Abstract
The formation and development of legumes nodules requires a lot of energy. Legumes must strictly control the number and activity of nodules to ensure efficient energy distribution. The AON system can limit the number of rhizobia infections and nodule numbers through the systemic signal pathway network that the aboveground and belowground parts participate in together. It can also promote the formation of nodules when plants are deficient in nitrogen. The currently known AON pathway includes four parts: soil NO3− signal and Rhizobium signal recognition and transmission, CLE-SUNN is the negative regulation pathway, CEP-CRA2 is the positive regulation pathway and the miR2111/TML module regulates nodule formation and development. In order to ensure the biological function of this important approach, plants use a variety of plant hormones, polypeptides, receptor kinases, transcription factors and miRNAs for signal transmission and transcriptional regulation. This review summarizes and discusses the research progress of the AON pathway in Legume nodule development.
1. Introduction
Legumes are distributed worldwide and are the third largest family of seed plants [1]. They comprise many species, and are an important source of starch, protein, and fat for humans. They can also be used as animal feed. Additionally, the symbiotic nitrogen fixation by legumes and rhizobia is vital for the sustainable development of agriculture. It is also one of the model systems for studying the symbiosis between prokaryotes and eukaryotes. Understanding the mechanisms regulating legumes symbiotic nitrogen fixation is both scientifically and agriculturally significant. The formation and development of functional nodules in legumes is energy intensive, and up to ~25% of the net photosynthetic products of legumes are used for nodule development [2]. Therefore, legumes must strictly regulate the number and activity of nodules to balance the ratio between (1) carbon invested, and nitrogen gained, and (2) plant growth and nodule growth, to ensure efficient energy partitioning. The nutrient acquisition by plants is regulated by resource availability and metabolic requirements, thus indicating communication between the plant roots and the above-ground parts. They exchange signal molecules and nutrients through the vascular network [3].
Evidence indicating the systemic signal control of nodule formation and activity was first found in the early 1950s. Using nodule removal, grafting, and split-root experiments, Kosslak et al. showed that host plants regulate and limit the number of nodules by integrating the stem and root signals [2,3,4,5,6,7,8,9]. This systemic regulation of nodulation is called autoregulation of nodulation (AON) [10]. The AON system limits both rhizobial infection and the number of root nodules. Recently, research on the AON pathway has become more in-depth. The CLAVATA3/EMBRYO-SURROUNDING REGION RELATED–Hypernodulation and Aberrant Root 1 SUper Numeric Nodules (CLE-HAR1/SUNN) negative regulation of nodulation system in Lotus japonicus and Medicago truncatula was discovered in 2009 and 2010, whereby the number of nodules was limited when the soil nitrogen supply was sufficient [11,12,13,14]. In 2016, a study by Mohd-Radzman et al. proved that the M. truncatula polypeptide C-terminally Encoded Peptide (MtCEP1) secreted by the plant root system when nitrogen-deficient was recognized by the receptor kinase COMPACT ROOT ARCHITECTURE 2 (MtCRA2) in the shoot bud, which promoted nodule formation [15]. In 2020, a study by Gautrat et al. found that the CLE/CEP pathways together affect the downstream miR2111/TML module to control the number of nodules and maintain the delicate plant energy balance [16]. The nitrate (NO3−) signals impact the secretion of CLE and CEP peptides. In 2018, Tsikou and other studies found that high soil NO3− concentration can inhibit rhizobial infection and legume nodulation, since NO3− uptake from soil is more cost-effective for plants than nitrogen-fixation via symbiosis with rhizobia, as a source of nitrogen [5]. The AON system also inhibits rhizobial infection by weakening the plant’s perception of the rhizobia signals [5,6,7]. Therefore, we have expanded the current research on the AON system to include NO3− signaling, as it inhibits nodule initiation and development.
The AON system involves local regulation in roots and long-distance regulation from the root to the aerial part and vice versa and involves important transcription factors such as plant hormones and NIN-mediated identification of rhizobia and NO3−, the SUNN negative regulatory pathway, the CEP-CRA2 positive regulatory pathway, and, finally, the miR2111/TML regulation of nodule formation and development. The present article summarizes the progress made in research on the AON system, including some of the main genes, proteins, and their mechanisms of action.
2. Phytohormones and Transcription Factors Mediate the Recognition of and Response to Rhizobia and the NO3− Signals
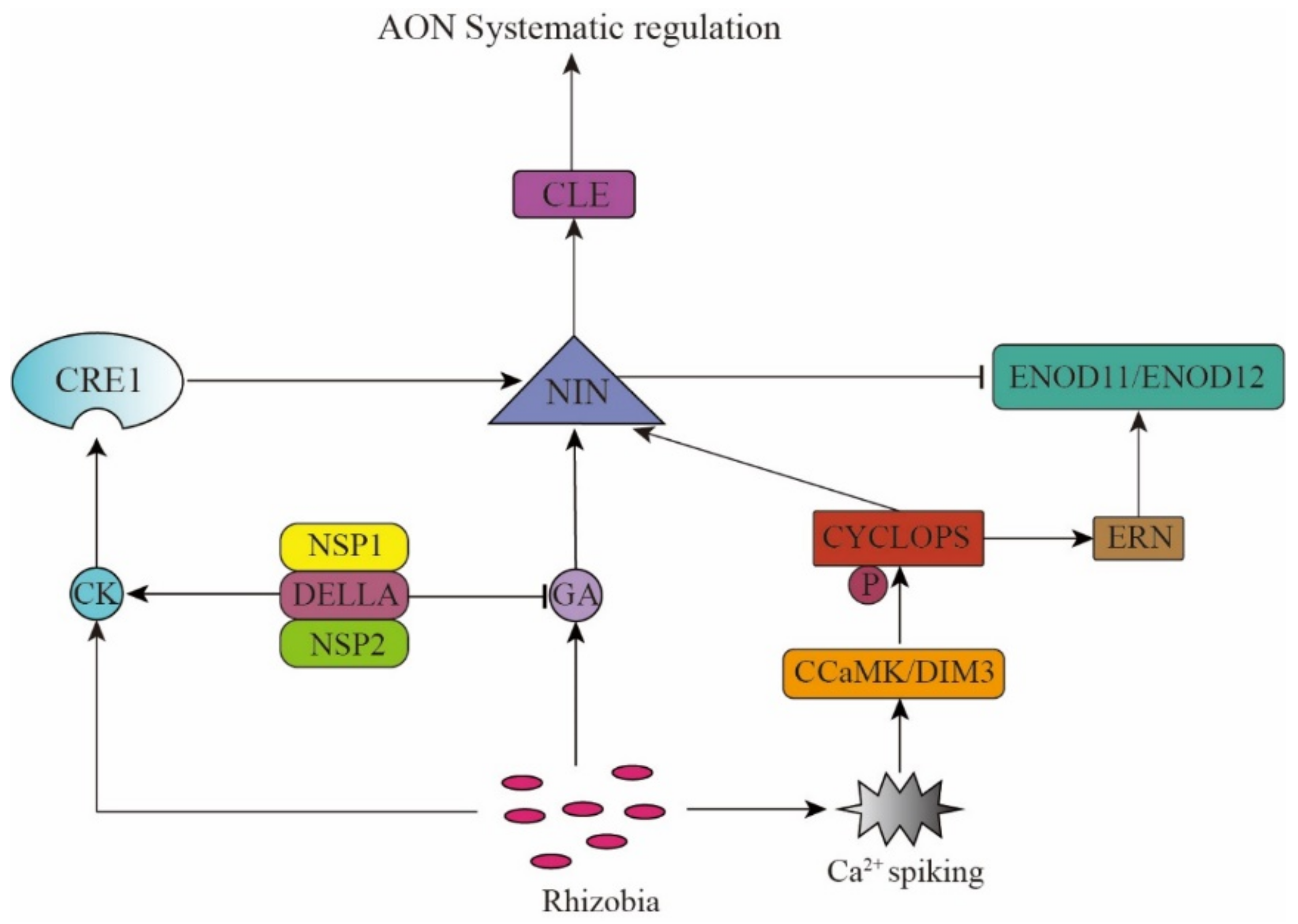
There is a symbiotic relationship between rhizobia and legume plants that forms via mutual recognition. Flavonoids secreted by the plant roots and the rhizobial nodulation factors results in Ca2+ oscillations in the plant cell nucleus, which are recognized by the Ca2+-calmodulin-dependent protein kinase, which helps phosphorylate the corresponding transcription factors, and subsequently affects the expression of specific genes, thereby ultimately resulting in nodule formation. The AON pathway controls the nodulation process according to soil rhizobia abundance and soil nitrogen content and also maintains the energy balance in the plant. Therefore, the AON pathway is inseparable from the recognition of and response to rhizobial and the NO3− signals. Ethylene (ET), cytokinin (CK), gibberellin (GA), and other plant hormones, along with the transcription factors, mediate the recognition of and response to these signals, together forming a complex regulatory network [17].
