Novel Nickel(II), Palladium(II), and Platinum(II) Complexes with O,S Bidendate Cinnamic Acid Ester Derivatives: An In Vitro Cytotoxic Comparison to Ruthenium(II) and Osmium(II) Analogues
Abstract
:1. Introduction
2. Results and Discussion
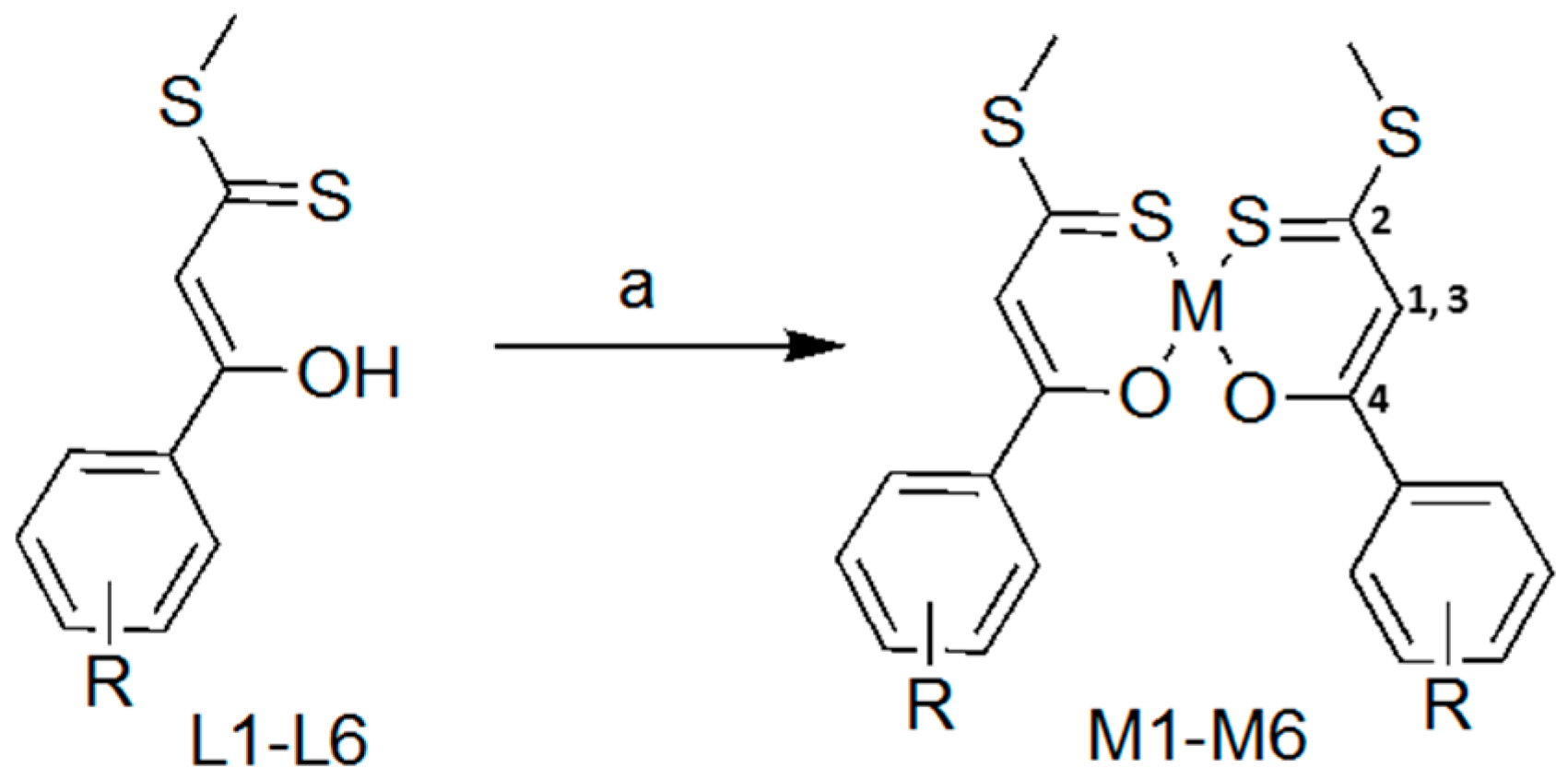
2.1. Synthesis and Characterisation
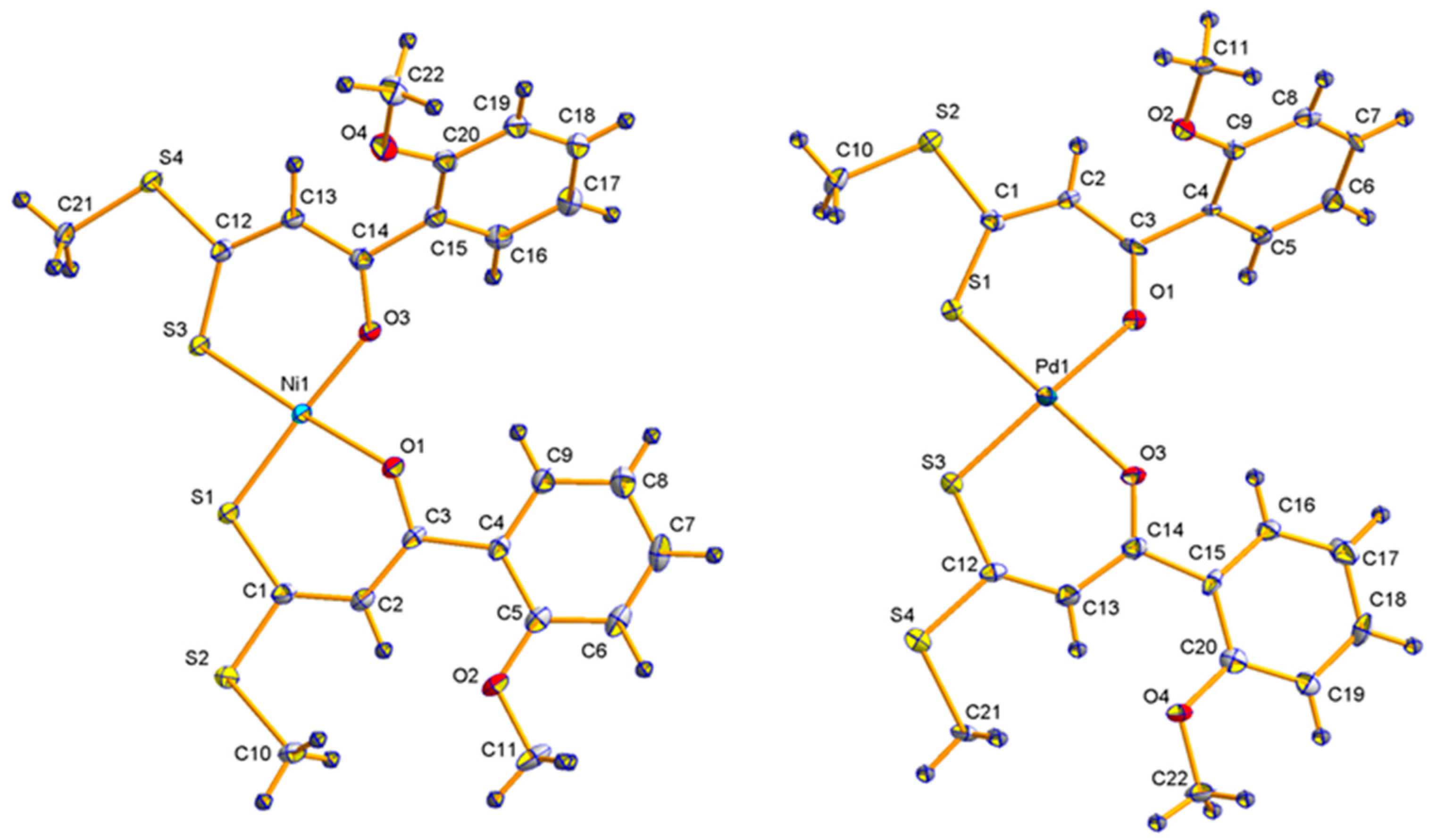
2.2. Molecular Structures
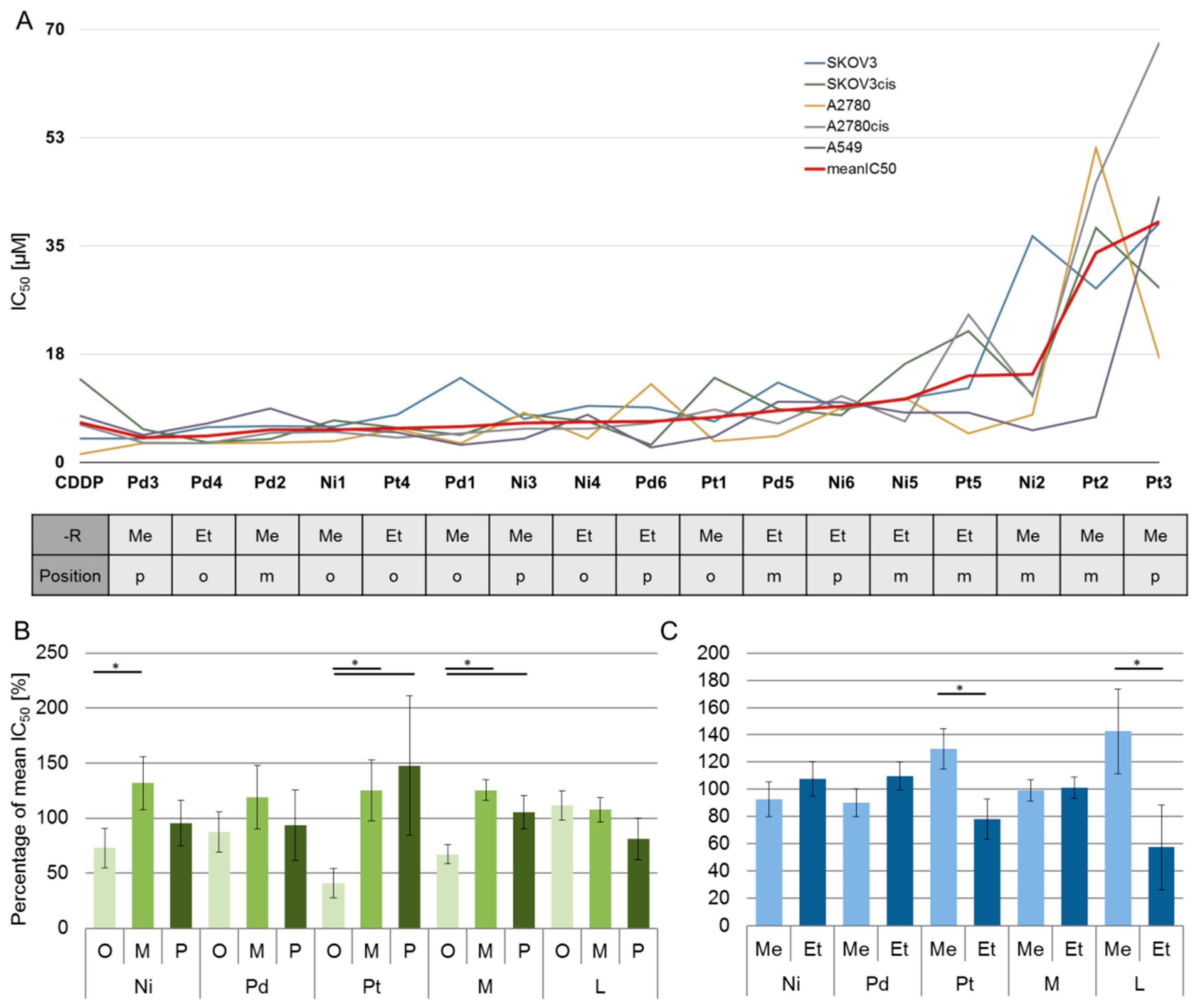
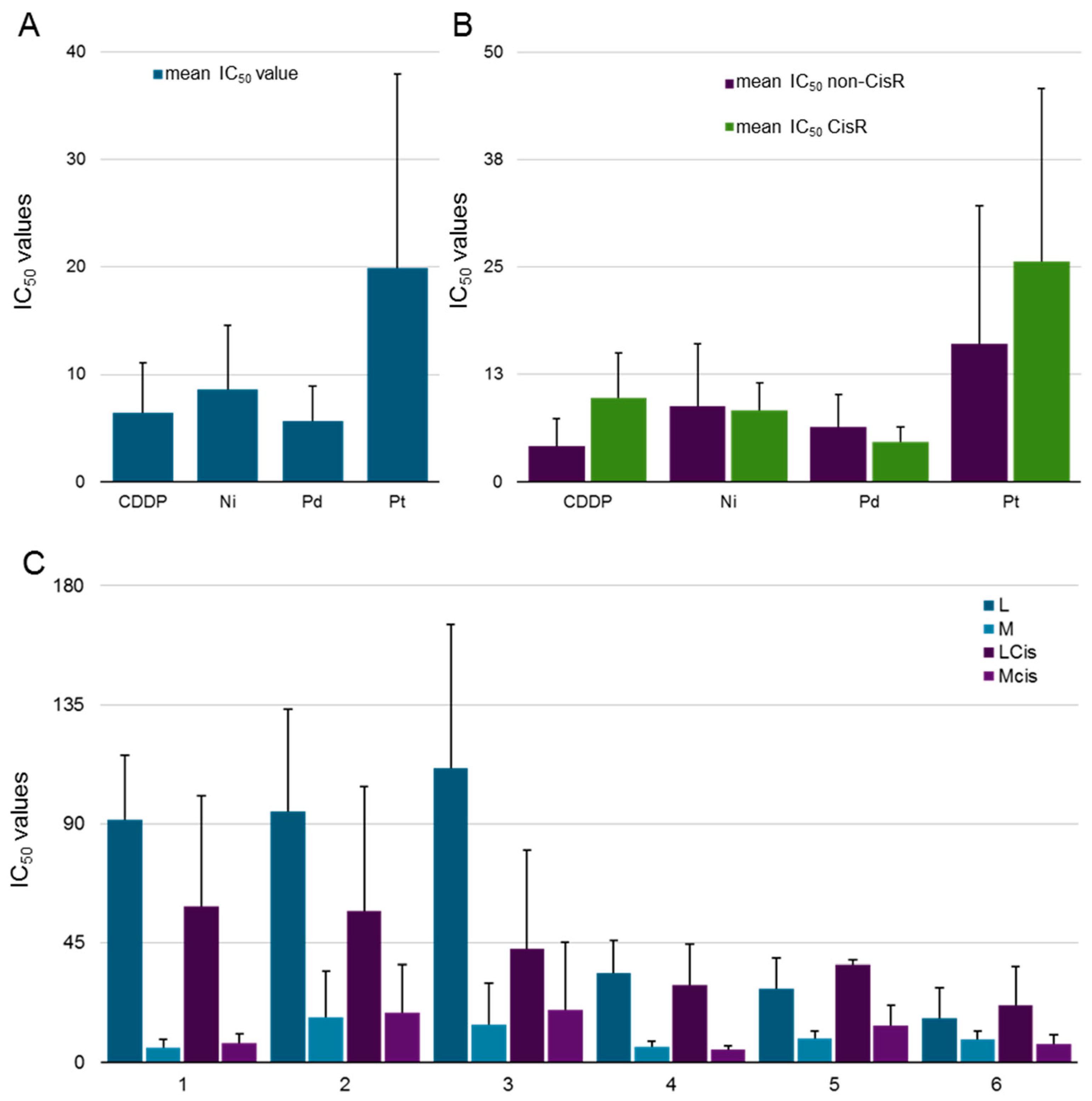
2.3. Biological Behavior
3. Materials and Methods
3.1. Materials and Techniques
3.2. Synthesis
3.3. Crystal Structure Determination
3.4. Stability Determinations
3.5. Biological Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosenberg, B.; Van Camp, L.; Krigas, T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef] [PubMed]
- Dilruba, S.; Kalayda, G.V. Platinum-based drugs: Past, present and future. Cancer Chemother. Pharmacol. 2016, 77, 1103–1124. [Google Scholar] [CrossRef] [PubMed]
- Brabec, V.; Hrabina, O.; Kasparkova, J. Cytotoxic platinum coordination compounds. DNA binding agents. Coord. Chem. Rev. 2017, 351, 2–31. [Google Scholar] [CrossRef]
- Jungwirth, U.; Xanthos, D.N.; Gojo, J.; Bytzek, A.K.; Korner, W.; Heffeter, P.; Abramkin, S.A.; Jakupec, M.A.; Hartinger, C.G.; Windberger, U.; et al. Anticancer Activity of Methyl-Substituted Oxaliplatin Analogs. Mol. Pharmacol. 2012, 81, 719–728. [Google Scholar] [CrossRef]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef]
- Messori, L.; Merlino, A. Cisplatin binding to proteins: A structural perspective. Coord. Chem. Rev. 2016, 315, 67–89. [Google Scholar] [CrossRef]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef]
- Gibson, D. Platinum(IV) anticancer prodrugs—Hypotheses and facts. Dalton Trans. 2016, 45, 12983–12991. [Google Scholar] [CrossRef]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalton Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef]
- Muggia, F.M.; Bonetti, A.; Hoeschele, J.D.; Rozencweig, M.; Howell, S.B. Platinum Antitumor Complexes: 50 Years Since Barnett Rosenberg’s Discovery. J. Clin. Oncol. 2015, 33, 4219–4226. [Google Scholar] [CrossRef]
- Fink, D.; Nebel, S.; Aebi, S.; Zheng, H.; Cenni, B.; Nehme, A.; Christen, R.D.; Howell, S.B. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996, 56, 4881–4886. [Google Scholar] [PubMed]
- Stordal, B.; Pavlakis, N.; Davey, R. Oxaliplatin for the treatment of cisplatin-resistant cancer: A systematic review. Cancer Treat. Rev. 2007, 33, 347–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu-Surrah, A.S.; Al-Sa’doni, H.H.; Abdalla, M.Y. Palladium-based chemotherapeutic agents: Routes toward complexes with good antitumor activity. Cancer Ther. 2008, 6, 1–10. [Google Scholar]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [Green Version]
- Hanif, M.; Hartinger, C.G. Anticancer metallodrugs: Where is the next cisplatin? Future Med. Chem. 2018, 10, 615–617. [Google Scholar] [CrossRef]
