Abstract
The diverse repertoires of cellular mechanisms that progress certain cancer types are being uncovered by recent research and leading to more effective treatment options. Ovarian cancer (OC) is among the most difficult cancers to treat. OC has limited treatment options, especially for patients diagnosed with late-stage OC. The dysregulation of miRNAs in OC plays a significant role in tumorigenesis through the alteration of a multitude of molecular processes. The development of OC can also be due to the utilization of endogenously derived reactive oxygen species (ROS) by activating signaling pathways such as PI3K/AKT and MAPK. Both miRNAs and ROS are involved in regulating OC angiogenesis through mediating multiple angiogenic factors such as hypoxia-induced factor (HIF-1) and vascular endothelial growth factor (VEGF). The NAPDH oxidase subunit NOX4 plays an important role in inducing endogenous ROS production in OC. This review will discuss several important miRNAs, NOX4, and ROS, which contribute to therapeutic resistance in OC, highlighting the effective therapeutic potential of OC through these mechanisms.
Keywords:
ovarian cancer; miRNA dysregulation; ROS; NOX4; HIF1-α; VEGF; angiogenesis; HER3; therapeutic resistance 1. Ovarian Cancer
Known as the silent killer, ovarian cancer (OC) has the lowest survival rate and the worst prognosis among all gynecologic malignancies in the US; and is the eighth most common cancer in women worldwide [1,2]. In 2022, the American Cancer Society estimates about 21,000 new cases of OC will be diagnosed, and approximately 14,000 women will die from this type of cancer. The overall 5-year survival rate is only 48% due to OC’s ambiguous symptoms and inadequate screening capabilities at the early stages of the disease. Due to late detection, about 60% of new cases are diagnosed when the disease has already progressed to the advanced stage [2]. OC is a heterogeneous disease with several subtypes that differ in their gene expression, tumor origin, pathway alterations, and pathogenesis. The majority of OC originates from three main cell types: epithelial cells (90%), stromal cells (7%), and germ cells (3%) [1,3,4]. In general, epithelial OC can be further divided into five histotypes: high-grade serous (HGSOC; 70%), endometrioid (ENOC; 10%), clear cell (CCOC; 10%), mucinous (MOC; 5%), and low-grade serous (LGSOC; less than 5%) OC [4]. In addition, another classification system was introduced a decade ago that divided OC into type I and II tumors. Type I tumors are low-grade neoplasms, including mucinous carcinomas, endometrioid carcinomas, malignant Brenner tumors, and clear cell carcinomas. Type I tumors are typically characterized by mutations in BRAF, KRAS, and PTEN with DNA instability. Type II tumors are high-grade serous carcinoma, carcinosarcoma, and undifferentiated carcinoma, which are frequently observed with mutations in p53, BRCA1/2, HER-2/HER-3 overexpression, and p16 inactivation [5,6,7,8]. Depending on the specific subtype and histopathology, OC treatment involves a combination of surgery and chemotherapy. For patients with advanced-stage tumors, debulking surgery is recommended; however, large tumors or residual tumors may show negative side effects leading to blockage of the perfusion area and the possibility of developing drug resistance [1,9]. Platinum-based chemotherapy is the standard line of treatment for OC, either in conjunction with or following surgery [10,11,12]. The combination of paclitaxel/carboplatin has been recognized as the standard postoperative chemotherapy for many years [13]. In recent years, PARP inhibitors have been incorporated into clinical treatment as a recommended maintenance drug [14]. However, due to the aggressive growth rates and the propensity of advanced tumors to evade treatment, there are critical limitations to the current lines of therapy. A better understanding of the molecular biology of OC is allowing more research efforts to establish new effective treatment options for advanced-stage tumors.
2. ROS
Reactive oxygen species (ROS) have remained a highly relevant topic over the last few decades due to their expansive effects on normal cellular function. Oxidative stress is generated through the accumulation of ROS, either through exogenous exposure or endogenous production. ROS are oxygen ions with unpaired electrons (singlet oxygen 1O2, superoxide O2·−) or oxygen-containing molecules, such as hydroxyl radicals (OH·−), hydrogen peroxide (H2O2), nitric oxide (NO), and nitrogen dioxide (NO2) [15]. Superoxide radicals are converted into H2O2 by the enzyme superoxide dismutase (SOD). However, superoxide can also react with nitric oxide to produce peroxynitrite (ONOO−), a strong oxidizer with damaging cellular effects [16]. The accumulation of H2O2 has detrimental effects on nuclear and mitochondrial DNA, which may lead to genetic instability to drive cancer progression with increased expression of oncogenes and decreased expression of tumor suppressors [17,18]. Several enzymes work in conjunction to convert H2O2 into the water, including catalase, glutathione peroxidases 1 and 4, and peroxiredoxins 3 and 5 [19,20,21,22]. Furthermore, H2O2 can also participate in the Fenton reaction, in which free iron Fe(II) reacts with H2O2, generating highly reactive hydroxyl radicals (·OH)(shown below). The production of hydroxyl radicals (·OH) by the Haber–Weiss reaction (shown below) further perpetuates the damaging effects of the accumulation of ROS.
Fenton Reaction:
Fe(II) + H2O2 ⟷ Fe(III) + ·OH + OH−
Haber–Weiss Reaction:
O2·− + H2O2 ⟷ OH + OH− + O2
Original studies implicated the mitochondria as primary endogenous sources of superoxide through the process of cellular respiration, a process dependent on the availability of O2 [23,24,25]. Based on this view, the production of ROS was thought to be a harmful by-product of intracellular metabolism. Then a family of transmembrane enzymes known as NADPH oxidase (NOX) proteins was identified, whose primary function was the production of endogenous ROS. NOX2, the first NOX protein discovered, was the primary producer of endogenous ROS in leukocytes to generate an oxidative burst, an essential process for the neutralization of pathogens [26,27,28,29]. The characterization of a disease called chronic granulomatous disease (CGD) caused by a mutation in the phagocytic NOX gene provided insight into the emerging role of endogenous ROS production on cellular functionality [30,31]. Subsequent work demonstrated a pivotal role of NOX proteins in mammalian cell transformation through the production of superoxide radicals and H2O2 [32,33]. Our group demonstrated that the accumulation of ROS in OC cells was attributed to H2O2 increased levels induced by NOX4 [34], identifying an endogenous mechanism for the overproduction of ROS and alteration of intracellular signaling in OC tumor development.
Under normal cellular conditions, low levels of endogenous ROS activate several signaling pathways involved in cell proliferation. However, the accumulation of ROS causes extensive damage to DNA, RNA, proteins, and lipids, thus causing a significant hindrance to normal cellular functions and contributing to the development of multiple human pathologies [35,36,37,38]. The damage can induce cell death pathways or trigger the mutation of DNA, as commonly found in cancer [39,40]. In addition to the endogenous production of ROS and oxidative stress, external or environmental exposure to ROS can have detrimental effects on mammals [41]. For instance, many chemotherapeutic agents induce oxidative stress as a means of inducing cellular damage and cell death pathways [42]. However, as demonstrated by more recent findings, ROS play an important role in the progression and advancement of human diseases. The counterweight for endogenous ROS is the genetically programmed redox system. This includes groups of genes coding for antioxidant proteins such as superoxide dismutase (SOD), catalase, and the glutathione system, which neutralize the ROS produced in cells [43,44,45]. The failure to neutralize endogenous ROS leads to a build-up of harmful oxygen species and, consequently, oxidative stress. In normal cells, oxidative stress leads to deleterious cellular effects, such as protein, lipid, and DNA damage, organelle dysfunction, and cell cycle arrest [46]. Higher levels of oxidative stress cause the activation of cell death pathways such as apoptosis and necrosis [46], which may be mitigated in cancer cells by an increase in antioxidant production. The upregulation of nuclear factor erythroid 2-related factor 2 (NRF2), a master transcriptional regulator of antioxidant genes, contributes to the neutralization of endogenous ROS in OC cells [47,48,49], making NRF2 a viable target for chemotherapeutic treatment in certain cases of OC. In addition, the genetic mutation of cellular pathways that induce cell death mechanisms in response to increased oxidative stress allows cancer cells to evade the activation of cell death pathways [50], thus providing cancer cells the ability to continue continuous proliferation in the presence of adverse cellular conditions, such as oxidative stress.
3. ROS in the Development of Ovarian Cancer
There is an established link between an increase in ROS production and cancer development in humans [51]. As secondary cellular signaling molecules, ROS are involved in the activation of several signaling pathways involved in cell proliferation and growth. Consequently, these pathways are constitutively activated in cancer cells with increased ROS levels that contribute to tumorigenesis [51]. For example, endogenously derived ROS activate the ERK1/2 MAPK signaling pathway and the AKT signaling pathway in OC, both of which promote cell proliferation [52,53]. The increased ROS generation also contributes to a genetic mutation in cancer cells, further contributing to cell transformation [54,55]. As opposed to the traditional view of ROS generation in cancer as a harmful secondary by-product, the increasing knowledge of cancer cell metabolism and signal transduction is exposing ROS as a positive contributing factor in tumorigenesis and cancer development. The increased metabolic activity of cancer cells was originally thought to be responsible for the accumulation of ROS as a byproduct of increased glycolytic metabolism and mitochondrial respiration [56]. However, the discovery of the role of NOX proteins in endogenous ROS production revealed a more important role for ROS production in non-phagocytic cells, particularly in cancer [57,58,59]. The endogenous production of ROS by NOX1 was found to be responsible for increased viability and proliferation in colon cancer [60,61]. Similarly, the role of NOX2-mediated ROS production was discovered to be critical for cell viability and proliferation in breast, colorectal, myelomonocytic leukemia, gastric, and prostate cancers [62,63,64,65,66,67]. NOX4 overexpression contributed to an oncogenic proliferation in renal cell carcinoma, melanoma, glioblastoma, ovarian, prostate, and lung cancers [34,68,69,70,71,72]. In OC cell lines, there is a significant increase in ROS production, which contributes to tumorigenesis [34]. The increase in ROS is a result of NADPH oxidase activity and mitochondrial metabolism, as this increase is diminished by NADPH oxidase and mitochondrial complex I inhibitors [34]. Moreover, the increased levels of ROS result from the upregulation of the NADPH oxidase subunit NOX4, which serves as the main contributor to ROS production in OC cells to promote tumor growth and angiogenesis [34]. Furthermore, the activation of NOX4 is positively correlated with TGF-β1 and NF-κB activity, which is suppressed by their inhibitors [34]. This system demonstrates that endogenous NOX4-derived ROS are a driving force in OC development. Moreover, NOX4 is a potential target for the therapeutic resistance of OC which is dependent on ROS production for an increase in oncogenic signaling.