The mutual recognition between rhizobia and legumes is necessary for nodule formation, with each of them releasing specific chemical signals into the rhizosphere [18]. Legume plants release flavonoids and activate soil rhizobia to produce nod factors (NFs). NFs are lipochitosanoligosaccharides [19], which are perceived by Nod Factor Receptor 1 (LjNFR1) and LjNFR5 as signal molecules in the L. japonicus root [20] and combine with the Symbiotic Receptor-like Kinase (SYMRK) on plasma membrane [21]. The signal from SYMRK is transmitted to the nucleus, causing Ca2+ oscillation [18,22,23,24]. These Ca2+ oscillations are recognized by the Calcium- and Calmodulin-dependent Protein Kinase Does not Make Infections 3 (CCaMK/DMI3) [25,26] that mediates the phosphorylation of the transcription factor CYCLOPS. Phosphorylated CYCLOPS activates the expression of Nodule Inception (NIN) [27,28,29], thereby resulting in nodule formation.
NIN is a very important transcription factor in the process of root nodule formation and development and is important in the endodermis cell division (root nodule primordia initiation) and infection thread formation (tubular structure in root cells that promotes rhizobia colonization) [30], which are necessary for nodule formation. The nin mutant has defective bacterial identification and shows the phenotype of excessive root hair curling and no nodule formation [27]. NIN encodes a transcription factor containing the RWP-RK domain, which participates in many signal regulation pathways at different stages of nodule development [31].
The root nodule organogenesis and the infection thread formation are regulated by plant hormone ET [32,33,34]. It was found that when legume plants were exposed to ET gas or ET precursors (such as ACC), the rhizobial infection and the cell division capacity of the root nodule primordia were inhibited and showed a reduced number of root nodules [33,35,36]. The L. japonicus ethylene receptor transgenics (Ljetr) and M. truncatula ethylene insensitive 2 (Mtein2) mutant plants, which are important elements in the ET signaling pathway, showed increasing nodule numbers. These findings show that ET inhibits nodule formation [37]. Cerri et al. (2016) found that the Ethylene Responsive Factor (ERF) transcription factor Ethylene Response Factor1 (ERN1) required for nodulation was necessary for nodulation, and the ern1 mutant could not form nodules, but showed symbiotic reaction in the early stage of rhizobial infection. MtERN1 is induced by the CCaMK/CYCLOPS complex and controls the expression of the cell wall-related Early Nodulin 11 (MtENOD11) and MtENOD12 [38,39,40,41], whereas MtNIN regulates the degree of infection by restricting the expression of MtENOD11 in root epidermis, which antagonizes ERN1 and plays a negative regulatory role in rhizobial infection.
In the AON pathway, CK, GA, and NIN help identify the rhizobial and nitrate (NO3−) signals and trigger the generation of the CLE and CEP system signals. On the one hand, the rhizobia signal promotes the CK synthesis. The CK receptor CRE1 recognizes the CK signal and promotes the NIN gene expression. The transcription factor, NIN, then binds to the promoters of MtCLE13 and LjCLE-RS1/2 (LjCLE-Root Signal1/2), triggering their expression [12,42,43,44,45,46,47]. On the other hand, rhizobia can also promote MtCEP7 expression via the CK/CRE1 and NIN pathway and rely on MtCRA2 to promote nodulation [48]. Hence, the positive or negative regulation of nodulation depends on the changes in the environment and the plant’s needs. The nitrate (NO3−) signal can also trigger the CLE signal transduction by affecting the activity of transcription factor NIN-Like Protein (NLP) [49,50,51,52], thus indicating that NIN activates the CLE gene expression. It is speculated that the CLE peptide inhibits the NIN expression via negative feedback regulation, thus promoting nodulation in plants when they are nitrogen deficient, while inhibiting it when nitrogen is sufficient, thereby maintaining the dynamic and delicate balance of plant energy partitioning (Figure 1).
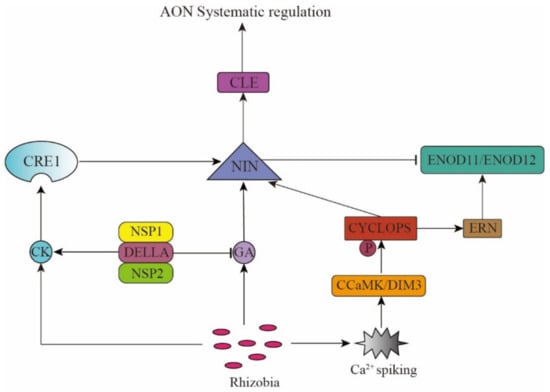
Figure 1.
Phytohormones and the transcription factor NIN mediate the rhizobium signal recognition.
Akamatsu and other researchers found that the GA signal plays a key regulatory role in the AON pathway and inhibits nodule formation. In the process of nodule formation, GA biosynthesis in the vascular bundle of the nodule is activated, causing its accumulation in the nodule. The NIN promoter contains a GA-responsive cis-acting element, which activates the NIN in the vascular bundle and stimulates the differentiation of the nodule cells and inhibits the rhizobial infection. The cis-acting deletion mutant of NIN is more susceptible to rhizobial infection, with the GA-inducible expression of CLE-RS1 and CLE-RS2 in the mutant being reduced. This result indicates that the GA inhibition happens through the AON pathway, where GA induces NIN to activate the AON system to regulate nodule formation [53,54]. Mayoshik et al. found that when a high concentration of GA3 or GA4 (10−7–10−4 M) were used to treat the roots of L. japonicus and other leguminous plants without rhizobia, the cells in the pericycle would divide and form nodule-like structures [53]. However, due to the trace amounts of phytohormones in plants and the multiple functions of GAs, the exact role of GA has not yet been clarified. Fonouni-Farde and others found that the DELLA protein (a negative regulator of GA signal) activates the CK signal in the cortex by regulating the effects of the Nodulation Signaling Pathway 2 (NSP2) and Nuclear Factor-Y 1 (NF-YA1), thereby promoting nodule formation [55,56]. The transcription factors CYCLOPS, NSP1, and NSP2 are functionally conserved in legumes, non-legumes, and mycorrhizal symbiosis. They interact with the DELLA protein to induce the NIN expression in symbiotic nitrogen fixation [57,58]. In peas, DELLA promotes the rhizobial infection of the epidermal cells [59] (Figure 1).
3. CLE-SUNN Negatively Regulates the Nodulation Signaling Pathway
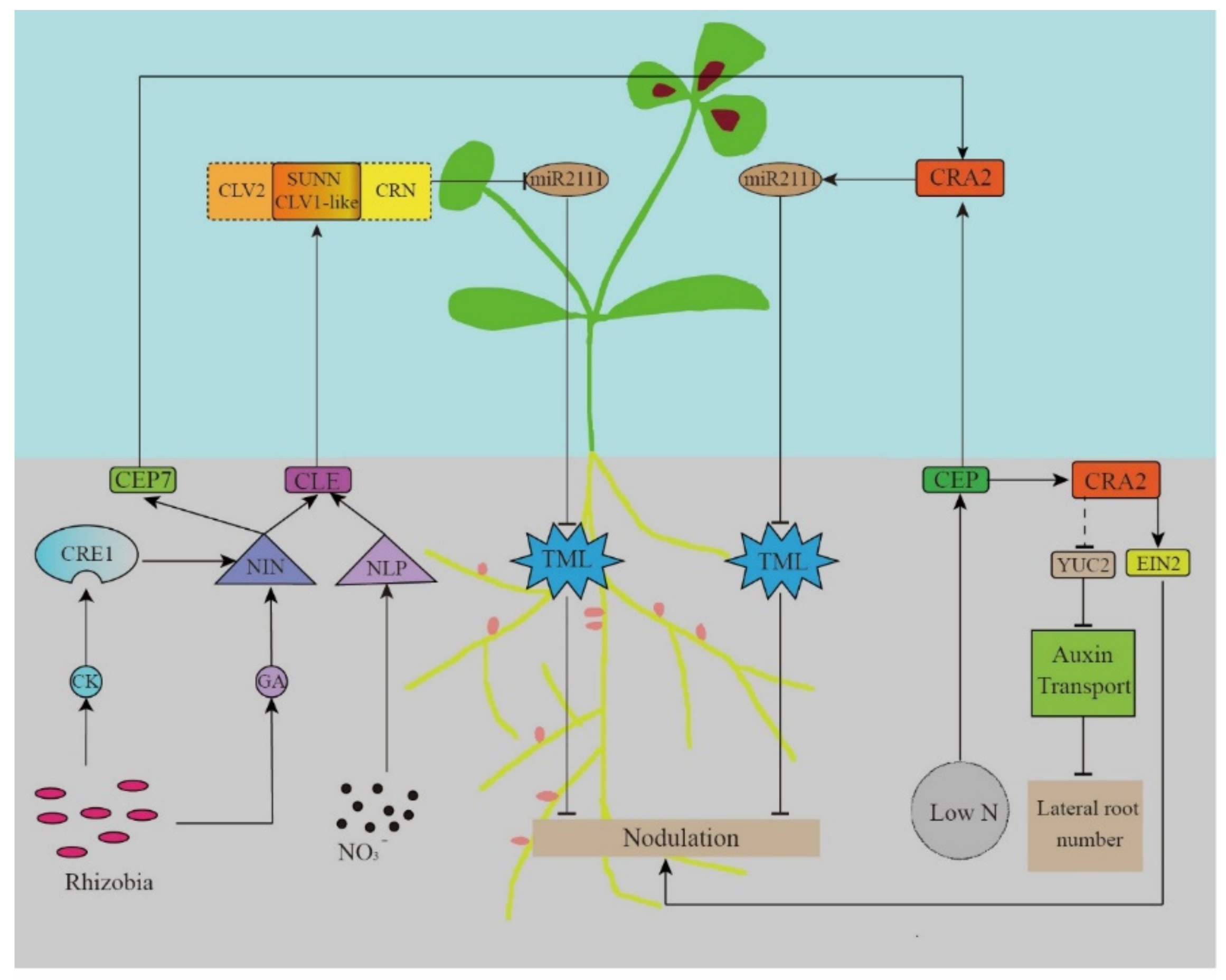
The long-distance signal transmission of the AON system starts with the secretion of specific peptides. CLE is a small 12-amino acid-long highly-conserved peptide in legumes. Rhizobia induce its expression in the roots, which depends on the soil nitrogen (NO3−) concentration. Then, it is transported to the above-ground parts through the xylem. CLE is recognized by the receptor kinases Nodule Autoregulation Receptor Kinase (GmNARK)/LjHAR1/MtSUNN, and negatively regulates the nodule formation (Figure 2).
