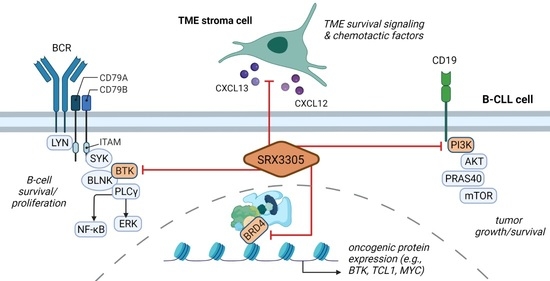
A Novel Triple-Action Inhibitor Targeting B-Cell Receptor Signaling and BRD4 Demonstrates Preclinical Activity in Chronic Lymphocytic Leukemia
Abstract
1. Introduction
2. Results
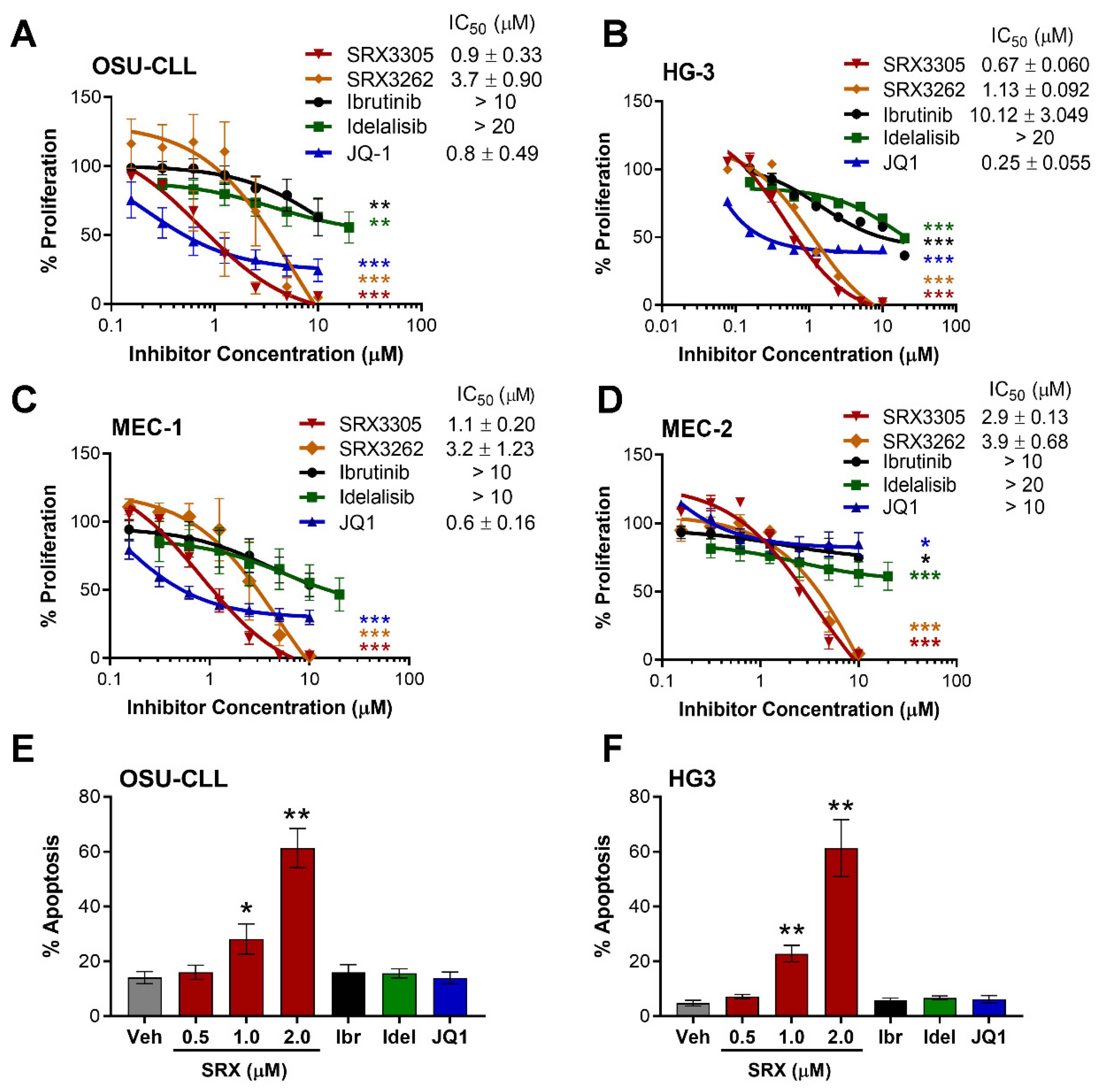
2.1. SRX3305 Inhibits Proliferation and Induces Apoptosis of Malignant B-Cell Lines
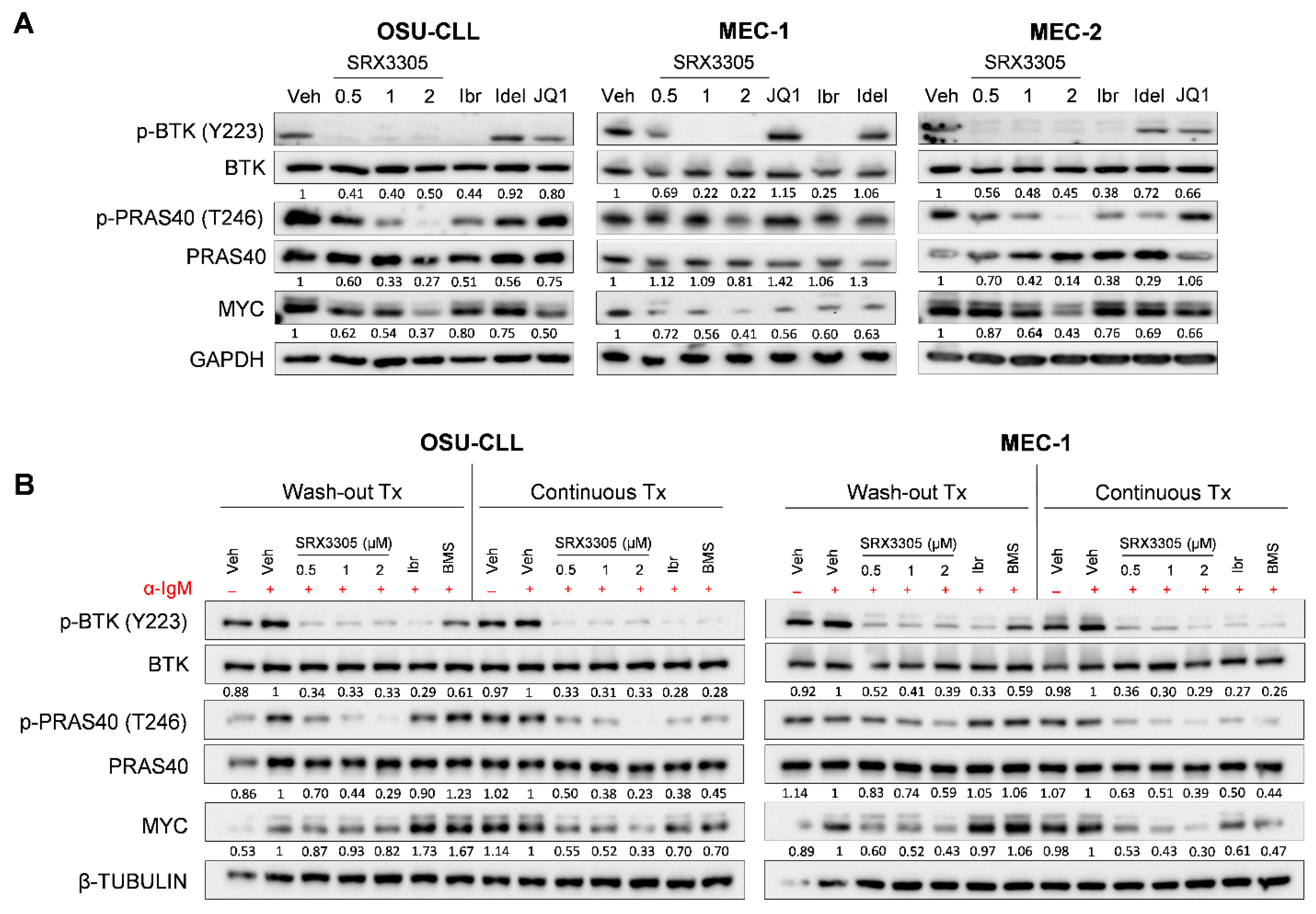