4. miRNA Dysregulation in Cancer
The progression of cancer is often associated with dysregulation of non-coding RNAs, including microRNAs (miRNAs) [73,74]. miRNAs are 18–25 nucleotide long, non-coding single-stranded RNA molecules that regulate the expression of messenger RNA (mRNA) [75]. The discovery of miRNA in 1993 by Ambros and colleagues in the nematode C. Elegans revealed the critical role of miRNAs in the post-transcriptional regulation of mRNA [76,77]. In these studies, the miRNA lin-4 was found to regulate the expression of the critical developmental transcription factor, lin-14 [76,77]. The primary transcripts of miRNA (pri-miRNA) are modified within the nucleus by the RNase III DROSHA and its cofactor DGCR8 before being exported to the cytoplasm as pre-miRNA [78,79]. Mature miRNA molecules are the result of the cleavage of pre-miRNA at the terminal loop by the RNase III endonuclease, DICER [80,81]. The regulation of miRNA processing can have expansive effects on cellular processes, as demonstrated by the gain-of-function mutation of DICER as a contributory factor in cancer development [82]. As transcriptional regulatory molecules, miRNAs typically recognize and bind the 3′-UTR of target mRNAs to repress expression or induce degradation [83]. The activation of genes by miRNAs occurs through association with the promoter region and upstream regulatory regions of target genes [84]. The search for the role of miRNA in humans yielded a plethora of data that are still accumulating, particularly the dysregulation of miRNAs in oncogenesis. The original studies identifying the role of miRNAs in human oncogenesis demonstrated the effect of miR-15a/16a repression on promoting the oncogenic protein Bcl-2 in chronic lymphocytic leukemia [85]. Most human miRNAs function as tumor suppressors by directly targeting and inhibiting oncogenes, such as RAS and MYC. For instance, the downregulation of Let-7 family of miRNAs, which target KRAS and C-MYC, is found in OC, which induces tumor growth and development [86,87]. However, some miRNAs function as oncogenes by directly targeting and inhibiting tumor suppressors such as p53 [88,89]. For example, miR-25 and miR-30d target p53 for degradation and contribute to colon cancer development; the downregulation of both miR-25 and miR-30d led to an increase in p53 protein expression and increased apoptosis in multiple cancer types [90]. Many miRNAs are dysregulated in multiple cancers, including the upregulation of miR-155 in lymphomas and colorectal cancers [91,92], indicating a commonality in the mode of miRNA dysregulation in multiple cancer/tissue types. The molecular effects of miRNA dysregulation include feedback mechanisms, such as the miR-17-92 cluster/E2F family/c-MYC loop. In this feedback mechanism, miR-17-92 is activated by c-MYC and inhibits E2F family protein translation [93,94]. The E2F family of proteins (E2F1, E2F2, E2F3) are critical cell-cycle regulated inducers of proliferation, therefore proper regulation of these proteins is necessary under normal conditions [95]. Further investigation revealed that c-MYC activation of E2F family proteins activates miR-17-92, leading to a feedback loop to tightly control the expression of E2F proteins in healthy cells [96,97]. However, in cancer cells the amplification and overexpression of miR-17-92 disrupts this feedback loop and contributes to high cell proliferation and tumorigenesis [98]. In another example, miR-221/222 upregulation in cancer cells contributes to oncogenesis through the inhibition of cell cycle regulating protein p27 [99,100,101]. The dysregulation of particular miRNAs can differ between subtypes of OC. For instance, the overexpression of miR-483 occurs in serous epithelial ovarian cancer (EOC), but does not occur in non-serous EOC [102]. As demonstrated in voluminous publications, miRNA dysregulation affects a variety of cellular processes that contribute to oncogenesis in a wide variety of cancers. The complex role of miRNAs in cancer development highlights the potential for therapies targeting specific miRNAs that are dysregulated in different cancers.
5. ROS and miRNA Dysregulation in Angiogenesis and Ovarian Cancer Development
The development of tumors involves a wide variety of cellular processes. In this regard, ROS contribute to critical cellular processes that occur within tumors, including angiogenesis and micro-RNA (miRNA) dysregulation. Angiogenesis is the creation of new blood vessels within existing vasculature, which is essential for processes such as embryogenesis, tissue repair, and organ regeneration [103]. Unsurprisingly, angiogenesis plays a pivotal role in cancer development through the establishment of nutrients and blood supply to newly formed tumors [104]. A significant contribution to angiogenic signaling is made by vascular endothelial growth factor (VEGF), which is highly upregulated in developing embryonic cells and tumor cells [105,106,107]. The limited oxygen availability in tumors often leads to hypoxic conditions, in which signaling pathways are activated to initiate tumor growth and angiogenesis [108]. The hypoxia-inducible factor 1 alpha (HIF-1α) plays a vital role in the hypoxic response in tumor cells, partially through the upregulation of VEGF [109,110,111]. The dysregulation of HIF1-α occurs in a wide variety of cancers which contributes to tumorigenesis [112]. The upregulation of HIF-1α and VEGF are positively correlated with NOX4-derived ROS production in OC cells and promotes angiogenesis and tumor growth [34]. In turn, HIF-1α induces the expression of VEGF; and promotes the production of NOX4 through an alternative splicing mechanism [34,113]. This positive feedback system demonstrates the capacity of OC cells to utilize the overproduction of NOX4-derived ROS to activate HIF-1α and VEGF and promote angiogenesis and tumor growth.
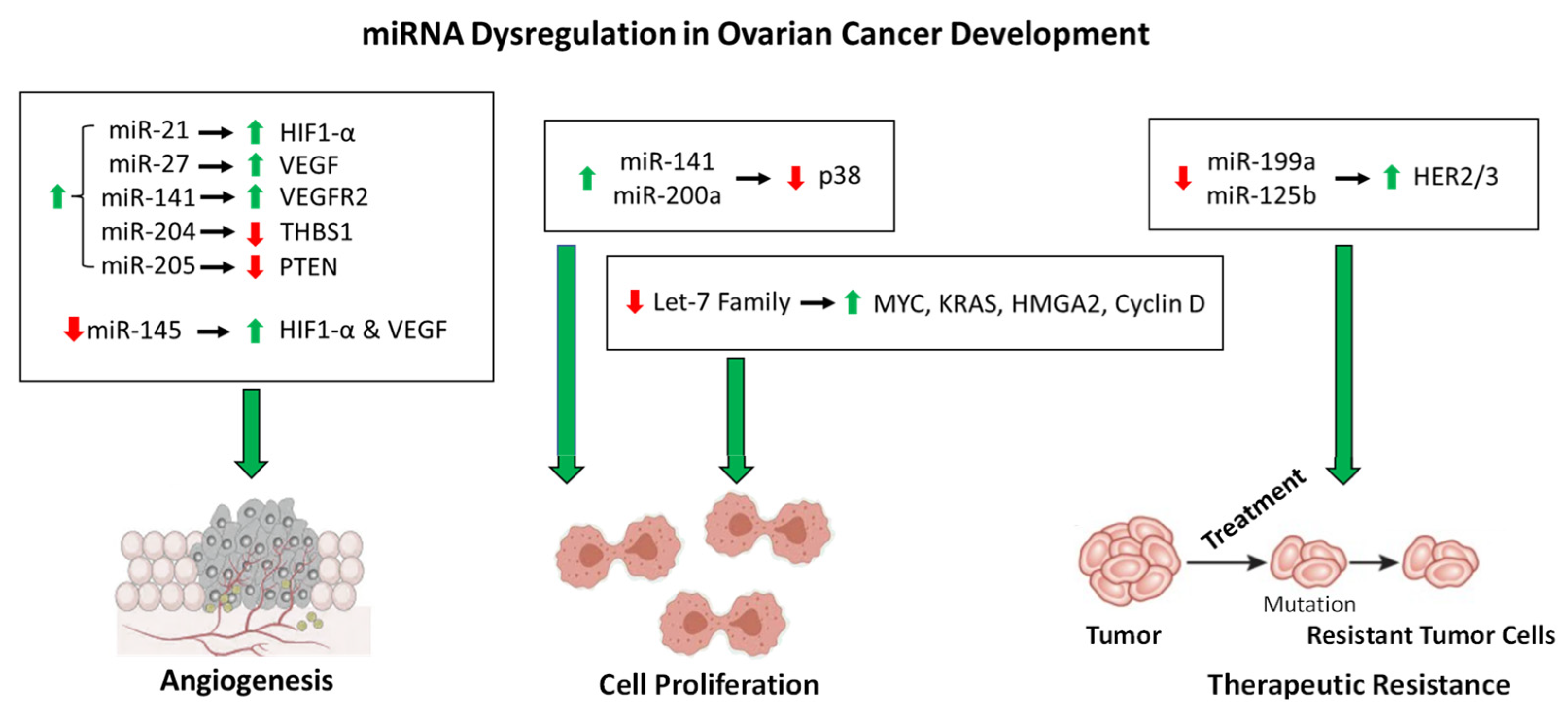
This regulation system is complicated more by the dysregulation of miRNAs by increased intracellular ROS, which contributes to OC tumorigenesis. For instance, miR-199a and miR-125b downregulate the expression of the oncogenic proteins HER2 and HER3 under normal cellular conditions [114]. However, increased ROS in OC cells results in the downregulation of miR-199a and miR-125b through DNA hypermethylation, thereby increasing the expression of HER2/3 and contributing to tumorigenesis [114]. More importantly, this demonstrates the role of epigenetic regulation of miRNA expression. Another example of epigenetic regulation of miRNA expression is the increased acetylation of miR-466-5p in response to increased ROS, thereby inducing the expression of miR-466-5p, which then activates pro-apoptotic genes [115,116]. The expression of some miRNAs is regulated by ROS-stress-responsive transcription factors, which contribute to molecular signaling cascades. The tumor suppressor p53 is induced in response to ROS and subsequently activates the miR-200 family of miRNAs [117]. The miR-200 family members have been implicated as tumor suppressors, and overexpression of these miRNAs inhibits tumor development in OC [118,119]. However, two members of this family of miRNAs, miR-141 and miR-200a, directly target p38α in response to increased levels of ROS, resulting in evasion of apoptosis induction and upregulation in antioxidant production [120,121], thus demonstrating the complexity of molecular roles of a single family of miRNAs in OC progression. As contributing factors to the pathway described above, miR-21 and miR-27a induce angiogenesis in OC through the upregulation of HIF1-α and VEGF, respectively [122,123]. Additional pathways affected by miRNA dysregulation also contribute to angiogenesis in OC. For instance, miR-141 is upregulated in OC to induce the expression of VEGFR2, resulting in an increase in angiogenesis [124]. By a differing mechanism, the upregulation of miR-205 in OC results in an increase in angiogenesis through the downregulation of tumor suppressor PTEN and an increase in AKT signaling [125]. Similarly, miR-204 upregulation in OC contributes to angiogenesis through the downregulation of anti-angiogenic protein THBS1 [126,127]. The miRNAs listed in Table 1 are upregulated in OC cells and contribute positively to tumor growth, development, angiogenesis, and therapeutic resistance. However, there are substantial data demonstrating a tumor suppressor role for various miRNAs in OC, whose downregulation results in tumorigenesis, angiogenesis, and treatment resistance (Table 2) [128]. For instance, miR-145 acts as a tumor suppressor, and the downregulation of miR-145 in OC contributes to angiogenesis through the upregulation of HIF-1α and VEGF [129]. The overall role of the miRNAs in OC development described here is shown in Figure 1. Another important avenue of miRNA research involves the dysregulation of specific circulatory miRNAs, which has the inherent propensity to impact a multitude of tissue types. Recent work has shown that the expression levels of miR-200b, miR-200c, miR-141, and miR-1274A in OC patients’ circulatory systems are negatively correlated with survival [130]. Therefore, the roles of miRNA dysregulation in OC angiogenesis and development remain to be fully understood which warrants further investigation to provide therapeutic options and/or targets in the future.