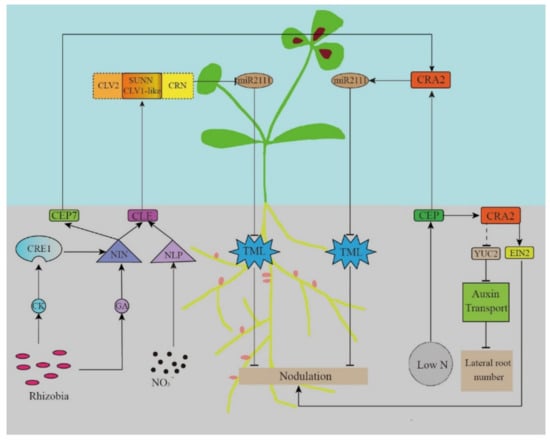
Figure 2.
The AON systemic regulatory pathway model.
The CLE peptide involved in the AON pathway in M. truncatula is encoded by the MtCLE12, MtCLE13, and MtCLE35 genes induced by the rhizobial signal [11,12,51,60]. Plants overexpressing MtCLE12 and MtCLE13 lose their nodulation ability [13,61]. In L. japonicus roots, CLE is encoded by LjCLE-RS1 (RS1), LjCLE-RS2, and LjCLE-RS3 (LjCLE-RS2 and LjCLE-RS3 can also be induced by the NO3− signal) [13,62]. Similarly, in soybeans, CLE is encoded by the GmRIC1 and GmRIC2 genes (GmCLE1 can also be induced by the NO3− signal) [14].
The stability and activity of polypeptides depend on post-translational modifications. Studies have shown that MtCLE12 is hydroxylated and triarabinosylated by Root Determined Nodation 1 (MtRDN1) in the root system [61,63,64], whereas LjCLE-RS2 and LjCLE-RS3 also undergo the same modifications by LjPLENTY [65,66,67]. Only the modified CLE peptide can inhibit nodule formation. The hydroxyproline arabinosyltransferase (HPAT) in the rdn1 mutant is damaged, thus showing hyper nodulation phenotype [68,69,70]. CLE can be detected in the xylem sap, suggesting the existence of a long-distance signaling pathway from the roots to the above-ground parts. Follow-up studies have found that CLE in the above-ground parts is regulated by the receptor kinase MtSUNN, LjHAR1 [66,68,69,70,71], or GmNARK, thus influencing signal recognition [14,72].
GmNARK/LjHAR1/MtSUNN encodes a class XI leucine-rich repeats receptor-like kinases (LRR-RLKs) located in the shoot buds, including the N-terminal cell membrane targeting signals, leucine-rich repetitive sequences, the transmembrane domain, and the cytoplasmic serine/threonine kinase domain that are homologous to the Arabidopsis thaliana gene CLAVATA1 (CLV1) involved in meristem maintenance [4,68,72,73]. Krusell et al. (2011) found that LjHAR1 (CLV1) interacts with LjCLV2 and LjCRN. The Tnt1 insertion mutant of MtCRN has more nodules, and the CLV2 mutant in pea and L. japonicus also shares a similar phenotype [74,75]. In Arabidopsis thaliana, a CLAVATA (CLV)-WUSCHEL (WUS) negative feedback loop is important in maintaining the proliferation and differentiation of the stem cells and the continuous initiation and growth of plant organs. Among them, WUSCHEL (WUS) is necessary for stem cell recognition, whereas the CLAVATA1, 2, and 3 (CLV) genes promote organ initiation. The CLV gene inhibits the WUS gene expression. WUS is a homeodomain transcription factor that directly activates the CLV3 expression of and promotes the activity of stem cells in the shoot apex meristem. These key regulatory factors interact with each other to establish a negative feedback pathway in the plant shoot apex meristem [76]. Subsequent studies found that a series of receptor kinases either participated in the CLV-WUS pathway independently or formed isoforms. RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2), CLV2, and pseudokinase CORYNE (CRN) participate in the perception of CLV3 signals to inhibit WUS expression [77,78]. Similar to the Arabidopsis CLV1, LjHAR1, LjCLV2, and LjCRN1 could also form isoforms, with an interaction between LjCRN and LjCLV2. Therefore, we speculate that CLV2 and CRN may play a role in the AON system by forming a multi-protein complex with CLV1 in the legumes as a co-receptor of CLE [74,75].
4. CEP-CRA2 Positively Regulates the Nodulation Signal Pathway
A CEP-CRA2 signal opposite to the CLE-SUNN signal also exists in the AON pathway. In the absence of rhizobia or plant nitrogen deficiency, the CEP1 produced by the root system is transported through the xylem and recognized by the receptor CRA2 in the shoot buds to promote nodulation [79,80].
CEP has been proven to be a peptide hormone that is important in regulating the nitrogen (N) demand signals, nodulation, and lateral root development [81,82,83]. Being a PTM (precursor derived, post translationally modified) peptide [84], the CEP gene encodes a non-functional peptide that is subsequently processed to produce smaller functional peptides. CEP has an N-terminal signal sequence, one or more conserved CEP domain, and one or more flanking variable region [85]. The proline residues in CEPS are usually hydroxylated, thereby influencing their biological activities [79,86,87,88,89,90]. There is also evidence that the hydroxyproline residues in some CEP peptides have been triacetylated [86]. Despite hydroxylation and triacetylation being well-known for their importance in the activity of CLE peptides, very few studies can be found on how CEP modification affects its activity and nodulation functions (Figure 2).
In A. thaliana, AtCEP1 is expressed under nitrogen deficiency. Using the long-distance root-stem signal transmission, AtCEP1 is recognized by its receptor CEPR1 (CEP Receptor 1) [91]. AtCEP1 affects the transcription of the nitrate transporters NRT1.1, NRT2.1, and NRT3.1, thereby regulating the nitrogen uptake and nitrogen status [90,91,92,93,94,95]. Ohkubo et al. found that the CEP-CEPR1 signal affected the NRT2.1 transcription, contingent on the transport of CEPD1 (CEP Downstream 1) and CEPD2 from the phloem to the roots. Interestingly, CEPD only affected transcription of NRT2.1, but not of NRT1.1 and NRT3.1 [92,94].
In 2016, Radzman et al. demonstrated that the peptide MtCEP1 secreted by roots positively regulates nodulation through the receptor kinase MtCRA2 in shoot buds [15]. MtCRA2 is homologous to A. thaliana CEPR1, belonging to the LRR-RLKs receptor kinase family, which participates in the AON pathway as a CEP receptor. Unlike the CEP-CEPR signal in A. thaliana, the CEP-CRA2 signal depends on miRNA transmission to the root (see the next section). CRA2 was also expressed in roots and nodules similarly to MtSUNN, a receptor kinase in CLE-SUNN signaling. The study found that compared with wild-type plants, the cra2 mutants showed shorter taproots, more lateral roots, and fewer nodules, thereby indicating that CRA2 negatively regulates lateral root formation, while positively regulating the nodule development of [72]. The by CRA2 regulation of the development of lateral roots is independent from that in nodule formation. The regulation of lateral root development is local. Contrastingly, regulating nodule development involves not only the systemic AON pathway, but also the CRA2 expressed in the root. In the roots, MtCRA2 inhibited the MtYCU2 expression and the auxin biosynthesis via unknown pathways, thus regulating the formation of lateral roots. Under nitrogen-deficient conditions, MtCEP1 can also promote MtCRA2 activity and phosphorylate MtEIN2 in roots, which partly relieves the ET inhibition of rhizobial infection and the promotion of nodulation [96].
