.; Orme-Johnson, M.; Liu, M.; Walz, T.; Hur, S. Cooperative Assembly and Dynamic Disassembly of MDA5 Filaments for Viral dsRNA Recognition. Proc. Natl. Acad. Sci. USA 2011, 108, 21010–21015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Chen, Z.J. The cGAS–cGAMP–STING Pathway Connects DNA Damage to Inflammation, Senescence, and Cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Rapp, A.; Berndt, N.; Staroske, W.; Schuster, M.; Dobrick-Mattheuer, M.; Kretschmer, S.; König, N.; Kurth, T.; Wieczorek, D.; et al. RPA and Rad51 Constitute a Cell Intrinsic Mechanism to Protect the Cytosol from Self DNA. Nat. Commun. 2016, 7, 11752. [Google Scholar] [CrossRef]
- Yang, Y.-G.; Lindahl, T.; Barnes, D.E. Trex1 Exonuclease Degrades ssDNA to Prevent Chronic Checkpoint Activation and Autoimmune Disease. Cell 2007, 131, 873–886. [Google Scholar] [CrossRef] [Green Version]
- Dickey, C.A.; Kamal, A.; Lundgren, K.; Klosak, N.; Bailey, R.M.; Dunmore, J.; Ash, P.; Shoraka, S.; Zlatkovic, J.; Eckman, C.B.; et al. The High-Affinity HSP90-CHIP Complex Recognizes and Selectively Degrades Phosphorylated Tau Client Proteins. J. Clin. Investig. 2007, 117, 648–658. [Google Scholar] [CrossRef] [Green Version]
- Murphy, G.; Pfeiffer, R.; Camargo, M.C.; Rabkin, C.S. Meta-Analysis Shows That Prevalence of Epstein–Barr Virus-Positive Gastric Cancer Differs Based on Sex and Anatomic Location. Gastroenterology 2009, 137, 824–833. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Rosa, M.D.; Gottlieb, E.; Lerner, M.R.; Steitz, J.A. Striking Similarities Are Exhibited by Two Small Epstein-Barr Virus-Encoded Ribonucleic Acids and the Adenovirus-Associated Ribonucleic Acids VAI and VAII. Mol. Cell. Biol. 1981, 1, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Lerner, M.R.; Andrews, N.C.; Miller, G.; Steitz, J.A. Two Small RNAs Encoded by Epstein-Barr Virus and Complexed with Protein Are Precipitated by Antibodies from Patients with Systemic Lupus Erythematosus. Proc. Natl. Acad. Sci. USA 1981, 78, 805–809. [Google Scholar] [CrossRef] [Green Version]
- Sharp, T.V.; Schwemmle, M.; Jeffrey, I.; Laing, K.; Mellor, H.; Proud, C.G.; Hilse, K.; Clemens, M.J. Comparative Analysis of the Regulation of the Interferon-Inducible Protein Kinase PKR by Epstein-Barr Virus RNAs EBER-1 and EBER-2 and Adenovirus VAI RNA. Nucleic Acids Res. 1993, 21, 4483–4490. [Google Scholar] [CrossRef] [PubMed]
- Samanta, M.; Iwakiri, D.; Kanda, T.; Imaizumi, T.; Takada, K. EB Virus-Encoded RNAs Are Recognized by RIG-I and Activate Signaling to Induce Type I IFN. EMBO J. 2006, 25, 4207–4214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Liu, M.; Tian, Y.; Wang, J.; Xu, Y. Cryo-EM Structure of Human DNA-PK Holoenzyme. Cell Res. 2017, 27, 1341–1350. [Google Scholar] [CrossRef] [Green Version]
- Nanduri, S.; Carpick, B.W.; Yang, Y.; Williams, B.R.G.; Qin, J. Structure of the Double-Stranded RNA-Binding Domain of the Protein Kinase PKR Reveals the Molecular Basis of Its dsRNA-Mediated Activation. EMBO J. 1998, 17, 5458–5465. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.-M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuzman, D.; Levy, Y. Intrinsically Disordered Regions as Affinity Tuners in Protein –DNA Interactions. Mol. Biosyst. 2012, 8, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lin, Y.; Eschmann, N.A.; Zhou, H.; Rauch, J.N.; Hernandez, I.; Guzman, E.; Kosik, K.S.; Han, S. RNA Stores Tau Reversibly in Complex Coacervates. PLOS Biol. 2017, 15, e2002183. [Google Scholar] [CrossRef] [Green Version]