Table 1.
miRNAs upregulated in OC cells that contribute to tumor growth and development, angiogenesis, and therapeutic resistance.

Table 2.
miRNAs downregulated in OC cells that function as tumor suppressors.
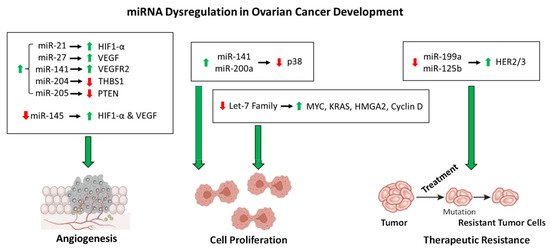
Figure 1.
The roles of miRNA dysfunction in OC development. As described in the text, the miRNAs shown above are dysregulated in OC that contribute to angiogenesis, cell proliferation, and therapeutic resistance. The dysregulation of each miRNA is denoted by an up arrow (upregulation) or a down arrow(downregulation). The regulation of proteins affected by the dysregulation miRNAs is denoted in the same manner.
6. Potential Mechanism of ROS in Therapeutic Resistance in Ovarian Cancer
The two major obstacles facing efficient treatment of OC are late detection/diagnosis and acquired therapeutic resistance. The standard treatment for OC includes preliminary debulking surgery followed by platinum-based (carboplatin and cisplatin) and/or taxane family-based (paclitaxel and docetaxel) chemotherapy [164,165]. The mode of action of platinum-based therapies is oxidative stress-induced cellular damage and initiation of cell death pathways, such as apoptosis, which is triggered by this class of chemotherapeutics [166]. The taxane family-based drugs are used to inhibit cell division through microtubule stabilization [167]. However, due to toxic side effects associated with high-dose treatment and acquired resistance to carboplatin and cisplatin treatment, this traditional route of therapy has critical limitations. The mechanisms of drug resistance to these treatment options include an increase in DNA damage repair and an increase in antioxidant production to detoxify cancer cells [168]. Regarding this, other chemotherapeutic agents are used to treat resistant tumors, including gemcitabine, doxorubicin, and bevacizumab [169,170,171]. There are additional combinations of chemotherapy used to treat OC, such as targeted treatment of anti-apoptotic proteins that are overexpressed in OC cells. For instance, the anti-apoptotic protein Bcl-2 is overexpressed in OC, and treatment of tumor cells with a combination of cisplatin or carboplatin and Bcl-2 inhibitors show an increased level of cancer cell death induction [172,173,174,175]. The class of VEGF regulators known as specific proteins (Sp) are also targeted by small molecule inhibitors to induce cell death [176]. Another important mechanism for cancer cells to evade treatment is the upregulation of the glycoproteins that form the molecular pumps to export chemotherapeutic agents out of the cancer cells, driving the process of multi-drug resistance [177,178]. The complexity of miRNA dysregulation in OC also contributes to treatment resistance. For instance, OC cells evade apoptosis in response to paclitaxel treatment through upregulation of miR-21 and miR-106a, that target and downregulate the pro-apoptotic proteins APAF1 and CASP7, respectively [131,132]. Similarly, miR-182 upregulation in OC results in evasion of apoptosis in response to cisplatin/paclitaxel treatment through the downregulation of pro-apoptotic protein PDCD4 [142]. Regarding therapeutic efficacy, the ROS-induced miRNAs mentioned previously, miR-200a and miR-141, although shown as oncogenic, can increase the sensitivity of OC to paclitaxel treatment through the downregulation of p38 [121,179]. Similarly, overexpression of miR-522 can increase the sensitivity of OC to paclitaxel treatment [180]. A better understanding of the molecular mechanisms driving treatment resistance in OC is of vital importance for the design of therapies that will effectively treat aggressive, resistant tumors.
The increase in intracellular ROS levels in OC has been shown to contribute to therapeutic resistance. For instance, an increase in ROS in OC results in the overexpression of dCTP pyrophosphatase I (DCTPP1), which has a role in DNA damage repair and plays a major contribution to cisplatin resistance [181]. By a differing mechanism, the upregulation of calcium/calmodulin-dependent protein kinase II gamma (CAMK2G) in response to increasing levels of ROS reprograms the cellular redox system through the phosphorylation of inositol triphosphate3-kinase B (ITPKB), resulting in adaptive redox homeostasis and increased resistance to cisplatin treatment [182]. Similarly, the upregulation of PGC1-α by increasing intracellular ROS contributes to chemotherapy resistance through the upregulation of drug resistance-related proteins, MDR1 and ABCG2, leading to increased antioxidant production and drug efflux [183]. The increase in ROS in OC downregulates miR-199a and miR-125b, resulting in the increased expression of HER2 and HER3 and therapeutic resistance [114]. Thus, another mode of treatment for OC is vaccines targeting human HER2 and HER3 [184,185]. In work highlighted here, the increase in NOX4-derived ROS contributes to therapeutic resistance in OC through the upregulation of HER3. The upregulation of HER3 is a clinical marker for OC, which is positively correlated with poor prognosis [186]. NOX4 directly activates HER3 and contributes to the increased resistance of OC cells to chemotherapy and radiation treatments [113]. The deletion of NOX4 results in a reduction in the therapeutic resistance of OC cells [113]. Similarly, inhibition of NOX4 acts synergistically with HER3 inhibition to decrease tumor growth in OC [113]. The knockdown of NOX4 using siRNA also results in enhanced sensitivity to radiation treatment in OC cells, proving this pathway relevant in multi-modal therapeutic resistance [113]. The NOX4-driven system of endogenous ROS production demonstrates a new mechanism in OC cells to promote tumor development, angiogenesis, and an increase in therapeutic resistance through the upregulation of HER3, reflecting a candidate for targeted therapy of treatment-resistant OC (Figure 2). These findings shed light on the importance of endogenous NOX4-derived ROS production in cell signaling and the progression of OC and the propensity of tumors to evade current lines of treatment.
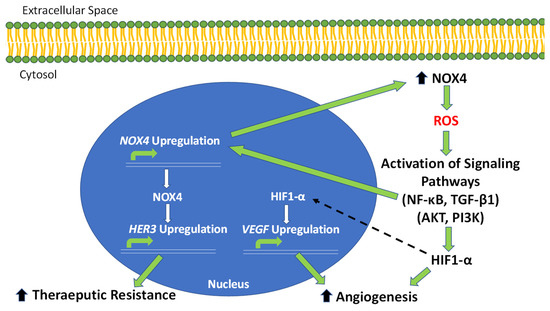
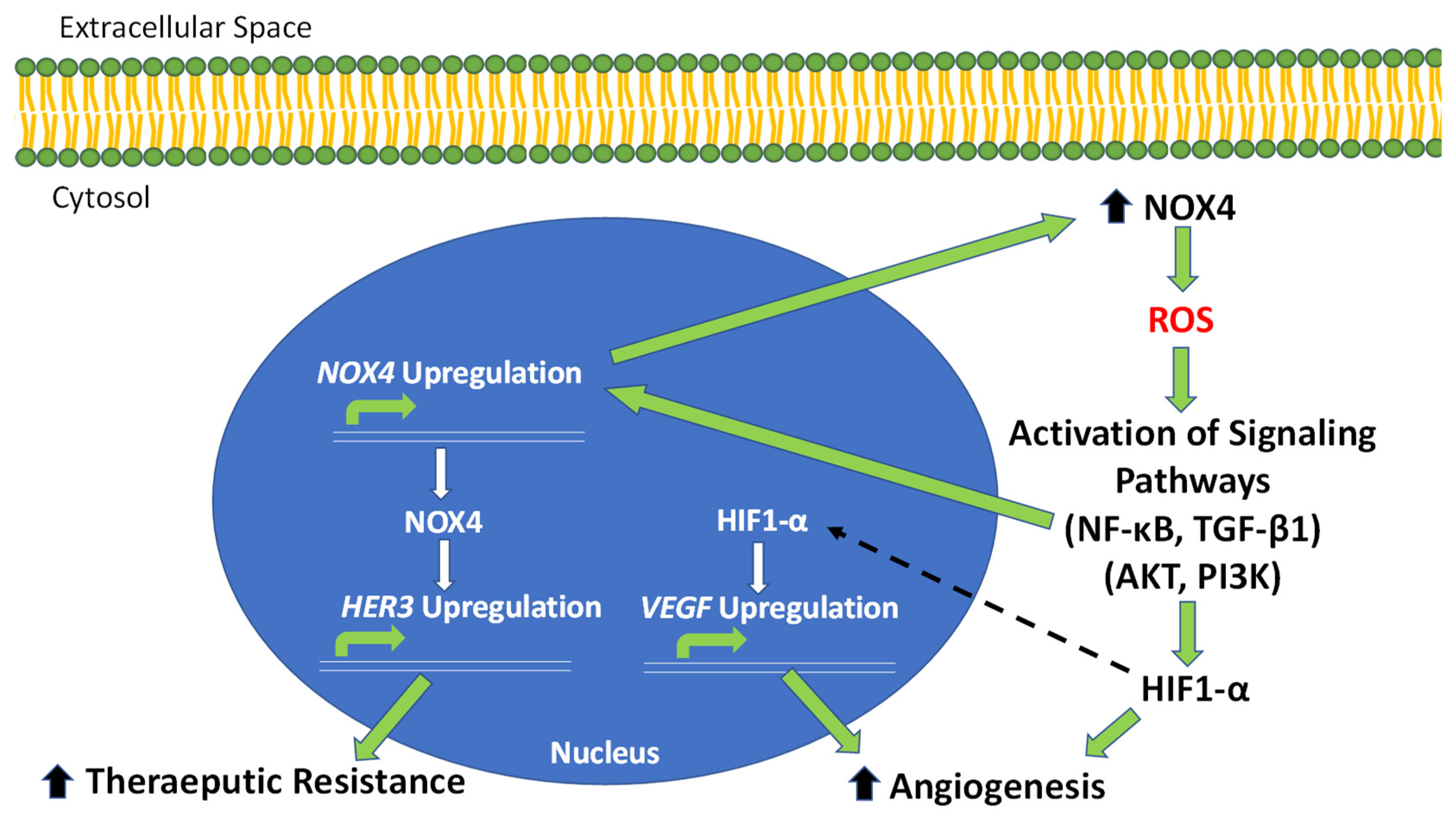
Figure 2.