Taken together, the CEP-CRA2 and CLE-SUNN signals are opposing and independent. They jointly improve the legume response to the soil rhizobia and nitrogen signals and are the key components of the AON pathway, which helps regulate nodule formation and energy balance [97].
5. miR2111/TML Responds to the AON Signal to Control Nodulation
As described previously, the plant roots secrete the CLE and CEP peptides to inhibit and promote nodulation, respectively. Both these peptides were transported by xylem and recognized by the receptor kinases SUNN and CRA2, respectively, in the above-ground plant parts. These signals will finally regulate the formation and development of nodules. After the signal is recognized by the receptors in the above-ground plant parts, new signals are produced and transmitted to the roots, thereby strengthening the response to the final effector and directly regulating nodulation [5,6].
The miR2111/TML regulatory module is a common effector of the CLE and CEP signals, which are critical for regulating both root nodulation and development in the AON pathway. Gautrat et al. found that miR2111 in shoot buds transmits signals via the phloem to negatively regulate TML expression in the roots [16]. TML was first found in Lotus roots, with the Ljtml mutant showing a super-nodulation phenotype [98]. Therefore, TML inhibits nodule formation, whereas miR2111 promotes nodule formation [99]. Under nitrogen-deficient conditions, the CRA2 recognizes the CEP1 signal to induce miR2111 and promotes nodule formation. When nitrogen is sufficient and rhizobia is abundant, the CLE-SUNN signal negatively regulates the miR2111 expression and inhibits nodulation [16] (Figure 2). However, the mechanism of the negative regulatory ability of TML is still unclear.
There are two copies of the TML gene in M. truncatula, MtTML1 and MtTML2 [6]. Sequence analysis revealed that the TML protein had two conserved domains (F-box and kelch repeat). The F-box domain that binds to the ASK family proteins to form the Skp1/cul1/F-box (SCF)-E3 ubiquitin ligase complex, while the kelch repeats domain is involved in protein–protein interaction. In the F-box protein containing the kelch repeats, the latter is necessary for physically interacting with its target protein. Therefore, we speculate that the TML protein may regulate nodulation by mediating the degradation of the downstream target proteins.
A. thaliana also possesses the miR2111/TML regulatory module, thereby indicating that it is evolutionarily conserved. Besides regulating nodulation, it may play a complementary role in regulating nitrogen utilization. In Arabidopsis thaliana, the MtTML homologous gene At3g27150 is also targeted and regulated by miR2111, which can be detected in Arabidopsis phloem sap [100]. Interestingly, when compared with legumes, it was phosphate (Pi) deficiency that induced miR2111 in A. thaliana, and not nitrogen deficiency [100]. Similarly, the E3 ubiquitin ligase Nitrogen Limitation Attachment (NLA) and E2 ubiquitin binding enzyme PHOSPHATE2 (PHO2) in A. thaliana were both negatively regulated by miR827 and miR399 transferred from shoot buds to roots under Pi deficiency, thereby promoting Pi acquisition [101,102,103]. Therefore, these results indicate that the miRNA/F-box regulatory module has multiple functions in the homeostatic regulation of N and P nutrition in higher plants.
6. Interplay of AON with Arbuscular Mycorrhizal Symbiosis and Phosphate
Phosphorus is a macronutrient essential for plant growth and development. It participates not only in the biosynthesis of various important organic compounds in plants, but also in various plant metabolic activities in various forms. Legumes, like other land plants, can only absorb phosphorus from the soil as an inorganic phosphate (i.e., orthophosphate; Pi) [104]. However, the soil phosphorus rapidly forms complexes with other ions, thereby making uptake impossible [105]. Most terrestrial plants can form mutualistic symbiosis with arbuscular mycorrhizal (AM) fungi, thereby increasing the plant’s phosphate (Pi) acquisition. The chitin oligomers secreted by AM are recognized by plants, which then activate the fungal symbiosis program via a symbiotic signaling pathway common to rhizobia. Similar to rhizobial symbiosis, AM symbiotic development is systematically regulated by plant Pi levels; high Pi concentrations inhibit both the initiation and further development of symbiosis and also the strigolactone synthesis and its export [106,107]. Since phosphorus promotes the synthesis and export of strigolactones, AM fungal colonization negatively regulates further symbiotic development and is independent of the plant’s phosphorus status [108,109]. This systemic regulation is like AON, and is therefore referred to as the autoregulation of mycorrhization (AOM).
The inhibitory effect of the AOM pathway on AM colonization is also regulated by the system involving the CLE peptide and LRR-RLK CLAVATA1. Mutants of the CLV1 homologs GmNARK, LjHAR1, PvSYM29, and MtSUNN in legumes all showed enhanced AM colonization [110]. Müller and Le Marquer et al. examined the transcript levels of CLEs in the AM-colonized roots of M. truncatula and showed that the expressions of MtCLE16, MtCLE45, and MtCLE53 were induced by AM, with MtCLE45 and MtCLE53 expression increasing the most [111,112]. Further studies found that MtCLE53 expression was induced in AM-colonized roots, which triggered a negative feedback loop that negatively affected colonization by reducing strigolactone levels. Although this effect depends on MtSUNN, other unknown receptors may also be involved [111]. As an early symbiotic signal of AM colonization, strigolactone promotes AM symbiosis, fungal energy metabolism, hyphal bifurcation and the secretion of fungal chitin oligomers, thereby activating the plant symbiotic signaling pathway [18,113]. Moreover, the strigolactone release is also partially regulated by Pi [106], which can directly affect the secretion of fungal chitin oligomers [113,114,115,116].
Additionally, multiple studies have found that Pi deficiency can not only significantly reduce the number of nodules in legumes, but also reduce their nitrogen fixation ability [117,118,119,120]. Physiological studies on different legumes have shown that phosphorus is preferentially transferred from other organs to the root nodules during Pi deficiency [117,119,120]. Isidra-Arellano et al. (2018) showed that Pi deficiency negatively affects early symbiotic events, i.e., the expression of symbiosis-related genes and infection thread formation [121]. The latest study found that the inhibitory effect of Pi deficiency on legume nodulation depends on the AON pathway. Isidra-Arellano et al. (2020) proposed a model for activating the AON pathway under Pi deficiency. During Pi absence, the master regulator of Pi perception and signal transduction, the MYB transcription factor PHR1 (PHOSPHATE STARVATION1), directly binds to NIN and the promoters of RIC1 and RIC2, which encode CLE peptides. These two AON-related CLE peptides are transported from the root to the stem. This activates the negative regulatory pathway mediated by CLE and its receptors in the AON pathway, thereby inhibiting nodulation. Furthermore, PHR1 can also directly bind to the TML promoter and promote its expression, thereby reducing nodule growth [122].
7. Future Perspectives
The formation of legume symbiotic nodules requires abundant energy. When the soil NO3− content is sufficient, it is obviously more economical to absorb nitrogen directly from the soil rather than from symbiotic nitrogen fixation. Therefore, legumes need to control the number of nodules for maintaining the energy and carbon-nitrogen balance.
The nodule number and its development in legumes are strictly limited by the negative feedback regulation of long-distance signals from the roots to shoot buds and back to roots, which is called nodule self-regulation (autoregulation or AON). The AON pathway involves systemic regulation in the root–soil–shoot continuum, including the CLE-SUNN negative regulatory pathway and the CEP-CRA2 positive regulatory pathway. When nitrogen and rhizobia were abundant in the soil, the rhizobial signal can affect the CK concentration, and CRE1 and NIN can trigger the CLE signal transduction. NO3− can also trigger the CLE signal transduction by affecting the activity of the NLP transcription factor. When plants are deficient in nitrogen, the rhizobial signal promotes the expression of MtCEP7 through CK/CRE1 and NIN signaling. After the CLE and CEP signals are recognized by SUNN and CRA2, respectively, the downstream signal from the shoot buds to the roots is conducted by the shoot bud-produced miR2111. Activated SUNN and CRA2 inhibit and promote the formation of miR2111, respectively, and then miR2111 negatively regulates the TML expression in the roots. This systemic regulatory pathway allows the nodulation capacity of the legume roots to be fine-tuned, depending on the nitrogen utilization.
Existing studies have shown that the understanding of the recognition of rhizobia in the early stage of nodule formation is basically complete. Transcription factors such as NIN and NLP, and plant hormones such as ET, CK, GA, and auxin have an important role in transcriptional regulation and signal transduction. However, some mechanisms in the AON pathway are still unclear.