- Kenny, R.G.; Marmion, C.J. Toward Multi-Targeted Platinum and Ruthenium Drugs—A New Paradigm in Cancer Drug Treatment Regimens? Chem. Rev. 2019, 119, 1058–1137. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Wilson, J.J.; Lippard, S.J. Monofunctional and Higher-Valent Platinum Anticancer Agents. Inorg. Chem. 2013, 52, 12234–12249. [Google Scholar] [CrossRef] [Green Version]
- Hildebrandt, J.; Häfner, N.; Görls, H.; Kritsch, D.; Ferraro, G.; Dürst, M.; Runnebaum, I.B.; Merlino, A.; Weigand, W. Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance. Dalton Trans. 2016, 45, 18876–18891. [Google Scholar] [CrossRef]
- Muegge, C.; Liu, R.; Goerls, H.; Gabbiani, C.; Michelucci, E.; Ruediger, N.; Clement, J.H.; Messori, L.; Weigand, W. Novel platinum(II) compounds with O,S bidentate ligands: Synthesis, characterization, antiproliferative properties and biomolecular interactions. Dalton Trans. 2014, 43, 3072–3086. [Google Scholar] [CrossRef]
- Muegge, C.; Marzo, T.; Massai, L.; Hildebrandt, J.; Ferraro, G.; Rivera-Fuentes, P.; Metzler-Nolte, N.; Merlino, A.; Messori, L.; Weigand, W. Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding. Inorg. Chem. 2015, 54, 8560–8570. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.N.; Huq, F. Comprehensive review on tumour active palladium compounds and structure-activity relationships. Coord. Chem. Rev. 2016, 316, 36–67. [Google Scholar] [CrossRef]
- Meier-Menches, S.M.; Gerner, C.; Berger, W.; Hartinger, C.G.; Keppler, B.K. Structure-activity relationships for ruthenium and osmium anticancer agents—Towards clinical development. Chem. Soc. Rev. 2018, 47, 909–928. [Google Scholar] [CrossRef] [PubMed]
- Notaro, A.; Gasser, G. Monomeric and dimeric coordinatively saturated and substitutionally inert Ru(II) polypyridyl complexes as anticancer drug candidates. Chem. Soc. Rev. 2017, 46, 7317–7337. [Google Scholar] [CrossRef] [PubMed]
- Thota, S.; Rodrigues, D.A.; Crans, D.C.; Barreiro, E.J. Ru(II) Compounds: Next-Generation Anticancer Metallotherapeutics? J. Med. Chem. 2018, 61, 5805–5821. [Google Scholar] [CrossRef]
- Nabiyeva, T.; Marschner, C.; Blom, B. Synthesis, structure and anti-cancer activity of osmium complexes bearing pi-bound arene substituents and phosphane Co-Ligands: A review. Eur. J. Med. Chem. 2020, 201, 112483. [Google Scholar] [CrossRef]
- Imberti, C.; Sadler, P.J. 150 years of the periodic table: New medicines and diagnostic agents. Med. Chem. 2020, 75, 3–56. [Google Scholar] [CrossRef]
- Anthony, E.J.; Bolitho, E.M.; Bridgewater, H.E.; Carter, O.W.L.; Donnelly, J.M.; Imberti, C.; Lant, E.C.; Lermyte, F.; Needham, R.J.; Palau, M.; et al. Metallodrugs are unique: Opportunities and challenges of discovery and development. Chem. Sci. 2020, 11, 12888–12917. [Google Scholar] [CrossRef]
- Konkankit, C.C.; Marker, S.C.; Knopf, K.M.; Wilson, J.J. Anticancer activity of complexes of the third row transition metals, rhenium, osmium, and iridium. Dalton Trans. 2018, 47, 9934–9974. [Google Scholar] [CrossRef]