NOX4-driven pathways of OC tumor progression, angiogenesis and therapeutic resistance. The overexpression of NOX4 in OC results in an increase in intracellular ROS production. Increased ROS leads to an increase in HIF1-A through activating PI3K and AKT signaling. HIF1-α then activates the critical angiogenic factor, VEGF. Increased ROS also activates NF-κB and TGF-β1 signaling, which lead to the direct upregulation of NOX4. The increase in NOX4 contributes, in a positive-feedback manner, to increased ROS production. NOX4 also activates the expression of HER3, contributing to therapeutic resistance in OC tumors.
7. Future Directions
The key to effective treatment of OC is the understanding of the molecular mechanisms that drive tumor development and resistance to current treatments. In the system described above, the increased levels of endogenous ROS produced by NOX4 is utilized by OC cells to stimulate tumorigenesis, angiogenesis, and treatment resistance (Figure 2). This adaptation in cellular signaling allows OC tumors to proliferate and develop resistance to chemotherapeutics through ROS production and upregulation of HER3, thus identifying this NOX4-driven pathway as a potential target for the treatment of chemoresistant tumors. In support of this, clinical trial studies show HER3 upregulation is associated with poor prognosis in OC, which serves as a clinical marker of tumor development, and HER3 expression is induced in response to current chemotherapeutics agents [186,187]. Therefore, this pathway provides an explanation for the ineffectiveness of traditional therapies for advanced OC and the development of therapeutic resistance. The implications of the findings reviewed here include the potential for NOX4 overexpression and increased levels of ROS to be utilized as a diagnostic biomarker in OC. Furthermore, there is clinical relevance for identifying new treatable targets in OC affected by this NOX4-driven system, particularly in resistant tumors.
As a significant mediator of miRNA dysregulation, ROS can have widespread effects on cellular processes. The roles of miRNA dysregulation in OC complicate the understanding of signaling pathways altered by tumors, with some acting as oncogenes and others acting as tumor suppressors. Similarly, the dysregulation of miRNAs in a cell-type-specific manner provides an opportunity to target specific miRNAs in different types of cancers. This could be accomplished by targeting the suppression of oncogenic miRNAs, which are typically upregulated in tumors, whereas the expression levels of tumor suppressor-like miRNAs are typically downregulated or lost in tumors [188]. The suppression of oncogenic miRNAs can be achieved with the use of anti-miRNA molecules targeting specific miRNA for inhibition or degradation [189]. For instance, anti-miR-21 treatment in breast cancer and glioblastoma induces apoptosis through the inhibition of PI3K signaling [190,191]. Alternatively, the upregulation of tumor suppressor miRNAs can be achieved with the use of miRNA mimics, which are delivered as mature miRNA molecules [192]. The use of miRNA mimics in combination with other forms of therapies improves treatment efficacy and the elimination of tumor cells. For example, the delivery of miR-204-5p in combination with oxaliplatin in colon cancer reduced tumor growth and induced apoptosis [193]. In further support of this, the treatment of relapsed, multidrug-resistant OC tumors with anti-Let-7 improved the efficacy of paclitaxel-induced cell death [194]. Since miRNAs are upstream regulators of a variety of cellular processes, the manipulation of their expression could cause adverse effects on surrounding tissues [195]. However, current research focusing on miRNA dysregulation is deciphering the mechanisms by which miRNAs affect different types of cancer. The increasing understanding of miRNA dysregulations in OC will allow for more direct targeting of the molecular pathways that are altered at each stage of tumor development. In addition, the up or downregulation of certain miRNAs in OC can also act as diagnostic biomarkers, as they have been demonstrated to have potential in many different cancer types [196]. Altogether, the altered molecular mechanisms driving OC development and treatment resistance are in part regulated by increased levels of endogenous ROS production and miRNA dysregulations. There are potentially new opportunities for more effective treatment of advanced OC by targeting the overlap in signaling pathways between these two mechanisms. However, limitations in our complete understanding of the roles of increased ROS and miRNA dysregulations in OC development necessitate more research efforts in these areas of study.
Author Contributions
Conceptualization, B.-H.J., L.-Z.L. and D.C.S.; writing—original draft preparation, D.C.S. and Y.W.; writing—review and editing, D.C.S., B.-H.J. and L.-Z.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institutes of Health grants (No. R01ES027901, R01CA232587, R01ES033197, R01CA263506 and K02ES029119), American Cancer Society Research Scholar (No. NEC-129306), and Commonwealth University Research Enhancement Program grant with the Pennsylvania Department of Health (SAP#41000).
Institutional Review Board Statement
Ethical review and approval were waived for this study, due to the use of already available published data.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study by the investigator of each published study included in the present review.
Data Availability Statement
The data presented in this study are openly available in Medline and Embase.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.W.; Ruiz, B.; Killeen, J.L.; Coté, T.R.; Wu, X.C.; Correa, C.N.; Howe, H.L. Pathology and classification of ovarian tumors. Cancer 2003, 97, 2631–2642. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of Ovarian Cancer: A Review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar]
- Banerjee, S.; Kaye, S.B. New Strategies in the Treatment of Ovarian Cancer: Current Clinical Perspectives and Future Potential. Clin. Cancer Res. 2013, 19, 961–968. [Google Scholar] [CrossRef]
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian Cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Shih, I.M.; Kurman, R.J. Ovarian Tumorigenesis: A Proposed Model Based on Morphological and Molecular Genetic Analysis. Am. J. Pathol. 2004, 164, 1511–1518. [Google Scholar] [CrossRef]
- Rocco, J.W.; Sidransky, D. P16(Mts-1/Cdkn2/Ink4a) in Cancer Progression. Exp. Cell Res. 2001, 264, 42–55. [Google Scholar] [CrossRef]
- Berek, J.S.; Kehoe, S.T.; Kumar, L.; Friedlander, M. Cancer of the Ovary, Fallopian Tube, and Peritoneum. Int. J. Gynecol. Obstet. 2018, 143 (Suppl. 2), 59–78. [Google Scholar] [CrossRef]
- Bookman, M.A.; McGuire, W.P., 3rd; Kilpatrick, D.; Keenan, E.; Hogan, W.M.; Johnson, S.W.; O’Dwyer, P.; Rowinsky, E.; Gallion, H.H.; Ozols, R.F. Carboplatin and Paclitaxel in Ovarian Carcinoma: A Phase I Study of the Gynecologic Oncology Group. J. Clin. Oncol. 1996, 14, 1895–1902. [Google Scholar] [CrossRef]
- Bookman, M.A.; Brady, M.F.; McGuire, W.P.; Harper, P.G.; Alberts, D.S.; Friedlander, M.; Colombo, N.; Fowler, J.M.; Argenta, P.A.; De Geest, K.; et al. Evaluation of New Platinum-Based Treatment Regimens in Advanced-Stage Ovarian Cancer: A Phase III Trial of the Gynecologic Cancer InterGroup. J. Clin. Oncol. 2009, 27, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Ozols, R.F.; Bundy, B.N.; Greer, B.E.; Fowler, J.M.; Clarke-Pearson, D.; Burger, R.A.; Mannel, R.S.; DeGeest, K.; Hartenbach, E.M.; Baergen, R.; et al. Phase III Trial of Carboplatin and Paclitaxel Compared With Cisplatin and Paclitaxel in Patients With Optimally Resected Stage III Ovarian Cancer: A Gynecologic Oncology Group Study. J. Clin. Oncol. 2003, 21, 3194–3200. [Google Scholar] [CrossRef] [PubMed]
- McGuire, W.P.; Hoskins, W.J.; Brady, M.F.; Kucera, P.R.; Partridge, E.E.; Look, K.Y.; Clarke-Pearson, D.L.; Davidson, M. Cyclophosphamide and cisplatin versus paclitaxel and cisplatin: A phase III randomized trial in patients with suboptimal stage III/IV ovarian cancer (from the Gynecologic Oncology Group). Semin. Oncol. 1996, 23 (Suppl. 2), 40–47. [Google Scholar] [PubMed]
- Armstrong, D.K.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.-M.; Cristea, M.; DeRosa, M.; Eisenhauer, E.L.; et al. Ovarian Cancer, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 191–226. [Google Scholar] [CrossRef] [PubMed]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef]
- Beckman, J.S.; Beckman, T.W.; Chen, J.; Marshall, P.A.; Freeman, B.A. Apparent hydroxyl radical production by peroxynitrite: Implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. USA 1990, 87, 1620–1624. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic Instability—An Evolving Hallmark of Cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Van Houten, B.; Woshner, V.; Santos, J.H. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair 2006, 5, 145–152. [Google Scholar] [CrossRef]
- Hurd, T.R.; Costa, N.J.; Dahm, C.; Beer, S.M.; Brown, S.E.; Filipovska, A.; Murphy, M. Glutathionylation of Mitochondrial Proteins. Antioxidants Redox Signal. 2005, 7, 999–1010. [Google Scholar] [CrossRef]