SUNN is the receptor of the CLE-SUNN system, which is homologous to the CLV1 in A. thaliana. In A. thaliana, the CLAVATA (CLV)-WUSCHEL (WUS) negative feedback loop is important in maintaining the proliferation and differentiation of stem cells and the continuous initiation and growth of plant organs, including the participation of a series of receptors such as kinases, including CLV1, 2, 3, CRN, and CIKs. 1. Do other receptor-like kinases involved in this pathway also have functions similar to CLV1 in nodulation regulation in legumes? 2. Does WUS also regulate nodule formation and/or development? 3. Given that ClV1, CLV2, and CRN can form homotypic and heteromorphic complexes between each other (whereby CRN, as a pseudokinase, needs to bind with other receptor kinases to play its role), can CLV2 or CLV2-CRN regulate the formation and development of nodules independently of CLV1?
In the CEP-CRA2 signaling pathway, CEP controls the N-demand signal and lateral root development. Interestingly, in A. thaliana, the downstream CEP signal is transmitted from CEPD to the roots, which differs from the miR2111/TML module downstream of the AON pathway. Additionally, the miR2111 and TML homologs in Arabidopsis affect Pi uptake. If CEPD exists in legumes, what kind of function does it have? Does CRA2 also have a CEPR1/CEPR2 co-receptor? It should be borne in mind that CRA2 also has a relatively independent function in legume roots, i.e., to suppress YUC2 expression. However, the mechanism by which it affects auxin biosynthesis and inhibits lateral root growth still remains unclear.
Downstream of the AON pathway, the miR2111/TML module acts as the common effector of CLE-SUNN and CEP-CRA2 to regulate the formation and development of nodules. However, the mechanism of negative regulation of nodules by TML is still unclear. As an F-box protein containing the kelch repeats domain, TML can theoretically form the Skp1/CUL1/F-box (SCF) E3 ubiquitin ligase complex. In A. thaliana, similarly to MtTML, the ZEITLUPE/Flavin-binding Kelch Repeat, F-BOX/LOV Kelch Protein2 (ZTL/FKF1/LKP2) protein family with the F-box and kelch repeats domains can participate in plant photomorphogenesis and stress response by mediating the ubiquitin-mediated proteosomal degradation of specific transcription factors (Timing of CAB Expression1, Cycling Dof Factor 1, etc.) [123]. Recent studies have shown that AtFKF1 can facilitate the ubiquitin-mediated proteosomal degradation of DELLA protein via the kelch repeat domain and the GRAS domain of DELLA protein, thereby promoting the flowering of Arabidopsis thaliana under long-day conditions [124]. Does MtTML also negatively regulate nodule formation by mediating DELLA or other transcription factors (such as NLP, NSP, etc.) involved in the process of nodule formation? Moreover, miR827/AtNLA, miR399/AtPHO2, and miR2111/MtTML have similar functions in Arabidopsis thaliana. Are miRNAs, such as miR827, miR399, etc., also involved in regulating nodule development?
Recently, there have been many studies on the formation and development of legume nodules. As an important regulatory pathway to balance nodule energy consumption with other plant energy demands, the AON pathway is a hot research topic. Summarizing the existing research results, we found that not only many important AON pathway genes have been reported, but also their homologs in A. thaliana have similar functions, such as regulating nitrogen and phosphorus uptake and meristem differentiation. This is because the AON pathway is closely related to plant energy partitioning, and the selective meristem differentiation is crucial to the formation of nodules. Due to the existence of nodulation mechanism in legumes, these genes have been endowed with new functions related to regulating nodulation in legumes.
In conclusion, while exploring new genes regulating nodule development, we can also refer to their research results in A. thaliana and focus on the genes related to energy partitioning and hormone regulation, thus providing new ideas for further studying the nodulation mechanism of legumes in the future.
Author Contributions
Conceptualization: Y.C., Y.L. and Y.P.; Data curation: Y.L. and Y.P.; Writing-orginal draft preparation: Y.C., Y.L. and Y.P.; Supervision: Y.C.; Writing-Review: Y.C., Y.S. and R.Z.; Writing-Editing: Y.C., M.K., Y.M. and D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the foundation of the Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations (lzujbky-2019-kb05; lzujbky-2021-kb05).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available in a publicly accessible repository.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Doyle, J.J. The promise of genomics for a “next generation” of advances in higher-level legume molecular systematics. S. Afr. J. Bot. 2013, 89, 10–18. [Google Scholar] [CrossRef]
- Oono, R.; Anderson, C.G.; Denison, R.F. Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc. R. Soc. B-Biol. Sci. 2011, 278, 2698–2703. [Google Scholar] [CrossRef] [PubMed]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.R.; Helariutta, Y.; He, X.Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The Plant Vascular System: Evolution, Development and Functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, E.; Journet, E.P.; de Carvalho-Niebel, F.; Duc, G.; Frugoli, J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005, 58, 809–822. [Google Scholar] [CrossRef]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 2018, 362, 233–235. [Google Scholar] [CrossRef]
- Gautrat, P.; Mortier, V.; Laffont, C.; De Keyser, A.; Fromentin, J.; Frugier, F.; Goormachtig, S. Unraveling new molecular players involved in the autoregulation of nodulation in Medicago truncatula. J. Exp. Bot. 2019, 70, 1407–1417. [Google Scholar] [CrossRef]
- Yoro, E.; Suzaki, T.; Kawaguchi, M. CLE-HAR1 Systemic Signaling and NIN-Mediated Local Signaling Suppress the Increased Rhizobial Infection in the daphne Mutant of Lotus japonicus. Mol. Plant-Microbe Interact. 2020, 33, 320–327. [Google Scholar] [CrossRef]
- Kosslak, R.M.; Bohlool, B.B. Suppression of Nodule Development of One Side of a Split-Root System of Soybeans Caused by Prior Inoculation of the Other Side. Plant Physiol. 1984, 75, 125–130. [Google Scholar] [CrossRef]
- Lawn, R.J.; Brun, W.A. Symbiotic Nitrogen Fixation in Soybeans. III. Effect of Supplemental Nitrogen and Intervarietal Grafting1. Crop Sci. 1974, 14, 22–25. [Google Scholar] [CrossRef]
- Caetanoanolles, G.; Gresshoff, P.M. Plant Genetic-Control of Nodulation. Annu. Rev. Microbiol. 1991, 45, 345–382. [Google Scholar] [CrossRef]
- Mortier, V.; De Wever, E.; Vuylsteke, M.; Holsters, M.; Goormachtig, S. Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J. 2012, 70, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Mortier, V.; Den Herder, G.; Whitford, R.; Van de Velde, W.; Rombauts, S.; D’haeseleer, K.; Holsters, M.; Goormachtig, S. CLE Peptides Control Medicago truncatula Nodulation Locally and Systemically. Plant Physiol. 2010, 153, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Ohnishi, E.; Sato, S.; Takahashi, H.; Nakazono, M.; Tabata, S.; Kawaguchi, M. Nod Factor/Nitrate-Induced CLE Genes that Drive HAR1-Mediated Systemic Regulation of Nodulation. Plant Cell Physiol. 2009, 50, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.E.; Ferguson, B.J.; Gresshoff, P.M. Inoculation- and Nitrate-Induced CLE Peptides of Soybean Control NARK-Dependent Nodule Formation. Mol. Plant-Microbe Interact. 2011, 24, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Mohd-Radzman, N.A.; Laffont, C.; Ivanovici, A.; Patel, N.; Reid, D.; Stougaard, J.; Frugier, F.; Imin, N.; Djordjevic, M.A. Different Pathways Act Downstream of the CEP Peptide Receptor CRA2 to Regulate Lateral Root and Nodule Development. Plant Physiol. 2016, 171, 2536–2548. [Google Scholar] [CrossRef]