- Andreeva, L.; Hiller, B.; Kostrewa, D.; Lässig, C.; de Oliveira Mann, C.C.; Drexler, D.J.; Maiser, A.; Gaidt, M.; Leonhardt, H.; Hornung, V.; et al. cGAS Senses Long and HMGB/TFAM-Bound U-Turn DNA by Forming Protein–DNA Ladders. Nature 2017, 549, 394–398. [Google Scholar] [CrossRef]
- Beghini, A.; Magnani, I.; Roversi, G.; Piepoli, T.; Terlizzi, S.D.; Moroni, R.F.; Pollo, B.; Conti, A.M.F.; Cowell, J.K.; Finocchiaro, G.; et al. The Neural Progenitor-Restricted Isoform of the MARK4 Gene in 19q13.2 Is Upregulated in Human Gliomas and Overexpressed in a Subset of Glioblastoma Cell Lines. Oncogene 2003, 22, 2581–2591. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Satoh, S.; Okabe, H.; Kitahara, O.; Ono, K.; Kihara, C.; Tanaka, T.; Tsunoda, T.; Yamaoka, Y.; Nakamura, Y.; et al. Isolation of a Novel Human Gene, MARKL1, Homologous to MARK3 and Its Involvement in Hepatocellular Carcinogenesis. Neoplasia 2001, 3, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Pardo, O.E.; Castellano, L.; Munro, C.E.; Hu, Y.; Mauri, F.; Krell, J.; Lara, R.; Pinho, F.G.; Choudhury, T.; Frampton, A.E.; et al. MiR-515-5p Controls Cancer Cell Migration through MARK 4 Regulation. EMBO Rep. 2016, 17, 570–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Murata-Kamiya, N.; Saito, Y.; Hatakeyama, M. Role of Partitioning-Defective 1/Microtubule Affinity-Regulating Kinases in the Morphogenetic Activity of Helicobacter Pylori CagA. J. Biol. Chem. 2009, 284, 23024–23036. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishikawa, H.; Christiany, P.; Hayashi, T.; Iizasa, H.; Yoshiyama, H.; Hatakeyama, M. Kinase Activity of PAR1b, Which Mediates Nuclear Translocation of the BRCA1 Tumor Suppressor, Is Potentiated by Nucleic Acid-Mediated PAR1b Multimerization. Int. J. Mol. Sci. 2022, 23, 6634. https://doi.org/10.3390/ijms23126634
Nishikawa H, Christiany P, Hayashi T, Iizasa H, Yoshiyama H, Hatakeyama M. Kinase Activity of PAR1b, Which Mediates Nuclear Translocation of the BRCA1 Tumor Suppressor, Is Potentiated by Nucleic Acid-Mediated PAR1b Multimerization. International Journal of Molecular Sciences. 2022; 23(12):6634. https://doi.org/10.3390/ijms23126634
Chicago/Turabian StyleNishikawa, Hiroko, Priscillia Christiany, Takeru Hayashi, Hisashi Iizasa, Hironori Yoshiyama, and Masanori Hatakeyama. 2022. "Kinase Activity of PAR1b, Which Mediates Nuclear Translocation of the BRCA1 Tumor Suppressor, Is Potentiated by Nucleic Acid-Mediated PAR1b Multimerization" International Journal of Molecular Sciences 23, no. 12: 6634. https://doi.org/10.3390/ijms23126634
APA StyleNishikawa, H., Christiany, P., Hayashi, T., Iizasa, H., Yoshiyama, H., & Hatakeyama, M. (2022). Kinase Activity of PAR1b, Which Mediates Nuclear Translocation of the BRCA1 Tumor Suppressor, Is Potentiated by Nucleic Acid-Mediated PAR1b Multimerization. International Journal of Molecular Sciences, 23(12), 6634. https://doi.org/10.3390/ijms23126634