- Rhee, S.G.; Kang, S.W.; Chang, T.-S.; Jeong, W.; Kim, K. Peroxiredoxin, a Novel Family of Peroxidases. IUBMB Life 2001, 52, 35–41. [Google Scholar] [CrossRef]
- Rhee, S.G.; Yang, K.S.; Kang, S.W.; Woo, H.A.; Chang, T.S. Controlled Elimination of Intracellular H2O2: Regulation of Peroxiredoxin, Catalase, and Glutathione Peroxidase Via Post-Translational Modification. Antioxid. Redox Signal. 2005, 7, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Imai, H.; Nakagawa, Y. Biological Significance of Phospholipid Hydroperoxide Glutathione Peroxidase (Phgpx, Gpx4) in Mammalian Cells. Free. Radic. Biol. Med. 2003, 34, 145–169. [Google Scholar] [CrossRef]
- Loschen, G.; Flohe, L.; Chance, B. Respiratory Chain Linked H2O2 Production in Pigeon Heart Mitochondria. FEBS Lett. 1971, 18, 261–264. [Google Scholar] [CrossRef]
- Skulachev, V.P. Role of uncoupled and non-coupled oxidations in maintenance of safely low levels of oxygen and its one-electron reductants. Q. Rev. Biophys. 1996, 29, 169–202. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Rossi, F.; Zatti, M. Biochemical aspects of phagocytosis in poly-morphonuclear leucocytes. NADH and NADPH oxidation by the granules of resting and phagocytizing cells. Experientia 1964, 20, 21–23. [Google Scholar] [CrossRef]
- Quie, P.G.; White, J.G.; Holmes, B.; Good, R.A. In Vitro Bactericidal Capacity of Human Polymorphonuclear Leukocytes: Diminished Activity in Chronic Granulomatous Disease of Childhood *. J. Clin. Investig. 1967, 46, 668–679. [Google Scholar] [CrossRef]
- Baehner, R.L.; Nathan, D.G. Leukocyte Oxidase: Defective Activity in Chronic Granulomatous Disease. Science 1967, 155, 835–836. [Google Scholar] [CrossRef]
- Holmes, B.; Page, A.R.; Good, R.A. Studies of the Metabolic Activity of Leukocytes from Patients with a Genetic Abnormality of Phagocytic Function*. J. Clin. Investig. 1967, 46, 1422–1432. [Google Scholar] [CrossRef]
- Berendes, H.; Bridges, R.A.; Good, R.A. A fatal granulomatosus of childhood: The clinical study of a new syndrome. Minn. Med. 1957, 40, 309–312. [Google Scholar]
- Royer-Pokora, B.; Kunkel, L.M.; Monaco, A.; Goff, S.C.; Newburger, P.; Baehner, R.L.; Cole, F.S.; Curnutte, J.T.; Orkin, S.H. Cloning the gene for an inherited human disorder—Chronic granulomatous disease—On the basis of its chromosomal location. Nature 1986, 322, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.-A.; Arnold, R.S.; Lassegue, B.; Shi, J.; Xu, X.X.; Sorescu, D.; Chung, A.B.; Griendling, K.K.; Lambeth, J.D. Cell transformation by the superoxide-generating oxidase Mox1. Nature 1999, 401, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, J.; Cheng, G.; Arnold, R.S.; Edens, W.A. Novel homologs of gp91phox. Trends Biochem. Sci. 2000, 25, 459–461. [Google Scholar] [CrossRef]
- Xia, C.; Meng, Q.; Liu, L.-Z.; Rojanasakul, Y.; Wang, X.-R.; Jiang, B.-H. Reactive Oxygen Species Regulate Angiogenesis and Tumor Growth through Vascular Endothelial Growth Factor. Cancer Res. 2007, 67, 10823–10830. [Google Scholar] [CrossRef]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Paravicini, T.M.; Touyz, R.M. Nadph Oxidases, Reactive Oxygen Species, and Hypertension: Clinical Implications and Therapeutic Possibilities. Diabetes Care 2008, 31 (Suppl. 2), S170–S180. [Google Scholar] [CrossRef]
- Martínez, M.C.; Andriantsitohaina, R. Reactive Nitrogen Species: Molecular Mechanisms and Potential Significance in Health and Disease. Antioxidants Redox Signal. 2009, 11, 669–702. [Google Scholar] [CrossRef]
- Cross, C.E.; Halliwell, B.; Borish, E.T.; Pryor, W.A.; Ames, B.N.; Saul, R.L.; Mccord, J.M.; Harman, D. Oxygen Radicals and Human Disease. Ann. Intern. Med. 1987, 107, 526–545. [Google Scholar] [CrossRef]
- Johnson, T.M.; Yu, Z.X.; Ferrans, V.J.; Lowenstein, R.A.; Finkel, T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc. Natl. Acad. Sci. USA 1996, 93, 11848–11852. [Google Scholar] [CrossRef]
- Dizdaroglu, M. Oxidatively Induced DNA Damage and Its Repair in Cancer. Mutat. Res. Rev. Mutat. Res. 2015, 763, 212–245. [Google Scholar] [CrossRef]
- Peluso, M.; Russo, V.; Mello, T.; Galli, A. Oxidative Stress and DNA Damage in Chronic Disease and Environmental Studies. Int. J. Mol. Sci. 2020, 21, 6936. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Villani, R.M.; Wang, H.; Simpson, M.J.; Roberts, M.S.; Tang, M.; Liang, X. The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Cancer Res. 2018, 37, 266. [Google Scholar] [CrossRef] [PubMed]
- Copin, J.-C.; Gasche, Y.; Chan, P.H. Overexpression of copper/zinc superoxide dismutase does not prevent neonatal lethality in mutant mice that lack manganese superoxide dismutase. Free Radic. Biol. Med. 2000, 28, 1571–1576. [Google Scholar] [CrossRef]
- Maiorino, F.M.; Brigelius-Flohé, R.; Aumann, K.; Roveri, A.; Schomburg, D.; Flohé, L. Diversity of Glutathione Peroxidases. Methods Enzymol. 1995, 252, 38–53. [Google Scholar]
- Arnér, E.S.J.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. BioChem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef]
- Ray, P.D.; Huang, B.-W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef]
- Khalil, H.S.; Goltsov, A.; Langdon, S.P.; Harrison, D.J.; Bown, J.; Deeni, Y. Quantitative analysis of NRF2 pathway reveals key elements of the regulatory circuits underlying antioxidant response and proliferation of ovarian cancer cells. J. Biotechnol. 2015, 202, 12–30. [Google Scholar] [CrossRef]
- Liew, P.-L.; Hsu, C.-S.; Liu, W.-M.; Lee, Y.-C.; Lee, Y.-C.; Chen, C.-L. Prognostic and predictive values of Nrf2, Keap1, p16 and E-cadherin expression in ovarian epithelial carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 5642–5649. [Google Scholar]
- Czogalla, B.; Kahaly, M.; Mayr, D.; Schmoeckel, E.; Niesler, B.; Kolben, T.; Burges, A.; Mahner, S.; Jeschke, U.; Trillsch, F. Interaction of Eralpha and Nrf2 Impacts Survival in Ovarian Cancer Patients. Int. J. Mol. Sci. 2018, 20, 112. [Google Scholar] [CrossRef]
- Thompson, C.B. Apoptosis in the Pathogenesis and Treatment of Disease. Science 1995, 267, 1456–1462. [Google Scholar] [CrossRef]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.W.; Liu, V.W.; Tsao, G.S.; Yao, K.-M.; Furukawa, T.; Chan, K.K.; Ngan, H.Y. Loss of MKP3 mediated by oxidative stress enhances tumorigenicity and chemoresistance of ovarian cancer cells. Carcinogenesis 2008, 29, 1742–1750. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-Z.; Hu, X.-W.; Xia, C.; He, J.; Zhou, Q.; Shi, X.; Fang, J.; Jiang, B.-H. Reactive oxygen species regulate epidermal growth factor-induced vascular endothelial growth factor and hypoxia-inducible factor-1α expression through activation of AKT and P70S6K1 in human ovarian cancer cells. Free Radic. Biol. Med. 2006, 41, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Tudek, B.; Winczura, A.; Janik, J.; Siomek, A.; Foksinski, M.; Oliński, R. Involvement of oxidatively damaged DNA and repair in cancer development and aging. Am. J. Transl. Res. 2010, 2, 254–284. [Google Scholar]
- Fridovich, I. The biology of oxygen radicals. Science 1978, 201, 875–880. [Google Scholar] [CrossRef]
- Meier, B.; Cross, A.R.; Hancock, J.T.; Kaup, F.J.; Jones, O.T.G. Identification of a superoxide-generating NADPH oxidase system in human fibroblasts. Biochem. J. 1991, 275, 241–245. [Google Scholar] [CrossRef]
- Szatrowski, T.P.; Nathan, C.F. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991, 51, 794–798. [Google Scholar]
- Griendling, K.K.; Minieri, C.A.; Ollerenshaw, J.D.; Alexander, R.W. Angiotensin II stimulates NADH and NADPH oxidase activity in cultured vascular smooth muscle cells. Circ. Res. 1994, 74, 1141–1148. [Google Scholar] [CrossRef]
- De Carvalho, D.D.; Sadok, A.; Bourgarel-Rey, V.; Gattacceca, F.; Penel, C.; Lehmann, M.; Kovacic, H. Nox1 downstream of 12-lipoxygenase controls cell proliferation but not cell spreading of colon cancer cells. Int. J. Cancer 2008, 122, 1757–1764. [Google Scholar] [CrossRef]
- Kajla, S.; Mondol, A.S.; Nagasawa, A.; Zhang, Y.; Kato, M.; Matsuno, K.; Matsuno, K.; Yabe-Nishimura, C.; Kamata, T. A Crucial Role for Nox 1 in Redox-Dependent Regulation of Wnt-Beta-Catenin Signaling. FASEB J. 2012, 26, 2049–2059. [Google Scholar] [CrossRef] [PubMed]
- Mukawera, E.; Chartier, S.; Williams, V.; Pagano, P.J.; Lapointe, R.; Grandvaux, N. Redox-Modulating Agents Target Nox2-Dependent Ikkepsilon Oncogenic Kinase Expression and Proliferation in Human Breast Cancer Cell Lines. Redox Biol. 2015, 6, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Banskota, S.; Regmi, S.C.; Kim, J.-A. NOX1 to NOX2 switch deactivates AMPK and induces invasive phenotype in colon cancer cells through overexpression of MMP-7. Mol. Cancer 2015, 14, 123. [Google Scholar] [CrossRef] [PubMed]
- Wiktorin, H.G.; Nilsson, T.; Aydin, E.; Hellstrand, K.; Palmqvist, L.; Martner, A. Role of NOX2 for leukaemic expansion in a murine model of BCR-ABL1 + leukaemia. Br. J. Haematol. 2018, 182, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Hole, P.S.; Zabkiewicz, J.; Munje, C.; Newton, Z.; Pearn, L.; White, P.; Marquez, N.; Hills, R.; Burnett, A.K.; Tonks, A.; et al. Overproduction of NOX-derived ROS in AML promotes proliferation and is associated with defective oxidative stress signaling. Blood 2013, 122, 3322–3330. [Google Scholar] [CrossRef] [PubMed]