- Gautrat, P.; Laffont, C.; Frugier, F. Compact Root Architecture 2 Promotes Root Competence for Nodulation through the miR2111 Systemic Effector. Curr. Biol. 2020, 30, 1339–1345. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Foo, E.; Ross, J.J.; Reid, J.B. Relationship between gibberellin, ethylene and nodulation in Pisum sativum. New Phytol. 2011, 189, 829–842. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Denarie, J.; Debelle, F.; Prome, J.C. Rhizobium lipo-chitooligosaccharide nodulation factors: Signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 1996, 65, 503–535. [Google Scholar] [CrossRef]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Miwa, H.; Sun, J.; Oldroyd, G.E.D.; Downie, J.A. Analysis of nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant-Microbe Interact. 2006, 19, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi-Anraku, H.; Takeda, N.; Charpentier, M.; Perry, J.; Miwa, H.; Umehara, Y.; Kouchi, H.; Murakami, Y.; Mulder, L.; Vickers, K.; et al. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 2005, 433, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, M.; Bredemeier, R.; Wanner, G.; Takeda, N.; Schleiff, E.; Parniske, M. Lotus japonicus CASTOR and POLLUX Are Ion Channels Essential for Perinuclear Calcium Spiking in Legume Root Endosymbiosis. Plant Cell 2008, 20, 3467–3479. [Google Scholar] [CrossRef] [PubMed]
- Mitra, R.M.; Gleason, C.A.; Edwards, A.; Hadfield, J.; Downie, J.A.; Oldroyd, G.E.D.; Long, S.R. A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 2004, 101, 4701–4705. [Google Scholar] [CrossRef]
- Miller, J.B.; Pratap, A.; Miyahara, A.; Zhou, L.; Bornemann, S.; Morris, R.J.; Oldroyd, G.E.D. Calcium/Calmodulin-Dependent Protein Kinase Is Negatively and Positively Regulated by Calcium, Providing a Mechanism for Decoding Calcium Responses during Symbiosis Signaling. Plant Cell 2013, 25, 5053–5066. [Google Scholar] [CrossRef]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef]
- Marsh, J.F.; Rakocevic, A.; Mitra, R.M.; Brocard, L.; Sun, J.; Eschstruth, A.; Long, S.R.; Schultze, M.; Ratet, P.; Oldroyd, G.E.D. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007, 144, 324–335. [Google Scholar] [CrossRef]
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M. CYCLOPS, A DNA-Binding Transcriptional Activator, Orchestrates Symbiotic Root Nodule Development. Cell Host Microbe 2014, 15, 139–152. [Google Scholar] [CrossRef]
- Soyano, T.; Kouchi, H.; Hirota, A.; Hayashi, M. NODULE INCEPTION Directly Targets NF-Y Subunit Genes to Regulate Essential Processes of Root Nodule Development in Lotus japonicus. PLoS Genet. 2013, 9, e1003352. [Google Scholar] [CrossRef]
- Vernie, T.; Kim, J.; Frances, L.; Ding, Y.L.; Sun, J.; Guan, D.; Niebel, A.; Gifford, M.L.; de Carvalho-Niebel, F.; Oldroyd, G.E.D. The NIN Transcription Factor Coordinates Diverse Nodulation Programs in Different Tissues of the Medicago truncatula Root. Plant Cell 2015, 27, 3410–3424. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.H.; Cardoza, V.; Mitchell, D.M.; Bright, L.; Oldroyd, G.; Harris, J.M. Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J. 2006, 46, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Mathesius, U. Signaling interactions during nodule development. J. Plant Growth Regul. 2003, 22, 47–72. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Engstrom, E.M.; Long, S.R. Ethylene inhibits the nod factor signal transduction pathway of Medicago truncatula. Plant Cell 2001, 13, 1835–1849. [Google Scholar] [CrossRef]
- Penmetsa, R.V.; Cook, D.R. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 1997, 275, 527–530. [Google Scholar] [CrossRef]
- Penmetsa, R.V.; Uribe, P.; Anderson, J.; Lichtenzveig, J.; Gish, J.C.; Nam, Y.W.; Engstrom, E.; Xu, K.; Sckisel, G.; Pereira, M.; et al. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 2008, 55, 580–595. [Google Scholar] [CrossRef]
- Chan, P.K.; Biswas, B.; Gresshoff, P.M. Classical Ethylene Insensitive Mutants of the Arabidopsis EIN2 Orthologue Lack the Expected ‘hypernodulation’ Response in Lotus japonicus. J. Integr. Plant Biol. 2013, 55, 395–408. [Google Scholar] [CrossRef]
- Cerri, M.R.; Frances, L.; Kelner, A.; Fournier, J.; Middleton, P.H.; Auriac, M.C.; Mysore, K.S.; Wen, J.Q.; Erard, M.; Barker, D.G.; et al. The Symbiosis-Related ERN Transcription Factors Act in Concert to Coordinate Rhizobial Host Root Infection. Plant Physiol. 2016, 171, 1037–1054. [Google Scholar] [CrossRef]
- Kawaharada, Y.; Nielsen, M.W.; Kelly, S.; James, E.K.; Andersen, K.R.; Rasmussen, S.R.; Fuchtbauer, W.; Madsen, L.H.; Heckmann, A.B.; Radutoiu, S.; et al. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat. Commun. 2017, 8, 14534. [Google Scholar] [CrossRef]
- Journet, E.P.; El-Gachtouli, N.; Vernoud, V.; de Billy, F.; Pichon, M.; Dedieu, A.; Arnould, C.; Morandi, D.; Barker, D.G.; Gianinazzi-Pearson, V. Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant-Microbe Interact. 2001, 14, 737–748. [Google Scholar] [CrossRef]
- Andriankaja, A.; Boisson-Demier, A.; Frances, L.; Sauviac, L.; Jauneau, A.; Barker, D.G.; de Carvalho-Niebel, F. AP2-ERF transcription factors mediate nod factor-dependent mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 2007, 19, 2866–2885. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Hirakawa, H.; Sato, S.; Hayashi, M.; Kawaguchi, M. Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc. Natl. Acad. Sci. USA 2014, 111, 14607–14612. [Google Scholar] [CrossRef] [PubMed]
- Laffont, C.; Ivanovici, A.; Gautrat, P.; Brault, M.; Djordjevic, M.A.; Frugier, F. The NIN transcription factor coordinates CEP and CLE signaling peptides that regulate nodulation antagonistically. Nat. Commun. 2020, 11, 3167. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.L.; Kalo, P.; Yendrek, C.; Sun, J.H.; Liang, Y.; Marsh, J.F.; Harris, J.M.; Oldroyd, G.E.D. Abscisic Acid Coordinates Nod Factor and Cytokinin Signaling during the Regulation of Nodulation in Medicago truncatula. Plant Cell 2008, 20, 2681–2695. [Google Scholar] [CrossRef]
- Gonzalez-Rizzo, S.; Crespi, M.; Frugier, F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 2006, 18, 2680–2693. [Google Scholar] [CrossRef]
- Murray, J.D.; Karas, B.J.; Sato, S.; Tabata, S.; Amyot, L.; Szczyglowski, K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 2007, 315, 101–104. [Google Scholar] [CrossRef]
- Heckmann, A.B.; Sandal, N.; Bek, A.S.; Madsen, L.H.; Jurkiewicz, A.; Nielsen, M.W.; Tirichine, L.; Stougaard, J. Cytokinin Induction of Root Nodule Primordia in Lotus japonicus Is Regulated by a Mechanism Operating in the Root Cortex. Mol. Plant-Microbe Interact. 2011, 24, 1385–1395. [Google Scholar] [CrossRef]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.P.; Daniel-Vedele, F.; Fernandez, E.; et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009, 57, 426–435. [Google Scholar] [CrossRef]
- Nishida, H.; Tanaka, S.; Handa, Y.; Ito, M.; Sakamoto, Y.; Matsunaga, S.; Betsuyaku, S.; Miura, K.; Soyano, T.; Kawaguchi, M.; et al. A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat. Commun. 2018, 9, 499. [Google Scholar] [CrossRef]
- Lin, J.S.; Li, X.L.; Luo, Z.P.; Mysore, K.S.; Wen, J.Q.; Xie, F. NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula. Nat. Plants 2018, 4, 1125. [Google Scholar] [CrossRef]
- Moreau, C.; Gautrat, P.; Frugier, F. Nitrate-induced CLE35 signaling peptides inhibit nodulation through the SUNN receptor and miR2111 repression. Plant Physiol. 2021, 185, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Niu, Y.J.; Konishi, M.; Wu, Y.; Du, H.; Chung, H.S.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; ImaizumiAnraku, H.; Fukai, S.; Syono, K. Unusual branching in the seedlings of Lotus japonicus—Gibberellins reveal the nitrogen-sensitive cell divisions within the pericycle on roots. Plant Cell Physiol. 1996, 37, 461–470. [Google Scholar] [CrossRef][Green Version]