- Durig, J.R.; Danneman, J.; Behnke, W.D.; Mercer, E.E. Induction of Filamentous Growth in Escherichia-Coli by Ruthenium and Palladium Complexes. Chem. Biol. Interact. 1976, 13, 287–294. [Google Scholar] [CrossRef]
- Livingstone, S.E.; Mihkelson, A.E. Metal Chelates of Biologically Important Compounds.2. Nickel Complexes of Dialkyldithiophosphates and Their Adducts with Nitrogen Heterocycles. Inorg. Chem. 1970, 9, 2545–2551. [Google Scholar] [CrossRef]
- Butour, J.L.; Wimmer, S.; Wimmer, F.; Castan, P. Palladium(II) compounds with potential antitumour properties and their platinum analogues: A comparative study of the reaction of some orotic acid derivatives with DNA in vitro. Chem. Biol. Interact. 1997, 104, 165–178. [Google Scholar] [CrossRef]
- Wimmer, F.L.; Wimmer, S.; Castan, P.; Cros, S.; Johnson, N.; Colaciorodrigez, E. The Antitumor-Activity of Some Palladium(Ii) Complexes with Chelating Ligands. Anticancer Res. 1989, 9, 791–793. [Google Scholar] [PubMed]
- Zhao, G.H.; Lin, H.K.; Yu, P.; Sun, H.W.; Zhu, S.R.; Su, X.C.; Chen, Y.T. Ethylenediamine-palladium (II) complexes with pyridine and its derivatives: Synthesis, molecular structure and initial antitumor studies. J. Inorg. Biochem. 1999, 73, 145–149. [Google Scholar] [CrossRef]
- Icsel, C.; Yilmaz, V.T.; Kaya, Y.; Durmus, S.; Sarimahmut, M.; Buyukgungor, O.; Ulukaya, E. Cationic Pd(II)/Pt(II) 5,5-diethylbarbiturate complexes with bis(2-pyridylmethyl)amine and terpyridine: Synthesis, structures, DNA/BSA interactions, intracellular distribution, cytotoxic activity and induction of apoptosis. J. Inorg. Biochem. 2015, 152, 38–52. [Google Scholar] [CrossRef]
- Massai, L.; Pratesi, A.; Bogojeski, J.; Banchini, M.; Pillozzi, S.; Messori, L.; Bugarcic, Z.D. Antiproliferative properties and biomolecular interactions of three Pd(II) and Pt(II) complexes. J. Inorg. Biochem. 2016, 165, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, A.G.; Perez, J.M.; Montero, E.I.; Masaguer, J.R.; Alonso, C.; Navarro-Ranninger, C. Palladated and platinated complexes derived from phenylacetaldehyde thiosemicarbazone with cytotoxic activity in cis-DDP resistant tumor cells. Formation of DNA interstrand cross-links by these complexes. J. Inorg. Biochem. 1998, 70, 117–123. [Google Scholar] [CrossRef]
- Serrano, F.A.; Matsuo, A.L.; Monteforte, P.T.; Bechara, A.; Smaili, S.S.; Santana, D.P.; Rodrigues, T.; Pereira, F.V.; Silva, L.S.; Machado, J.; et al. A cyclopalladated complex interacts with mitochondrial membrane thiol-groups and induces the apoptotic intrinsic pathway in murine and cisplatin-resistant human tumor cells. BMC Cancer 2011, 11, 296. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hu, J.; Cai, Y.P.; Xu, S.M.; Weng, B.X.; Peng, K.S.; Wei, X.Y.; Wei, T.; Zhou, H.P.; Li, X.K.; et al. An Oxygen-Chelate Complex, Palladium Bis-acetylacetonate, Induces Apoptosis in H460 Cells via Endoplasmic Reticulum Stress Pathway Rather than Interacting with DNA. J. Med. Chem. 2013, 56, 9601–9611. [Google Scholar] [CrossRef]