- Harrison, I.P.; Vinh, A.; Johnson, I.; Luong, R.; Drummond, G.R.; Sobey, C.G.; Tiganis, T.; Williams, E.; O’Leary, J.; Brooks, D.; et al. NOX2 oxidase expressed in endosomes promotes cell proliferation and prostate tumour development. Oncotarget 2018, 9, 35378–35393. [Google Scholar] [CrossRef]
- Kim, S.-M.; Hur, D.Y.; Hong, S.-W.; Kim, J.H. EBV-encoded EBNA1 regulates cell viability by modulating miR34a-NOX2-ROS signaling in gastric cancer cells. Biochem. Biophys. Res. Commun. 2017, 494, 550–555. [Google Scholar] [CrossRef]
- Block, K.; Gorin, Y.; Hoover, P.; Williams, P.; Chelmicki, T.; Clark, R.A.; Yoneda, T.; Abboud, H.E. NAD(P)H Oxidases Regulate HIF-2α Protein Expression. J. Biol. Chem. 2007, 282, 8019–8026. [Google Scholar] [CrossRef]
- Brar, S.S.; Kennedy, T.P.; Sturrock, A.B.; Huecksteadt, T.P.; Quinn, M.T.; Whorton, A.R.; Hoidal, J.R. An NAD(P)H oxidase regulates growth and transcription in melanoma cells. Am. J. Physiol. Physiol. 2002, 282, C1212–C1224. [Google Scholar] [CrossRef]
- Diaz, B.; Shani, G.; Pass, I.; Anderson, D.; Quintavalle, M.; Courtneidge, S.A. Tks5-Dependent, Nox-Mediated Generation of Reactive Oxygen Species Is Necessary for Invadopodia Formation. Sci. Signal. 2009, 2, ra53. [Google Scholar] [CrossRef]
- Ogrunc, M.; Di Micco, R.; Liontos, M.; Bombardelli, L.; Mione, M.; Fumagalli, M.; Gorgoulis, V.G.; Daddadifagagna, F. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell Death Differ. 2014, 21, 998–1012. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lan, T.; Hou, J.; Li, J.; Fang, R.; Yang, Z.; Zhang, M.; Liu, J.; Liu, B. NOX4 promotes non-small cell lung cancer cell proliferation and metastasis through positive feedback regulation of PI3K/Akt signaling. Oncotarget 2014, 5, 4392–4405. [Google Scholar] [CrossRef]
- Simone, N.L.; Soule, B.P.; Ly, D.; Saleh, A.D.; Savage, J.E.; DeGraff, W.; Cook, J.; Harris, C.C.; Gius, D.; Mitchell, J.B. Ionizing Radiation-Induced Oxidative Stress Alters miRNA Expression. PLoS ONE 2009, 4, e6377. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Heravi-Moussavi, A.; Anglesio, M.S.; Cheng, S.-W.G.; Senz, J.; Yang, W.; Prentice, L.; Fejes, A.P.; Chow, C.; Tone, A.; Kalloger, S.E.; et al. Recurrent Somatic DICER1 Mutations in Nonepithelial Ovarian Cancers. N. Engl. J. Med. 2012, 366, 234–242. [Google Scholar] [CrossRef]
- Bartel, D.P. Micrornas: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ramchandran, R.; Chaluvally-Raghavan, P. Mirna-Mediated Rna Activation in Mammalian Cells. Adv. Exp. Med. Biol. 2017, 983, 81–89. [Google Scholar] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Bussing, I.; Slack, F.J.; Grosshans, H. Let-7 Micrornas in Development, Stem Cells and Cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef]
- Wang, X.; Cao, L.; Wang, Y.; Wang, X.; Liu, N.; You, Y. Regulation of let-7 and its target oncogenes (Review). Oncol. Lett. 2012, 3, 955–960. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Garofalo, M.; Leva, G.D.; Croce, C.M. Micrornas as Anti-Cancer Therapy. Curr. Pharm. Des. 2014, 20, 5328–5335. [Google Scholar] [CrossRef]
- Kumar, M.; Lu, Z.; Takwi, A.A.L.; Chen, W.; Callander, N.S.; Ramos, K.S.; Young, K.H.; Li, Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 2011, 30, 843–853. [Google Scholar] [CrossRef]
- Costinean, S.; Zanesi, N.; Pekarsky, Y.; Tili, E.; Volinia, S.; Heerema, N.; Croce, C.M. Pre-B Cell Proliferation and Lymphoblastic Leukemia/High-Grade Lymphoma in E(Mu)-Mir155 Transgenic Mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7024–7029. [Google Scholar] [CrossRef]
- Valeri, N.; Gasparini, P.; Fabbri, M.; Braconi, C.; Veronese, A.; Lovat, F.; Adair, B.; Vannini, I.; Fanini, F.; Bottoni, A.; et al. Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. USA 2010, 107, 6982–6987. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre, Y.; De Guire, V.; Querido, E.; Mukhopadhyay, U.K.; Bourdeau, V.; Major, F.; Ferbeyre, G.; Chartrand, P. An E2F/miR-20a Autoregulatory Feedback Loop. J. Biol. Chem. 2007, 282, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
- Trimarchi, J.; Lees, J.A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002, 3, 11–20. [Google Scholar] [CrossRef]
- Coller, H.A.; Forman, J.J.; Legesse-Miller, A. Myc’ed Messages: Myc Induces Transcription of E2f1 While Inhibiting Its Translation Via a Microrna Polycistron. PLoS Genet. 2007, 3, e146. [Google Scholar] [CrossRef] [PubMed]
- Woods, K.; Thomson, J.M.; Hammond, S.M. Direct Regulation of an Oncogenic Micro-RNA Cluster by E2F Transcription Factors. J. Biol. Chem. 2007, 282, 2130–2134. [Google Scholar] [CrossRef]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef]
- Galardi, S.; Mercatelli, N.; Giorda, E.; Massalini, S.; Frajese, G.V.; Ciafrè, S.A.; Farace, M.G. miR-221 and miR-222 Expression Affects the Proliferation Potential of Human Prostate Carcinoma Cell Lines by Targeting p27Kip1. J. Biol. Chem. 2007, 282, 23716–23724. [Google Scholar] [CrossRef]
- Le Sage, C.; Nagel, R.; Egan, D.A.; Schrier, M.; Mesman, E.; Mangiola, A.; Anile, C.; Maira, G.; Mercatelli, N.; Ciafrè, S.A.; et al. Regulation of the p27Kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007, 26, 3699–3708. [Google Scholar] [CrossRef]
- Visone, R.; Russo, L.; Pallante, P.; De Martino, I.; Ferraro, A.; Leone, V.; Borbone, E.; Petrocca, F.; Alder, H.; Croce, C.M.; et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr.-Relat. Cancer 2007, 14, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Rattanapan, Y.; Korkiatsakul, V.; Kongruang, A.; Siriboonpiputtana, T.; Rerkamnuaychoke, B.; Chareonsirisuthigul, T. MicroRNA Expression Profiling of Epithelial Ovarian Cancer Identifies New Markers of Tumor Subtype. MicroRNA 2020, 9, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Hoeben, A.; Landuyt, B.; Highley, M.S.; Wildiers, H.; Van Oosterom, A.T.; De Bruijn, E.A. Vascular Endothelial Growth Factor and Angiogenesis. Pharmacol. Rev. 2004, 56, 549–580. [Google Scholar] [CrossRef]
- Chung, A.S.; Ferrara, N. Developmental and Pathological Angiogenesis. Annu. Rev. Cell Dev. Biol. 2011, 27, 563–584. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.W.; Cachianes, G.; Kuang, W.-J.; Goeddel, D.V.; Ferrara, N. Vascular Endothelial Growth Factor Is a Secreted Angiogenic Mitogen. Science 1989, 246, 1306–1309. [Google Scholar] [CrossRef] [PubMed]
- Breier, G.; Albrecht, U.; Sterrer, S.; Risau, W. Expression of vascular endothelial growth factor during embryonic angiogenesis and endothelial cell differentiation. Development 1992, 114, 521–532. [Google Scholar] [CrossRef]
- Berse, B.; Brown, L.F.; Van De Water, L.; Dvorak, H.F.; Senger, D.R. Vascular Permeability Factor (Vascular Endothelial Growth Factor) Gene Is Expressed Differentially in Normal Tissues, Macrophages, and Tumors. Mol. Biol. Cell 1992, 3, 211–220. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The Role of Hypoxia in Cancer Progression, Angiogenesis, Metastasis, and Resistance to Therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef]
- Semenza, G.L.; Wang, G.L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 1992, 12, 5447–5454. [Google Scholar]
- Wang, G.L.; Semenza, G.L. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J. Biol. Chem. 1993, 268, 21513–21518. [Google Scholar] [CrossRef]
- Forsythe, J.A.; Jiang, B.H.; Iyer, N.V.; Agani, F.; Leung, S.W.; Koos, R.D.; Semenza, G.L. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 1996, 16, 4604–4613. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010, 29, 625–634. [Google Scholar] [CrossRef]
- Liu, W.-J.; Huang, Y.-X.; Wang, W.; Zhang, Y.; Liu, B.-J.; Qiu, J.-G.; Jiang, B.-H.; Liu, L.-Z. NOX4 Signaling Mediates Cancer Development and Therapeutic Resistance through HER3 in Ovarian Cancer Cells. Cells 2021, 10, 1647. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xu, Q.; Jing, Y.; Agani, F.; Qian, X.; Carpenter, R.; Li, Q.; Wang, X.-R.; Peiper, S.S.; Lu, Z.; et al. Reactive oxygen species regulate ERBB2 and ERBB3 expression via miR-199a/125b and DNA methylation. EMBO Rep. 2012, 13, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Druz, A.; Chu, C.; Majors, B.; Santuary, R.; Betenbaugh, M.; Shiloach, J. A Novel Microrna Mmu-Mir-466h Affects Apoptosis Regulation in Mammalian Cells. Biotechnol. Bioeng. 2011, 108, 1651–1661. [Google Scholar] [CrossRef]
- Druz, A.; Betenbaugh, M.; Shiloach, J. Glucose Depletion Activates Mmu-Mir-466h-5p Expression through Oxidative Stress and Inhibition of Histone Deacetylation. Nucleic Acids Res. 2012, 40, 7291–7302. [Google Scholar] [CrossRef]
- Liu, B.; Chen, Y.; Clair, D.K.S. ROS and p53: A versatile partnership. Free Radic. Biol. Med. 2008, 44, 1529–1535. [Google Scholar] [CrossRef]
- Feng, X.; Wang, Z.; Fillmore, R.; Xi, Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014, 344, 166–173. [Google Scholar] [CrossRef]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef]
- Gutierrez-Uzquiza, A.; Arechederra, M.; Bragado, P.; Aguirre-Ghiso, J.A.; Porras, A. P38alpha Mediates Cell Survival in Response to Oxidative Stress Via Induction of Antioxidant Genes: Effect on the P70s6k Pathway. J. Biol. Chem. 2012, 287, 2632–2642. [Google Scholar] [CrossRef]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; De Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.-P.; Cottu, P.; Sastre-Garau, X.; et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef]