- Akamatsu, A.; Nagae, M.; Nishimura, Y.; Montero, D.R.; Ninomiya, S.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Kawaguchi, M.; Takeda, N. Endogenous gibberellins affect root nodule symbiosis via transcriptional regulation of NODULE INCEPTION in Lotus japonicus. Plant J. 2021, 105, 1507–1520. [Google Scholar] [CrossRef] [PubMed]
- Fonouni-Farde, C.; Tan, S.; Baudin, M.; Brault, M.; Wen, J.Q.; Mysore, K.S.; Niebel, A.; Frugier, F.; Diet, A. DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 2016, 7, 12636. [Google Scholar] [CrossRef]
- Fonouni-Farde, C.; Kisiala, A.; Brault, M.; Emery RJ, N.; Diet, A.; Frugier, F. DELLA1-Mediated Gibberellin Signaling Regulates Cytokinin-Dependent Symbiotic Nodulation. Plant Physiol. 2017, 175, 1795–1806. [Google Scholar] [CrossRef]
- Diedhiou, I.; Diouf, D. Transcription factors network in root endosymbiosis establishment and development. World J. Microbiol. Biotechnol. 2018, 34, 37. [Google Scholar] [CrossRef]
- Cerri, M.R.; Frances, L.; Laloum, T.; Auriac, M.C.; Niebel, A.; Oldroyd GE, D.; Barker, D.G.; Fournier, J.; de Carvalho-Niebel, F. Medicago truncatula ERN Transcription Factors: Regulatory Interplay with NSP1/NSP2 GRAS Factors and Expression Dynamics throughout Rhizobial Infection. Plant Physiol. 2012, 160, 2155–2172. [Google Scholar] [CrossRef]
- McAdam, E.L.; Reid, J.B.; Foo, E. Gibberellins promote nodule organogenesis but inhibit the infection stages of nodulation. J. Exp. Bot. 2018, 69, 2117–2130. [Google Scholar] [CrossRef]
- Mens, C.; Hastwell, A.H.; Su, H.N.; Gresshoff, P.M.; Mathesius, U.; Ferguson, B.J. Characterisation of Medicago truncatula CLE34 and CLE35 in nitrate and rhizobia regulation of nodulation. New Phytol. 2021, 229, 2525–2534. [Google Scholar] [CrossRef]
- Imin, N.; Patel, N.; Corcilius, L.; Payne, R.J.; Djordjevic, M.A. CLE peptide tri-arabinosylation and peptide domain sequence composition are essential for SUNN-dependent autoregulation of nodulation in Medicago truncatula. New Phytol. 2018, 218, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Handa, Y.; Tanaka, S.; Suzaki, T.; Kawaguchi, M. Expression of the CLE-RS3 gene suppresses root nodulation in Lotus japonicus. J. Plant Res. 2016, 129, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, E.L.; Kassaw, T.K.; Smith, L.S.; Marsh, J.F.; Oldroyd, G.E.; Long, S.R.; Frugoli, J.A. The ROOT DETERMINED NODULATION1 Gene Regulates Nodule Number in Roots of Medicago truncatula and Defines a Highly Conserved, Uncharacterized Plant Gene Family. Plant Physiol. 2011, 157, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Kassaw, T.; Schnabel, S.N.E.; Frugoli, J. Root Determined Nodulation1 Is Required for M. truncatula CLE12, But Not CLE13, Peptide Signaling through the SUNN Receptor Kinase. Plant Physiol. 2017, 174, 2445–2456. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.; Funayama-Noguchi, S.; Kawaguchi, M. plenty, a Novel Hypernodulation Mutant in Lotus japonicus. Plant Cell Physiol. 2010, 51, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Shinohara, H.; Mori, T.; Matsubayashi, Y.; Kawaguchi, M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 2013, 4, 2191. [Google Scholar] [CrossRef]
- Yoro, E.; Nishida, H.; Ogawa-Ohnishi, M.; Yoshida, C.; Suzaki, T.; Matsubayashi, Y.; Kawaguchi, M. PLENTY, a hydroxyproline O-arabinosyltransferase, negatively regulates root nodule symbiosis in Lotus japonicus. J. Exp. Bot. 2019, 70, 507–517. [Google Scholar] [CrossRef]
- Krusell, L.; Madsen, L.H.; Sato, S.; Aubert, G.; Genua, A.; Szczyglowski, K.; Duc, G.; Kaneko, T.; Tabata, S.; de Bruijn, F.; et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 2002, 420, 422–426. [Google Scholar] [CrossRef]
- Nishimura, R.; Hayashi, M.; Wu, G.J.; Kouchi, H.; Imaizumi-Anraku, H.; Murakami, Y.; Kawasaki, S.; Akao, S.; Ohmori, M.; Nagasawa, M.; et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature 2002, 420, 426–429. [Google Scholar] [CrossRef]
- Okamoto, S.; Kawaguchi, M. Shoot HAR1 mediates nitrate inhibition of nodulation in Lotus japonicus. Plant Signal. Behav. 2015, 10, e1000138. [Google Scholar] [CrossRef]
- Wopereis, J.; Pajuelo, E.; Dazzo, F.B.; Jiang, Q.Y.; Gresshoff, P.M.; de Bruijn, F.J.; Stougaard, J.; Szczyglowski, K. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 2000, 23, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Searle, I.R.; Men, A.E.; Laniya, T.S.; Buzas, D.M.; Iturbe-Ormaetxe, I.; Carroll, B.J.; Gresshoff, P.M. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 2003, 299, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, A.; Betsuyaku, S.; Osakabe, Y.; Mizuno, S.; Nagawa, S.; Stahl, Y.; Simon, R.; Yamaguchi-Shinozaki, K.; Fukuda, H.; Sawa, S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 2010, 137, 3911–3920. [Google Scholar] [CrossRef] [PubMed]
- Crook, A.D.; Schnabel, E.L.; Frugoli, J.A. The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. Plant J. 2016, 88, 108–119. [Google Scholar] [CrossRef]
- Krusell, L.; Sato, N.; Fukuhara, I.; Koch BE, V.; Grossmann, C.; Okamoto, S.; Oka-Kira, E.; Otsubo, Y.; Aubert, G.; Nakagawa, T.; et al. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 2011, 65, 861–871. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jurgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Hu, C.; Zhu, Y.F.; Cui, Y.W.; Cheng, K.L.; Liang, W.; Wei, Z.Y.; Zhu, M.S.; Yin, H.J.; Zeng, L.; Xiao, Y.; et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 2018, 4, 205–211. [Google Scholar] [CrossRef]
- Kassaw, T.; Bridges, W., Jr.; Frugoli, J. Multiple Autoregulation of Nodulation (AON) Signals Identified through Split Root Analysis of Medicago truncatula sunn and rdn1 Mutants. Plants 2015, 4, 209–224. [Google Scholar] [CrossRef]
- Imin, N.; Mohd-Radzman, N.A.; Ogilvie, H.A.; Djordjevic, M.A. The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J. Exp. Bot. 2013, 64, 5395–5409. [Google Scholar] [CrossRef]
- Laffont, C.; Huault, E.; Gautrat, P.; Endre, G.; Kalo, P.; Bourion, V.; Duc, G.; Frugier, F. Independent Regulation of Symbiotic Nodulation by the SUNN Negative and CRA2 Positive Systemic Pathways. Plant Physiol. 2019, 180, 559–570. [Google Scholar] [CrossRef]
- Chapman, K.; Ivanovici, A.; Taleski, M.; Sturrock, C.J.; Ng, J.L.P.; Mohd-Radzman, N.A.; Frugier, F.; Bennett, M.J.; Mathesius, U.; Djordjevic, M.A. CEP receptor signalling controls root system architecture in Arabidopsis and Medicago. New Phytol. 2020, 226, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Delay, C.; Chapman, K.; Taleski, M.; Wang, Y.W.; Tyagi, S.; Xiong, Y.; Imin, N.; Djordjevic, M.A. CEP3 levels affect starvation-related growth responses of the primary root. J. Exp. Bot. 2019, 70, 4763–4773. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.; Smith, S.; De Rybel, B.; Van Den Broeke, J.; Smet, W.; De Cokere, S.; Mispelaere, M.; De Smet, I.; Beeckman, T. The CEP family in land plants: Evolutionary analyses, expression studies, and role in Arabidopsis shoot development. J. Exp. Bot. 2013, 64, 5371–5381. [Google Scholar] [CrossRef] [PubMed]
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B.P.A. The Plant Peptidome: An Expanding Repertoire of Structural Features and Biological Functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef]
- Ogilvie, H.A.; Imin, N.; Djordjevic, M.A. Diversification of the C-Terminally Encoded Peptide (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes. BMC Genom. 2014, 15, 870. [Google Scholar] [CrossRef]
- Mohd-Radzman, N.A.; Binos, S.; Truong, T.T.; Imin, N.; Mariani, M.; Djordjevic, M.A. Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula. J. Exp. Bot. 2015, 66, 5289–5300. [Google Scholar] [CrossRef]
- Ohyama, K.; Ogawa, M.; Matsubayashi, Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008, 55, 152–160. [Google Scholar] [CrossRef]
- Delay, C.; Imin, N.; Djordjevic, M.A. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J. Exp. Bot. 2013, 64, 5383–5394. [Google Scholar] [CrossRef]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Suzuki, T.; Kawaguchi, M.; Higashiyama, T.; Matsubayashi, Y. A comprehensive strategy for identifying long-distance mobile peptides in xylem sap. Plant J. 2015, 84, 611–620. [Google Scholar] [CrossRef]