- Dobrova, A.; Platzer, S.; Bacher, F.; Milunovic, M.N.M.; Dobrov, A.; Spengler, G.; Enyedy, E.A.; Novitchi, G.; Arion, V.B. Structure-antiproliferative activity studies on L-proline- and homoproline-4-N-pyrrolidine-3-thiosemicarbazone hybrids and their nickel(II), palladium(II) and copper(II) complexes. Dalton Trans. 2016, 45, 13427–13439. [Google Scholar] [CrossRef] [Green Version]
- Haribabu, J.; Jeyalakshmi, K.; Arun, Y.; Bhuvanesh, N.S.P.; Perumal, P.T.; Karvembu, R. Synthesis, DNA/protein binding, molecular docking, DNA cleavage and in vitro anticancer activity of nickel(II) bis(thiosemicarbazone) complexes. RSC Adv. 2015, 5, 46031–46049. [Google Scholar] [CrossRef]
- Banerjee, K.; Biswas, M.K.; Choudhuri, S.K. A newly synthesized nickel chelate can selectively target and overcome multidrug resistance in cancer through redox imbalance both in vivo and in vitro. J. Biol. Inorg. Chem. 2017, 22, 1223–1249. [Google Scholar] [CrossRef] [PubMed]
- Matkar, S.S.; Wrischnik, L.A.; Jones, P.R.; Hellmann-Blumberg, U. Two closely related nickel complexes have different effects on DNA damage and cell viability. Biochem. Biophs. Res. Commun. 2006, 343, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Saumweber, R.; Robl, C.; Weigand, W. Functionalized 1,1-ethenedithioles as ligands. Part 4. Synthesis and coordination properties of amphiphilic 3-oxodithiocarboxylic esters. Inorg. Chim. Acta 1998, 269, 83–90. [Google Scholar] [CrossRef]
- Schubert, K.; Alpermann, T.; Niksch, T.; Gorls, H.; Weigand, W. Synthesis and analytical characterization of functionalized beta-hydroxydithiocinnamic acids and their esters. Complex chemistry towards nickel(II), palladium(II), and platin(II). Zeitschrift fuer Anorganische und Allgemeine Chemie 2006, 632, 1033–1042. [Google Scholar] [CrossRef]
- Schubert, K.; Goerls, H.; Weigand, W. Synthesis and analytical characterization of novel pyridyl-substituted 1,1-ethenedithiolato complexes. Heteroat. Chem. 2005, 16, 369–378. [Google Scholar] [CrossRef]
- Schubert, K.; Goerls, H.; Weigand, W. Derivatives of β-hydroxydithiocinnamic acids as ligands. Syntheses and characterization of novel 1,1-ethenedithiolato and O,S-chelate complexes. Zeitschrift fur Naturforschung B 2007, 62, 475–482. [Google Scholar] [CrossRef]
- Schubert, K.; Saumweber, R.; Goerls, H.; Weigand, W. Functionalized derivatives of β-hydroxydithiocinnamic acids as ligands. Crystal structure of 4′-hydroxy-β-hydroxydithiocinnamic acid methyl ester. Zeitschrift fuer Anorganische und Allgemeine Chemie 2003, 629, 2091–2096. [Google Scholar] [CrossRef]
- Weigand, W.; Saumweber, R.; Schulz, P. Functionalized 1,1-ethenedithiolates as ligands. II. Nickel(II), palladium(II), and platinum(II) complexes of substituted β-keto dithio acid dianions. Zeitschrift fur Naturforschung B 1993, 48, 1080–1088. [Google Scholar] [CrossRef]