- Xie, Z.; Cao, L.; Zhang, J. miR-21 modulates paclitaxel sensitivity and hypoxia-inducible factor-1α expression in human ovarian cancer cells. Oncol. Lett. 2013, 6, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Zhang, X.; Zhang, Z.; Shu, Y.; Luo, X.; Yang, Y.; Wang, X.; Yang, G.; Li, L.; Feng, Y. The Microrna-27a: Zbtb10-Specificity Protein Pathway Is Involved in Follicle Stimulating Hormone-Induced Vegf, Cox2 and Survivin Expression in Ovarian Epithelial Cancer Cells. Int. J. Oncol. 2013, 42, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Masoumi-Dehghi, S.; Babashah, S.; Sadeghizadeh, M. Microrna-141-3p-Containing Small Extracellular Vesicles Derived from Epithelial Ovarian Cancer Cells Promote Endothelial Cell Angiogenesis through Activating the Jak/Stat3 and Nf-Kappab Signaling Pathways. J. Cell Commun. Signal. 2020, 14, 233–244. [Google Scholar] [CrossRef]
- He, L.; Zhu, W.; Chen, Q.; Yuan, Y.; Wang, Y.; Wang, J.; Wu, X. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics 2019, 9, 8206–8220. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Mangala, L.S.; Mooberry, L.; Bayraktar, E.; Dasari, S.K.; Ma, S.; Ivan, C.; Court, K.A.; Rodriguez-Aguayo, C.; Bayraktar, R.; et al. Identifying and targeting angiogenesis-Related microRNAs in ovarian cancer. Oncogene 2019, 38, 6095–6108. [Google Scholar] [CrossRef]
- Lawler, P.; Lawler, J. Molecular Basis for the Regulation of Angiogenesis by Thrombospondin-1 and -2. Cold Spring Harb. Perspect. Med. 2012, 2, a006627. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Taher, M. Mirna Profile in Ovarian Cancer. Exp. Mol. Pathol. 2020, 113, 104381. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, L.-Z.; Qian, X.; Chen, Q.; Jiang, Y.; Li, D.; Lai, L.; Jiang, B.-H. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012, 40, 761–774. [Google Scholar] [CrossRef]
- Halvorsen, A.R.; Kristensen, G.; Embleton, A.; Adusei, C.; Barretina-Ginesta, M.P.; Beale, P.; Helland, Å. Evaluation of Prognostic and Predictive Significance of Circulating MicroRNAs in Ovarian Cancer Patients. Dis. Markers 2017, 2017, 3098542. [Google Scholar] [CrossRef]
- An, Y.; Yang, Q. MiR-21 modulates the polarization of macrophages and increases the effects of M2 macrophages on promoting the chemoresistance of ovarian cancer. Life Sci. 2020, 242, 117162. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, C.L.; Co, N.-N.; Tsuruga, T.; Yeung, T.-L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.-K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [PubMed]
- Lenkala, D.; LaCroix, B.; Gamazon, E.; Geeleher, P.; Im, H.K.; Huang, R.S. The impact of microRNA expression on cellular proliferation. Qual. Life Res. 2014, 133, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qiu, C.; Lu, N.; Liu, Z.; Jin, C.; Sun, C.; Bu, H.; Yu, H.; Dongol, S.; Kong, B. FOXD1 is targeted by miR-30a-5p and miR-200a-5p and suppresses the proliferation of human ovarian carcinoma cells by promoting p21 expression in a p53-independent manner. Int. J. Oncol. 2018, 52, 2130–2142. [Google Scholar] [CrossRef]
- Chen, M.-W.; Yang, S.-T.; Chien, M.-H.; Hua, K.-T.; Wu, C.-J.; Hsiao, S.M.; Lin, H.; Hsiao, M.; Su, J.-L.; Wei, L.-H. The STAT3-miRNA-92-Wnt Signaling Pathway Regulates Spheroid Formation and Malignant Progression in Ovarian Cancer. Cancer Res. 2017, 77, 1955–1967. [Google Scholar] [CrossRef]
- Yoshimura, A.; Sawada, K.; Nakamura, K.; Kinose, Y.; Nakatsuka, E.; Kobayashi, M.; Miyamoto, M.; Ishida, K.; Matsumoto, Y.; Kodama, M.; et al. Exosomal miR-99a-5p is elevated in sera of ovarian cancer patients and promotes cancer cell invasion by increasing fibronectin and vitronectin expression in neighboring peritoneal mesothelial cells. BMC Cancer 2018, 18, 1065. [Google Scholar] [CrossRef]
- Huh, J.H.; Kim, T.H.; Kim, K.; Song, J.-A.; Jung, Y.J.; Jeong, J.-Y.; Lee, M.J.; Kim, Y.K.; Lee, D.H.; An, H.J. Dysregulation of miR-106a and miR-591 confers paclitaxel resistance to ovarian cancer. Br. J. Cancer 2013, 109, 452–461. [Google Scholar] [CrossRef]
- Van Jaarsveld, M.T.M.; Helleman, J.; Boersma, A.W.M.; van Kuijk, P.F.; van Ijcken, W.F.; Despierre, E.; Vergote, I.; Mathijssen, R.H.J.; Berns, E.M.J.J.; Verweij, J.; et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene 2013, 32, 4284–4293. [Google Scholar] [CrossRef]
- Parikh, A.; Lee, C.; Joseph, P.; Marchini, S.; Baccarini, A.; Kolev, V.; Romualdi, C.; Fruscio, R.; Shah, H.; Wang, F.; et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial–mesenchymal transition. Nat. Commun. 2014, 5, 2977. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Segura, M.F.; Shao, C.; Lee, P.; Gong, Y.; Hernando, E.; Wei, J.-J. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J. Pathol. 2012, 228, 204–215. [Google Scholar] [CrossRef]
- McMillen, B.D.; Aponte, M.M.; Liu, Z.; Helenowski, I.B.; Scholtens, D.M.; Buttin, B.M.; Wei, J.-J. Expression analysis of MIR182 and its associated target genes in advanced ovarian carcinoma. Mod. Pathol. 2012, 25, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Q.; Guo, R.-D.; Guo, R.-M.; Sheng, W.; Yin, L.-R. MicroRNA-182 promotes cell growth, invasion, and chemoresistance by targeting programmed cell death 4 (PDCD4) in human ovarian carcinomas. J. Cell. Biochem. 2013, 114, 1464–1473. [Google Scholar] [CrossRef] [PubMed]
- Xiaohong, Z.; Lichun, F.; Na, X.; Kejian, Z.; Xiaolan, X.; Shaosheng, W. MiR-203 promotes the growth and migration of ovarian cancer cells by enhancing glycolytic pathway. Tumor Biol. 2016, 37, 14989–14997. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.; Liang, A.; Jiang, A.; Zong, L. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol. Lett. 2018, 15, 7571–7578. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, K.; You, Y.; Fu, X.; Hu, L.; Song, L.; Meng, Y. Hypoxia-induced miR-210 in epithelial ovarian cancer enhances cancer cell viability via promoting proliferation and inhibiting apoptosis. Int. J. Oncol. 2014, 44, 2111–2120. [Google Scholar] [CrossRef]
- Yang, H.; Kong, W.; He, L.; Zhao, J.-J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA Expression Profiling in Human Ovarian Cancer: miR-214 Induces Cell Survival and Cisplatin Resistance by Targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shen, H.; Yin, X.; Yang, M.; Wei, H.; Chen, Q.; Feng, F.; Liu, Y.; Xu, W.; Li, Y. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J. Exp. Clin. Cancer Res. 2019, 38, 81. [Google Scholar] [CrossRef]
- Yang, L.; Wei, Q.-M.; Zhang, X.-W.; Sheng, Q.; Yan, X.-T. MiR-376a promotion of proliferation and metastases in ovarian cancer: Potential role as a biomarker. Life Sci. 2017, 173, 62–67. [Google Scholar] [CrossRef]
- Furlong, F.; Fitzpatrick, P.; O’Toole, S.; Phelan, S.; McGrogan, B.; Maguire, A.; O’Grady, A.; Gallagher, M.; Prencipe, M.; McGoldrick, A.; et al. Low MAD2 expression levels associate with reduced progression-free survival in patients with high-grade serous epithelial ovarian cancer. J. Pathol. 2012, 226, 746–755. [Google Scholar] [CrossRef]
- Chaluvally-Raghavan, P.; Jeong, K.J.; Pradeep, S.; Silva, A.M.; Yu, S.; Liu, W.; Moss, T.; Rodriguez-Aguayo, C.; Zhang, D.; Ram, P.; et al. Direct Upregulation of STAT3 by MicroRNA-551b-3p Deregulates Growth and Metastasis of Ovarian Cancer. Cell Rep. 2016, 15, 1493–1504. [Google Scholar] [CrossRef]
- Zhao, W.; Han, T.; Li, B.; Ma, Q.; Yang, P.; Li, H. miR-552 promotes ovarian cancer progression by regulating PTEN pathway. J. Ovarian Res. 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.E.; Meghani, K.; Brault, M.-E.; Leclerc, L.; He, Y.; Day, T.A.; Elias, K.M.; Drapkin, R.; Weinstock, D.M.; Dao, F.; et al. Platinum and PARP Inhibitor Resistance Due to Overexpression of MicroRNA-622 in BRCA1-Mutant Ovarian Cancer. Cell Rep. 2016, 14, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Li-Ya, Q.; Feng, Z.; Yin, W.; Ji-Hong, L. MiR-939 promotes the proliferation of human ovarian cancer cells by repressing APC2 expression. Biomed. Pharmacother. 2015, 71, 64–69. [Google Scholar] [CrossRef]
- Kanlikilicer, P.; Bayraktar, R.; Denizli, M.; Rashed, M.H.; Ivan, C.; Aslan, B.; Mitra, R.; Karagoz, K.; Bayraktar, E.; Zhang, X.; et al. Exosomal Mirna Confers Chemo Resistance Via Targeting Cav1/P-Gp/M2-Type Macrophage Axis in Ovarian Cancer. EBioMedicine 2018, 38, 100–112. [Google Scholar] [CrossRef]
- Creighton, C.J.; Fountain, M.D.; Yu, Z.; Nagaraja, A.K.; Zhu, H.; Khan, M.; Olokpa, E.; Zariff, A.; Gunaratne, P.H.; Matzuk, M.M.; et al. Molecular Profiling Uncovers a p53-Associated Role for MicroRNA-31 in Inhibiting the Proliferation of Serous Ovarian Carcinomas and Other Cancers. Cancer Res. 2010, 70, 1906–1915. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Wang, C.; Gao, Y.; Wang, J. Decreased expression of microRNA-148a predicts poor prognosis in ovarian cancer and associates with tumor growth and metastasis. Biomed. Pharmacother. 2016, 83, 58–63. [Google Scholar] [CrossRef]
- Lin, Z.; Li, D.; Cheng, W.; Wu, J.; Wang, K.; Hu, Y. Microrna-181 Functions as an Antioncogene and Mediates Nf-Kappab Pathway by Targeting Rtkn2 in Ovarian Cancers. Reprod. Sci. 2019, 26, 1071–1081. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Huang, Y.; Zhang, Q.; Zhou, J.; Zhang, X.; Wang, X. miR-200b and miR-200c co-contribute to the cisplatin sensitivity of ovarian cancer cells by targeting DNA methyltransferases. Oncol. Lett. 2019, 17, 1453–1460. [Google Scholar] [CrossRef]