- Chapman, K.; Taleski, M.; Ogilvie, H.A.; Imin, N.; Djordjevic, M.A. CEP-CEPR1 signalling inhibits the sucrose-dependent enhancement of lateral root growth. J. Exp. Bot. 2019, 70, 3955–3967. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 2017, 3, 17029. [Google Scholar] [CrossRef] [PubMed]
- Taleski, M.; Chapman, K.; Imin, N.; Djordjevic, M.A.; Groszmann, M. The Peptide Hormone Receptor CEPR1 Functions in the Reproductive Tissue to Control Seed Size and Yield. Plant Physiol. 2020, 183, 620–636. [Google Scholar] [CrossRef] [PubMed]
- Ota, R.; Ohkubo, Y.; Yamashita, Y.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 2020, 11, 641. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.; Walters, L.A.; Cooper, A.M.; Olvera, J.G.; Rosas, M.A.; Rasmusson, A.G.; Escobar, M.A. Nitrate-Regulated Glutaredoxins Control Arabidopsis Primary Root Growth. Plant Physiol. 2016, 170, 989–999. [Google Scholar] [CrossRef]
- Zhu, F.G.; Deng, J.; Chen, H.; Liu, P.; Zheng, L.H.; Ye, Q.Y.; Li, R.; Brault, M.; Wen, J.Q.; Frugier, F.; et al. A CEP Peptide Receptor-Like Kinase Regulates Auxin Biosynthesis and Ethylene Signaling to Coordinate Root Growth and Symbiotic Nodulation in Medicago truncatula. Plant Cell 2020, 32, 2855–2877. [Google Scholar] [CrossRef]
- Huault, E.; Laffont, C.; Wen, J.Q.; Mysore, K.S.; Ratet, P.; Duc, G.; Frugier, F. Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase. PLoS Genet. 2014, 10, e1004891. [Google Scholar] [CrossRef]
- Magori, S.; Oka-Kira, E.; Shibata, S.; Umehara, Y.; Kouchi, H.; Hase, Y.; Tanaka, A.; Sato, S.; Tabata, S.; Kawaguchi, M. TOO MUCH LOVE, a Root Regulator Associated with the Long-Distance Control of Nodulation in Lotus japonicus. Mol. Plant-Microbe Interact. 2009, 22, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Takahara, M.; Magori, S.; Soyano, T.; Okamoto, S.; Yoshida, C.; Yano, K.; Sato, S.; Tabata, S.; Yamaguchi, K.; Shigenobu, S.; et al. TOO MUCH LOVE, a Novel Kelch Repeat-Containing F-box Protein, Functions in the Long-Distance Regulation of the Legume-Rhizobium Symbiosis. Plant Cell Physiol. 2013, 54, 433–447. [Google Scholar] [CrossRef]
- Pant, B.D.; Musialak-Lange, M.; Nuc, P.; May, P.; Buhtz, A.; Kehr, J.; Walther, D.; Scheible, W.R. Identification of Nutrient-Responsive Arabidopsis and Rapeseed MicroRNAs by Comprehensive Real-Time Polymerase Chain Reaction Profiling and Small RNA Sequencing. Plant Physiol. 2009, 150, 1541–1555. [Google Scholar] [CrossRef]
- Kant, S.; Peng, M.S.; Rothstein, S.J. Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in Arabidopsis. PLoS Genet. 2011, 7, e1002021. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Seo, J.S.; Chua, N.H. Nitrogen Limitation Adaptation Recruits Phosphate2 to Target the Phosphate Transporter PT2 for Degradation during the Regulation of Arabidopsis Phosphate Homeostasis. Plant Cell 2014, 26, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Lopez, O.; Arenas-Huertero, C.; Ramirez, M.; Girard, L.; Sanchez, F.; Vance, C.P.; Luis Reyes, J.; Hernandez, G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008, 31, 1834–1843. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Liu, W.; Kohlen, W.; Lillo, A.; Op den Camp, R.; Ivanov, S.; Hartog, M.; Limpens, E.; Jamil, M.; Smaczniak, C.; Kaufmann, K.; et al. Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 2011, 23, 3853–3865. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341–344. [Google Scholar] [CrossRef]
- Grdisa, M.; White, M.K. Erythrocytic differentiation and glyceraldehyde-3-phosphate dehydrogenase expression are regulated by protein phosphorylation and cAMP in HD3 cells. Int. J. Biochem. Cell Biol. 2000, 32, 589–595. [Google Scholar] [CrossRef]
- Vierheilig, H. Further root colonization by arbuscular mycorrhizal fungi in already mycorrhizal plants is suppressed after a critical level of root colonization. J. Plant Physiol. 2004, 161, 339–341. [Google Scholar] [CrossRef]
- Meixner, C.; Ludwig-Muller, J.; Miersch, O.; Gresshoff, P.; Staehelin, C.; Vierheilig, H. Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta 2005, 222, 709–715. [Google Scholar] [CrossRef]
- Le Marquer, M.; Becard, G.; Frey, N.F.D. Arbuscular mycorrhizal fungi possess a CLAVATA3/embryo surrounding region-related gene that positively regulates symbiosis. New Phytol. 2019, 222, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.M.; Flokova, K.; Schnabel, E.; Sun, X.P.; Fei, Z.J.; Frugoli, J.; Bouwmeester, H.J.; Harrison, M.J. A CLE-SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat. Plants 2019, 5, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Besserer, A.; Puech-Pages, V.; Kiefer, P.; Gomez-Roldan, V.; Jauneau, A.; Roy, S.; Portais, J.C.; Roux, C.; Becard, G.; Sejalon-Delmas, N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006, 4, 1239–1247. [Google Scholar] [CrossRef]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pages, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Becard, G.; Bonfante, P.; et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013, 198, 179–189. [Google Scholar] [CrossRef]
- Kobae, Y.; Kameoka, H.; Sugimura, Y.; Saito, K.; Ohtomo, R.; Fujiwara, T.; Kyozuka, J. Strigolactone Biosynthesis Genes of Rice are Required for the Punctual Entry of Arbuscular Mycorrhizal Fungi into the Roots. Plant Cell Physiol. 2018, 59, 544–553. [Google Scholar] [CrossRef]
- Hernandez, G.; Valdes-Lopez, O.; Ramirez, M.; Goffard, N.; Weiller, G.; Aparicio-Fabre, R.; Fuentes, S.I.; Erban, A.; Kopka, J.; Udvardi, M.K.; et al. Global Changes in the Transcript and Metabolic Profiles during Symbiotic Nitrogen Fixation in Phosphorus-Stressed Common Bean Plants. Plant Physiol. 2009, 151, 1221–1238. [Google Scholar] [CrossRef]
- Sulieman, S.; Van Ha, C.; Schulze, J.; Tran, L.S.P. Growth and nodulation of symbiotic Medicago truncatula at different levels of phosphorus availability. J. Exp. Bot. 2013, 64, 2701–2712. [Google Scholar] [CrossRef] [PubMed]
- Cabeza, R.A.; Liese, R.; Lingner, A.; von Stieglitz, I.; Neumann, J.; Salinas-Riester, G.; Pommerenke, C.; Dittert, K.; Schulze, J. RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N-2 fixation before emerging P deficiency reaches the nodules. J. Exp. Bot. 2014, 65, 6035–6048. [Google Scholar] [CrossRef]
- Esfahani, M.N.; Inoue, K.; Chu, H.D.; Nguyen, K.H.; Ha, C.V.; Watanabe, Y.; Burritt, D.J.; Herrera-Estrella, L.; Mochida, K.; Tran, L.S.P. Comparative transcriptome analysis of nodules of two Mesorhizobium-chickpea associations with differential symbiotic efficiency under phosphate deficiency. Plant J. 2017, 91, 911–926. [Google Scholar] [CrossRef]
- Isidra-Arellano, M.C.; Reyero-Saavedra, M.D.; Sanchez-Correa, M.D.; Pingault, L.; Sen, S.; Joshi, T.; Girard, L.; Castro-Guerrero, N.A.; Mendoza-Cozatl, D.G.; Libault, M.; et al. Phosphate Deficiency Negatively Affects Early Steps of the Symbiosis between Common Bean and Rhizobia. Genes 2018, 9, 498. [Google Scholar] [CrossRef] [PubMed]
- Isidra-Arellano, M.C.; Pozas-Rodriguez, E.A.; Reyero-Saavedra, M.D.; Arroyo-Canales, J.; Ferrer-Orgaz, S.; Sanchez-Correa, M.D.; Cardenas, L.; Covarrubias, A.A.; Valdes-Lopez, O. Inhibition of legume nodulation by Pi deficiency is dependent on the autoregulation of nodulation (AON) pathway. Plant J. 2020, 103, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.J.; Yan, J.D.; Yang, P.; Zhou, J.; Li, X.M.; Zhao, X.Y. Progress of function studies of ZTL/FKF1/LKP2 proteins family in Arabidopsis. Chin. J. Bioinform. 2019, 17, 145–150. [Google Scholar]
- Yan, J.D.; Li, X.M.; Zeng, B.J.; Zhong, M.; Yang, J.X.; Yang, P.; Li, X.; He, C.S.; Lin, J.Z.; Liu, X.M.; et al. FKF1 F-box protein promotes flowering in part by negatively regulating DELLA protein stability under long-day photoperiod in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1717–1740. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).