- Hildebrandt, J.; Görls, H.; Häfner, N.; Ferraro, G.; Dürst, M.; Runnebaum, I.B.; Weigand, W.; Merlino, A. Unusual mode of protein binding by a cytotoxic pi-arene ruthenium(II) piano-stool compound containing an O,S-chelating ligand. Dalton Trans. 2016, 45, 12283–12287. [Google Scholar] [CrossRef]
- Hildebrandt, J.; Hafner, N.; Kritsch, D.; Gorls, H.; Durst, M.; Runnebaum, I.B.; Weigand, W. Highly Cytotoxic Osmium(II) Compounds and Their Ruthenium(II) Analogues Targeting Ovarian Carcinoma Cell Lines and Evading Cisplatin Resistance Mechanisms. Int. J. Mol. Sci. 2022, 23, 4976. [Google Scholar] [CrossRef]
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Heinze, K.; Kritsch, D.; Mosig, A.S.; Dürst, M.; Häfner, N.; Runnebaum, I.B. Functional Analyses of RUNX3 and CaMKIINalpha in Ovarian Cancer Cell Lines Reveal Tumor-Suppressive Functions for CaMKIINalpha and Dichotomous Roles for RUNX3 Transcript Variants. Int. J. Mol. Sci. 2018, 19, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kritsch, D.; Hoffmann, F.; Steinbach, D.; Jansen, L.; Mary Photini, S.; Gajda, M.; Mosig, A.S.; Sonnemann, J.; Peters, S.; Melnikova, M.; et al. Tribbles 2 mediates cisplatin sensitivity and DNA damage response in epithelial ovarian cancer. Int. J. Cancer 2017, 141, 1600–1614. [Google Scholar] [CrossRef] [PubMed]
- Espino, J.; Fernandez-Delgado, E.; Estirado, S.; de la Cruz-Martinez, F.; Villa-Carballar, S.; Vinuelas-Zahinos, E.; Luna-Giles, F.; Pariente, J.A. Synthesis and structure of a new thiazoline-based palladium(II) complex that promotes cytotoxicity and apoptosis of human promyelocytic leukemia HL-60 cells. Sci. Rep. 2020, 10, 16745. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.G.; Romero-Canelon, I.; Coverdale, J.P.C.; Maia, P.I.S.; Clarkson, G.J.; Deflon, V.M.; Sadler, P.J. Novel tetranuclear Pd(II) and Pt(II) anticancer complexes derived from pyrene thiosemicarbazones. Dalton Trans. 2020, 49, 9595–9604. [Google Scholar] [CrossRef]
- Matesanz, A.I.; Leitao, I.; Souza, P. Palladium(II) and platinum(II) bis(thiosemicarbazone) complexes of the 2,6-diacetylpyridine series with high cytotoxic activity in cisplatin resistant A2780cisR tumor cells and reduced toxicity. J. Inorg. Biochem. 2013, 125, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Kataoka, H.; Yano, S.; Ohi, H.; Kawamoto, K.; Shibahara, T.; Mizoshita, T.; Mori, Y.; Tanida, S.; Kamiya, T.; et al. Anti-cancer effects of newly developed chemotherapeutic agent, glycoconjugated palladium (II) complex, against cisplatin-resistant gastric cancer cells. BMC Cancer 2013, 13, 237. [Google Scholar] [CrossRef] [Green Version]
- Arenaza-Corona, A.; Couce-Fortunez, M.D.; de Blas, A.; Morales-Morales, D.; Santillan, R.; Hopfl, H.; Rodriguez-Blas, T.; Barba, V. Further Approaches in the Design of Antitumor Agents with Response to Cell Resistance: Looking toward Aza Crown Ether-dtc Complexes. Inorg. Chem. 2020, 59, 15120–15134. [Google Scholar] [CrossRef]