- Dai, C.; Xie, Y.; Zhuang, X.; Yuan, Z. Mir-206 Inhibits Epithelial Ovarian Cancer Cells Growth and Invasion Via Blocking C-Met/Akt/Mtor Signaling Pathway. Biomed. Pharmacother. 2018, 104, 763–770. [Google Scholar] [CrossRef]
- Zhou, F.; Chen, J.; Wang, H. MicroRNA-298 inhibits malignant phenotypes of epithelial ovarian cancer by regulating the expression of EZH2. Oncol. Lett. 2016, 12, 3926–3932. [Google Scholar] [CrossRef]
- Liu, J.; Gu, Z.; Tang, Y.; Hao, J.; Zhang, C.; Yang, X. Tumour-suppressive microRNA-424-5p directly targets CCNE1 as potential prognostic markers in epithelial ovarian cancer. Cell Cycle 2018, 17, 309–318. [Google Scholar] [CrossRef]
- Chen, K.; Zeng, J.; Tang, K.; Xiao, H.; Hu, J.; Huang, C.; Yao, W.; Yu, G.; Xiao, W.; Guan, W.; et al. miR-490-5p suppresses tumour growth in renal cell carcinoma through targeting PIK3CA. Biol. Cell 2016, 108, 41–50. [Google Scholar] [CrossRef]
- Hong, L.; Wang, Y.; Chen, W.; Yang, S. Microrna-508 Suppresses Epithelial-Mesenchymal Transition, Migration, and Invasion of Ovarian Cancer Cells through the Mapk1/Erk Signaling Pathway. J. Cell. Biochem. 2018, 119, 7431–7440. [Google Scholar] [CrossRef] [PubMed]
- Parmar, M.K.; Ledermann, J.A.; Colombo, N.; Du, B.A.; Delaloye, J.F.; Kristensen, G.B.; Wheeler, S.; Swart, A.M.; Qian, W.; Torri, V.; et al. Paclitaxel Plus Platinum-Based Chemotherapy Versus Conventional Platinum-Based Chemotherapy in Women with Relapsed Ovarian Cancer: The Icon4/Ago-Ovar-2.2 Trial. Lancet 2003, 361, 2099–2106. [Google Scholar] [PubMed]
- Neijt, J.P.; Engelholm, S.A.; Tuxen, M.K.; Sørensen, P.G.; Hansen, M.; Sessa, C.; de Swart, C.A.M.; Hirsch, F.R.; Lund, B.; van Houwelingen, H.C. Exploratory Phase III Study of Paclitaxel and Cisplatin Versus Paclitaxel and Carboplatin in Advanced Ovarian Cancer. J. Clin. Oncol. 2000, 18, 3084–3092. [Google Scholar] [CrossRef] [PubMed]
- Schoch, S.; Gajewski, S.; Rothfuß, J.; Hartwig, A.; Köberle, B. Comparative Study of the Mode of Action of Clinically Approved Platinum-Based Chemotherapeutics. Int. J. Mol. Sci. 2020, 21, 6928. [Google Scholar] [CrossRef]
- Schiff, P.; Fant, J.; Horwitz, S.B. Promotion of microtubule assembly in vitro by taxol. Nature 1979, 277, 665–667. [Google Scholar] [CrossRef]
- Johnson, S.W.; Laub, P.B.; Beesley, J.S.; Ozols, R.F.; Hamilton, T.C. Increased platinum-DNA damage tolerance is associated with cisplatin resistance and cross-resistance to various chemotherapeutic agents in unrelated human ovarian cancer cell lines. Cancer Res. 1997, 57, 850–856. [Google Scholar]
- Pfisterer, J.; Plante, M.; Vergote, I.; du Bois, A.; Hirte, H.; Lacave, A.J.; Wagner, U.; Stahle, A.; Stuart, G.; Kimmig, R.; et al. Gemcitabine Plus Carboplatin Compared with Carboplatin in Patients with Platinum-Sensitive Recurrent Ovarian Cancer: An Intergroup Trial of the Ago-Ovar, the Ncic Ctg, and the Eortc Gcg. J. Clin. Oncol. 2006, 24, 4699–4707. [Google Scholar] [CrossRef]
- Sehouli, J.; Camara, O.; Schmidt, M.; Mahner, S.; Seipelt, G.; Otremba, B.; Schmalfeldt, B.; Tesch, H.; Lorenz-Schlüter, C.; Oskay-Ozcelik, G.; et al. Pegylated Liposomal Doxorubicin (Caelyx) in Patients with Advanced Ovarian Cancer: Results of a German Multicenter Observational Study. Cancer Chemother. Pharmacol. 2009, 64, 585–591. [Google Scholar] [CrossRef]
- Ferrandina, G.; Ludovisi, M.; Lorusso, D.; Pignata, S.; Breda, E.; Savarese, A.; Del Medico, P.; Scaltriti, L.; Katsaros, D.; Priolo, D.; et al. Phase III Trial of Gemcitabine Compared with Pegylated Liposomal Doxorubicin in Progressive or Recurrent Ovarian Cancer. J. Clin. Oncol. 2008, 26, 890–896. [Google Scholar] [CrossRef]
- Witham, J.; Valenti, M.R.; Alexis, K.; Vidot, S.; Eccles, S.A.; Kaye, S.B.; Richardson, A. The Bcl-2/Bcl-Xl Family Inhibitor Abt-737 Sensitizes Ovarian Cancer Cells to Carboplatin. Clin. Cancer Res. 2007, 13, 7191–7198. [Google Scholar] [CrossRef]
- Eliopoulos, A.G.; Kerr, D.J.; Herod, J.; Hodgkins, L.; Krajewski, S.; Reed, J.C.; Young, L. The control of apoptosis and drug resistance in ovarian cancer: Influence of p53 and Bcl-2. Oncogene 1995, 11, 1217–1228. [Google Scholar]
- Wang, H.; Zhang, Z.; Wei, X.; Dai, R. Small-molecule inhibitor of Bcl-2 (TW-37) suppresses growth and enhances cisplatin-induced apoptosis in ovarian cancer cells. J. Ovarian Res. 2015, 8, 3. [Google Scholar] [CrossRef]
- Zeitlin, B.D.; Zeitlin, I.J.; Nör, J.E. Expanding Circle of Inhibition: Small-Molecule Inhibitors of Bcl-2 as Anticancer Cell and Antiangiogenic Agents. J. Clin. Oncol. 2008, 26, 4180–4188. [Google Scholar] [CrossRef]
- Safe, S.; Abdelrahim, M. Sp transcription factor family and its role in cancer. Eur. J. Cancer 2005, 41, 2438–2448. [Google Scholar] [CrossRef]
- Stavrovskaya, A.A. Cellular Mechanisms of Multidrug Resistance of Tumor Cells. Biochemistry 2000, 65, 95–106. [Google Scholar]
- Kruh, G.D.; Belinsky, M.G. The Mrp Family of Drug Efflux Pumps. Oncogene 2003, 22, 7537–7552. [Google Scholar] [CrossRef]
- Liu, N.; Zhong, L.; Zeng, J.; Zhang, X.; Yang, Q.; Liao, D.; Wang, Y.; Chen, G.; Wang, Y. Upregulation of Microrna-200a Associates with Tumor Proliferation, Cscs Phenotype and Chemosensitivity in Ovarian Cancer. Neoplasma 2015, 62, 550–559. [Google Scholar] [CrossRef][Green Version]
- Miyamoto, M.; Sawada, K.; Nakamura, K.; Yoshimura, A.; Ishida, K.; Kobayashi, M.; Shimizu, A.; Yamamoto, M.; Kodama, M.; Hashimoto, K.; et al. Paclitaxel exposure downregulates miR-522 expression and its downregulation induces paclitaxel resistance in ovarian cancer cells. Sci. Rep. 2020, 10, 16755. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, P.; Chen, X.; Gong, D.; Wu, Y.; Huang, L.; Chen, Y. ROS-Induced DCTPP1 Upregulation Contributes to Cisplatin Resistance in Ovarian Cancer. Front. Mol. Biosci. 2022, 9, 838006. [Google Scholar] [CrossRef]
- Li, J.; Zheng, C.; Wang, M.; Umano, A.D.; Dai, Q.; Zhang, C.; Huang, H.; Yang, Q.; Yang, X.; Lu, J.; et al. ROS-regulated phosphorylation of ITPKB by CAMK2G drives cisplatin resistance in ovarian cancer. Oncogene 2022, 41, 1114–1128. [Google Scholar] [CrossRef]
- Kim, B.; Jung, J.W.; Jung, J.; Han, Y.; Suh, D.H.; Kim, H.S.; Dhanasekaran, D.N.; Song, Y.S. PGC1α induced by reactive oxygen species contributes to chemoresistance of ovarian cancer cells. Oncotarget 2017, 8, 60299–60311. [Google Scholar] [CrossRef]
- Uusi-Kerttula, H.; Davies, J.A.; Thompson, J.M.; Wongthida, P.; Evgin, L.; Shim, K.G.; Bradshaw, A.; Baker, A.T.; Rizkallah, P.J.; Jones, R.; et al. Ad5null-A20: A Tropism-Modified, Alphavbeta6 Integrin-Selective Oncolytic Adenovirus for Epithelial Ovarian Cancer Therapies. Clin. Cancer Res. 2018, 24, 4215–4224. [Google Scholar] [CrossRef]
- Lanitis, E.; Dangaj, D.; Hagemann, I.; Song, D.-G.; Best, A.; Sandaltzopoulos, R.; Coukos, G.; Powell, D.J., Jr. Primary Human Ovarian Epithelial Cancer Cells Broadly Express HER2 at Immunologically-Detectable Levels. PLoS ONE 2012, 7, e49829. [Google Scholar] [CrossRef]
- Chung, Y.W.; Kim, S.; Hong, J.H.; Lee, J.K.; Lee, N.W.; Lee, Y.S.; Song, J.Y. Overexpression of Her2/Her3 and Clinical Feature of Ovarian Cancer. J. Gynecol. Oncol. 2019, 30, e75. [Google Scholar] [CrossRef]
- Mizuno, T.; Kojima, Y.; Yonemori, K.; Yoshida, H.; Sugiura, Y.; Ohtake, Y.; Okuma, H.S.; Nishikawa, T.; Tanioka, M.; Sudo, K.; et al. Neoadjuvant chemotherapy promotes the expression of HER3 in patients with ovarian cancer. Oncol. Lett. 2020, 20, 336. [Google Scholar] [CrossRef]
- Jansson, M.D.; Lund, A.H. Microrna and Cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef]
- Babar, I.A.; Cheng, C.J.; Booth, C.J.; Liang, X.; Weidhaas, J.B.; Saltzman, W.M.; Slack, F.J. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl. Acad. Sci. USA 2012, 109, E1695–E1704. [Google Scholar] [CrossRef]
- Corsten, M.F.; Miranda, R.; Kasmieh, R.; Krichevsky, A.M.; Weissleder, R.; Shah, K. MicroRNA-21 Knockdown Disrupts Glioma Growth In vivo and Displays Synergistic Cytotoxicity with Neural Precursor Cell–Delivered S-TRAIL in Human Gliomas. Cancer Res. 2007, 67, 8994–9000. [Google Scholar] [CrossRef]
- Yan, L.X.; Wu, Q.N.; Zhang, Y.; Li, Y.Y.; Liao, D.Z.; Hou, J.H.; Fu, J.; Zeng, M.S.; Yun, J.P.; Wu, Q.L.; et al. Knockdown of Mir-21 in Human Breast Cancer Cell Lines Inhibits Proliferation, in Vitro Migration and in Vivo Tumor Growth. Breast Cancer Res. 2011, 13, R2. [Google Scholar] [CrossRef]
- Ji, W.; Sun, B.; Su, C. Targeting MicroRNAs in Cancer Gene Therapy. Genes 2017, 8, 21. [Google Scholar] [CrossRef]
- Yang, H.; Liu, Y.; Qiu, Y.; Ding, M.; Zhang, Y. MiRNA-204-5p and oxaliplatin-loaded silica nanoparticles for enhanced tumor suppression effect in CD44-overexpressed colon adenocarcinoma. Int. J. Pharm. 2019, 566, 585–593. [Google Scholar] [CrossRef]
- Gandham, S.K.; Rao, M.; Shah, A.; Trivedi, M.S.; Amiji, M.M. Combination Microrna-Based Cellular Reprogramming with Paclitaxel Enhances Therapeutic Efficacy in a Relapsed and Multidrug-Resistant Model of Epithelial Ovarian Cancer. Mol. Ther.-Oncolytics 2022, 25, 57–68. [Google Scholar] [CrossRef]
- Aagaard, L.; Rossi, J.J. Rnai Therapeutics: Principles, Prospects and Challenges. Adv. Drug Deliv. Rev. 2007, 59, 75–86. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).