- Eskandari, A.; Kundu, A.; Johnson, A.; Karmakar, S.; Ghosh, S.; Suntharalingam, K. A tri-metallic palladium complex with breast cancer stem cell potency. Dalton Trans. 2020, 49, 4211–4215. [Google Scholar] [CrossRef]
- Romero-Canelon, I.; Sadler, P.J. Next-Generation Metal Anticancer Complexes: Multitargeting via Redox Modulation. Inorg. Chem. 2013, 52, 12276–12291. [Google Scholar] [CrossRef]
- Romero-Canelon, I.; Mos, M.; Sadler, P.J. Enhancement of Selectivity of an Organometallic Anticancer Agent by Redox Modulation. J. Med. Chem. 2015, 58, 7874–7880. [Google Scholar] [CrossRef] [PubMed]
- Ballester, F.J.; Ortega, E.; Porto, V.; Kostrhunova, H.; Davila-Ferreira, N.; Bautista, D.; Brabec, V.; Dominguez, F.; Santana, M.D.; Ruiz, J. New half-sandwich ruthenium(ii) complexes as proteosynthesis inhibitors in cancer cells. Chem. Commun. 2019, 55, 1140–1143. [Google Scholar] [CrossRef] [PubMed]
- Ortega, E.; Ballester, F.J.; Hernandez-Garcia, A.; Hernandez-Garcia, S.; Guerrero-Rubio, M.A.; Bautista, D.; Santana, M.D.; Gandia-Herrero, F.; Ruiz, J. Novel organo-osmium(ii) proteosynthesis inhibitors active against human ovarian cancer cells reduce gonad tumor growth in Caenorhabditis elegans. Inorg. Chem. Front. 2021, 8, 141–155. [Google Scholar] [CrossRef]
- Thota, S.; Crans, D.C. (Eds.) Metal Nanoparticles: Synthesis and Applications in Pharmaceutical Sciences; Wiley-VCH: Weinheim, Germany, 2018; p. 261. [Google Scholar]
- Soldevila-Barreda, J.J.; Metzler-Nolte, N. Intracellular Catalysis with Selected Metal Complexes and Metallic Nanoparticles: Advances toward the Development of Catalytic Metallodrugs. Chem. Rev. 2019, 119, 829–869. [Google Scholar] [CrossRef] [PubMed]
- Khoury, A.; Deo, K.M.; Aldrich-Wright, J.R. Recent advances in platinum-based chemotherapeutics that exhibit inhibitory and targeted mechanisms of action. J. Inorg. Biochem. 2020, 207, 111070. [Google Scholar] [CrossRef] [PubMed]
- Roque, J.A.; Barrett, P.C.; Cole, H.D.; Lifshits, L.M.; Shi, G.; Monro, S.; von Dohlen, D.; Kim, S.; Russo, N.; Deep, G.; et al. Breaking the barrier: An osmium photosensitizer with unprecedented hypoxic phototoxicity for real world photodynamic therapy. Chem. Sci. 2020, 11, 9784–9806. [Google Scholar] [CrossRef]
- Xue, X.; Fu, Y.; He, L.; Salassa, L.; He, L.F.; Hao, Y.Y.; Koh, M.J.; Soulie, C.; Needham, R.J.; Habtemariam, A.; et al. Photoactivated Osmium Arene Anticancer Complexes. Inorg. Chem. 2021, 60, 17450–17461. [Google Scholar] [CrossRef]
- Doyle, J.R.; Slade, P.E.; Jonassen, H.B. Metal-Diolefin Coordination Compounds. Inorg. Syn. 1960, 6, 216–219. [Google Scholar] [CrossRef]
- Eysel, H.H.; Guggolz, E.; Kopp, M.; Ziegler, M.L. Synthesis and Characterization of Cis-Bis(Benzonitrile)Dichloroplatinum(Ii) and Trans-Bis(Benzonitrile)Dichloroplatinum(Ii)—X-Ray Structure-Analysis of Both the Cis-Species and Trans-Species. Zeitschrift fuer Anorganische und Allgemeine Chemie 1983, 499, 31–43. [Google Scholar] [CrossRef]
- SADABS 2.10, Bruker-AXS Inc.: Billerica, MA, USA, 2002.
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van De Streek, J. Mercury: Visualization and analysis of crystal structures. J. Appl. Cryst. 2006, 39, 453–457. [Google Scholar] [CrossRef] [Green Version]

| No. * | Signal | L21 | Ni2 | Pd2 | Pt2 | Ptdmso22 | Ru21 | Os22 |
|---|---|---|---|---|---|---|---|---|
| 1 | =C-H | 6.97 | 7.16 | 7.16 | 7.14 | 7.35 | 6.64 | 6.87 |
| 2 | -C=S | 217.3 | 181.4 | 180.9 | 185.9 | 186.7 | ||
| 3 | =C-H | 112.9 | 115.1 | 113.0 | 112.5 | 112.9 | 113.4 | 112.7 |
| 4 | -C-O- | 169.1 | 178.1 | 178.1 | 174.2 | 179.0 | 174.9 |
| Ni1 | Ni3 | Ni4 | Ni6 | Pd1 | |
|---|---|---|---|---|---|
| O(1)-M(1) | 1.8466 (17) | 1.8492 (12) | 1.8695 (17) | 1.8720 (16) | 2.023 (4) |
| O(3)-M(1) | 1.8584 (17) | 1.8512 (12) | 1.8668 (18) | 1.8827 (15) | 2.049 (4) |
| S(1)-M(1) | 2.1429 (7) | 2.1434 (4) | 2.1309 (7) | 2.1426 (6) | 2.2307 (16) |
| S(3)-M(1) | 2.1426 (7) | 2.1406 (5) | 2.1396 (7) | 2.1510 (6) | 2.2348 (16) |
| O(1)-C(3) | 1.258 (3) | 1.270 (2) | 1.277 (3) | 1.269 (3) | 1.271 (7) |
| O(3)-C(14) | 1.264 (3) | 1.278 (2) | 1.279 (3) | 1.268 (3) | 1.254 (7) |
| C(1)-S(1) | 1.698 (2) | 1.7075 (18) | 1.708 (3) | 1.711 (2) | 1.703 (6) |
| C(12)-S(3) | 1.696 (2) | 1.7085 (18) | 1.704 (3) | 1.720 (2) | 1.707 (6) |
| O(1)-M(1)-S(1) | 96.42 (6) | 95.47 (4) | 95.22 (6) | 96.44 (5) | 96.78 (13) |
| O(3)-M(1)-S(3) | 97.07 (5) | 95.76 (4) | 95.96 (6) | 95.37 (5) | 94.93 (13) |
| O(1)-M(1)-S(3) | 176.48 (6) | 176.99 (5) | 177.73 (6) | 177.53 (5) | 176.20 (13) |
| S(1)-M(1)-O(3) | 175.71 (6) | 176.90 (4) | 177.55 (7) | 179.16 (6) | 179.22 (13) |
| O(1)-M(1)-O(3) | 80.14 (7) | 81.43 (5) | 84.06 (8) | 83.38 (7) | 83.26 (17) |
| S(1)-M(1)-S(3) | 86.46 (3) | 87.334 (17) | 84.86 (3) | 84.84 (2) | 85.08 (6) |
| Bond | L118 | Ni1 | Pd1 | Ptdmso118 |
|---|---|---|---|---|
| C(1)-S(1) | 1.681 (2) | 1.698 (2) | 1.703 (6) | 1.710 (3) |
| C(3)-O(1) | 1.334 (3) | 1.2583 (3) | 1.271 (7) | 1.274 (3) |
| O(1)-M(1) | 1.8466 (17) | 2.023 (4) | 2.015 (7) | |
| S(1)-M(1) | 2.1429 (7) | 2.2307 (16) | 2.251 (6) |
| Compound | SKOV3 | SKOV3cis | RF SKOV3 | A2780 | A2780cis | RF A2780 | A549 |
|---|---|---|---|---|---|---|---|
| Ni1 | 5.8 (±0.6) | 6.8 (±3.0) | 1.2 | 3.5 (±0.3) | 5.0 (±1.1) | 1.4 | 5.4 (±0.1) |
| Ni2 | 36.6 (±9.3) | 11.0 (±2.1) | 0.3 | 7.7 (±2.0) | 10.7 (±6.0) | 1.4 | 5.2 (±1.0) |
| Ni3 | 7.1 (±3.7) | 7.8 (±3.4) | 1.1 | 8.0 (±4.8) | 5.4 (±0.8) | 0.7 | 3.8 (±0.0) |
| Ni4 | 9.2 (±5.1) | 6.7 (±2.0) | 0.7 | 3.8 (±0.1) | 5.4 (±1.0) | 1.4 | 7.7 (±2.2) |
| Ni5 | 10.3 (±5.0) | 15.9 (±2.3) | 1.5 | 10.4 (±0.8) | 6.6 (±0.3) | 0.6 | 8.1 (±3.9) |
| Ni6 | 8.6 (±4.6) | 7.6 (±0.2) | 0.9 | 8.8 (±8.1) | 10.8 (±6.5) | 1.2 | 9.7 (±6.3) |
| Pd1 | 13.7 (±8.2) | 4.4 (±1.3) | 0.3 | 3.1 (±0.0) | 4.7 (±2.3) | 1.5 | 2.8 (±1.2) |
| Pd2 | 5.8 (±4.1) | 3.7 (±0.8) | 0.6 | 3.1 (±0.6) | 4.7 (±2.3) | 1.5 | 8.7 (±8.2) |
| Pd3 | 3.8 (±1.0) | 5.4 (±3.1) | 1.4 | 3.1 (±0.0) | 3.2 (±0.1) | 1.0 | 4.5 (±1.7) |
| Pd4 | 5.7 (±2.5) | 3.3 (±0.2) | 0.6 | 3.1 (±0.0) | 3.1 (±0.0) | 1.0 | 6.3 (±2.4) |
| Pd5 | 12.9 (±7.1) | 8.6 (±3.4) | 0.7 | 4.3 (±0.5) | 6.2 (±2.0) | 1.4 | 9.8 (±1.3) |
| Pd6 | 8.9 (±1.0) | 2.8 (±0.2) | 0.3 | 12.7 (±1.0) | 6.4 (±0.4) | 0.5 | 2.4 (±0.4) |
| Pt1 | 6.6 (±2.1) | 13.7 (±7.5) | 2.1 | 3.4 (±0.0) | 8.5 (±3.4) | 2.5 | 4.2 (±0.7) |
| Pt2 | 28.2 (±7.3) | 37.9 (±0.6) | 1.3 | 50.9 (±15.4) | 45.2 (±9.6) | 0.9 | 7.4 (±2.5) |
| Pt3 | 38.7 (±5.1) | 28.2 (±6.6) | 0.7 | 16.8 (±18.9) | 67.9 (±28.4) | 4.0 | 43.0 (±2.9) |
| Pt4 | 7.7 (±0.6) | 5.6 (±1.2) | 0.7 | 5.4 (±3.0) | 4.0 (±0.9) | 0.7 | 4.8 (±1.3) |
| Pt5 | 12.0 (±7.2) | 21.2 (±8.0) | 1.8 | 4.7 (±1.0) | 24.0 (±0.6) | 5.1 | 8.0 (±0.8) |
| CDDP | 3.8 (±2.8) | 13.5 (±4.4) | 3.6 | 1.3 (±0.2) | 6.1 (±2.1) | 4.7 | (±2.6) |
| Cell Line | Ni1 [μM] | Pd3 [μM] | Pt4 [μM] | CDDP [μM] |
|---|---|---|---|---|
| Keratinocytes | 87.0 (±0) | 55.0 (±6.7) | >100 | 5.7 (±3.1) |
| Fibroblasts | 84.3 (±8.9) | 42.9 (±10.2) | >100 | 4.1 (±1.1) |
| MCF10A | 19.6 (±2.6) | 28.3 (±16.8) | 41.5 (±25.8) | 3.3 (±0.6) |
| Compound | SKOV3 | SKOV3cis | RF SKOV3 | A2780 | A2780cis | RF A2780 | A549 |
|---|---|---|---|---|---|---|---|
| L213 | 101.2 (±9.2) | 90.2 (±3.1) | 0.9 | 53.0 (±12.4) | 24.1 (±7.2) | 0.5 | 129.7 (±13.6) |
| Ni2 | 36.6 (±9.3) | 11.0 (±2.1) | 0.3 | 7.7 (±2.0) | 10.7 (±6.0) | 1.4 | 5.2 (±1.0) |
| Pd2 | 5.8 (±4.1) | 3.7 (±0.8) | 0.6 | 3.1 (±0.6) | 4.7 (±2.3) | 1.5 | 8.7 (±8.2) |
| Pt2 | 28.2 (±7.3) | 37.9 (±0.6) | 1.3 | 50.9 (±15.4) | 45.2 (±9.6) | 0.9 | 7.4 (±2.5) |
| Ptdmso213 | 28.8 (±4.9) | 20.1 (±3.0) | 0.7 | 19.8 (±1.6) | 21.0 (±3.3) | 1.1 | 24.2 (±2.0) |
| Ru242 | 22.4 (±9.6) | 17.8 (±0.9) | 0.8 | 16.4 (±3.3) | 15.0 (±2.9) | 0.9 | 15.3 (±8.1) |
| Os242 | 8.8 (±4.4) | 0.6 (±0.5) | 0.1 | 0.4 (±0.1) | 2.1 (±1.5) | 5.3 | 6.2 (±5.8) |
| CDDP | 3.8 (±2.8) | 13.5 (±4.4) | 3.6 | 1.3 (±0.2) | 6.1 (±2.1) | 4.7 | 7.6 (±2.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hildebrandt, J.; Häfner, N.; Görls, H.; Barth, M.-C.; Dürst, M.; Runnebaum, I.B.; Weigand, W. Novel Nickel(II), Palladium(II), and Platinum(II) Complexes with O,S Bidendate Cinnamic Acid Ester Derivatives: An In Vitro Cytotoxic Comparison to Ruthenium(II) and Osmium(II) Analogues. Int. J. Mol. Sci. 2022, 23, 6669. https://doi.org/10.3390/ijms23126669
Hildebrandt J, Häfner N, Görls H, Barth M-C, Dürst M, Runnebaum IB, Weigand W. Novel Nickel(II), Palladium(II), and Platinum(II) Complexes with O,S Bidendate Cinnamic Acid Ester Derivatives: An In Vitro Cytotoxic Comparison to Ruthenium(II) and Osmium(II) Analogues. International Journal of Molecular Sciences. 2022; 23(12):6669. https://doi.org/10.3390/ijms23126669
Chicago/Turabian StyleHildebrandt, Jana, Norman Häfner, Helmar Görls, Marie-Christin Barth, Matthias Dürst, Ingo B. Runnebaum, and Wolfgang Weigand. 2022. "Novel Nickel(II), Palladium(II), and Platinum(II) Complexes with O,S Bidendate Cinnamic Acid Ester Derivatives: An In Vitro Cytotoxic Comparison to Ruthenium(II) and Osmium(II) Analogues" International Journal of Molecular Sciences 23, no. 12: 6669. https://doi.org/10.3390/ijms23126669
APA StyleHildebrandt, J., Häfner, N., Görls, H., Barth, M.-C., Dürst, M., Runnebaum, I. B., & Weigand, W. (2022). Novel Nickel(II), Palladium(II), and Platinum(II) Complexes with O,S Bidendate Cinnamic Acid Ester Derivatives: An In Vitro Cytotoxic Comparison to Ruthenium(II) and Osmium(II) Analogues. International Journal of Molecular Sciences, 23(12), 6669. https://doi.org/10.3390/ijms23126669