2.2. SRX3305 Inhibits Critical BCR Survival Signaling in CLL
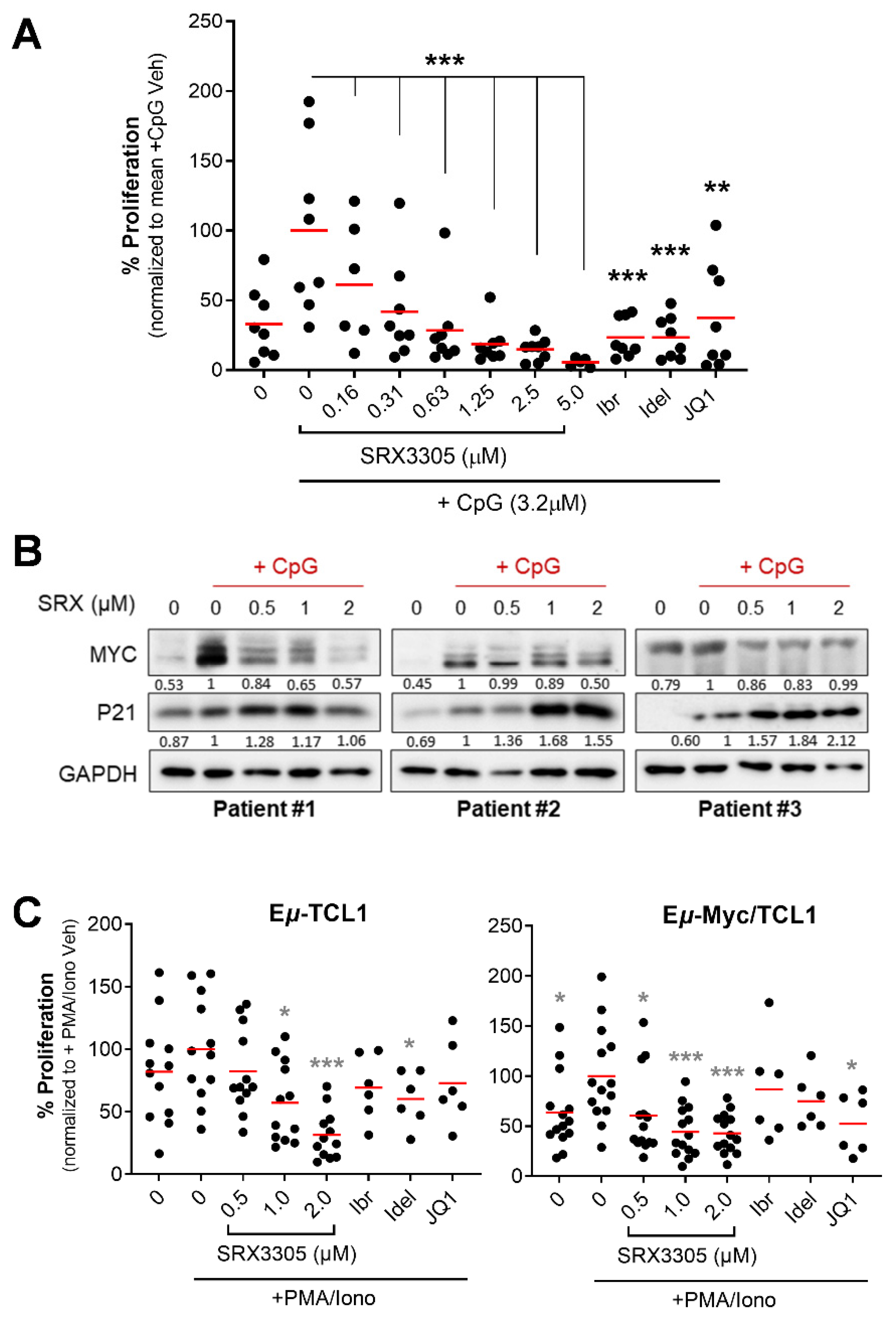
2.3. SRX3305 Inhibits Primary Malignant B-Cell Survival and Proliferation
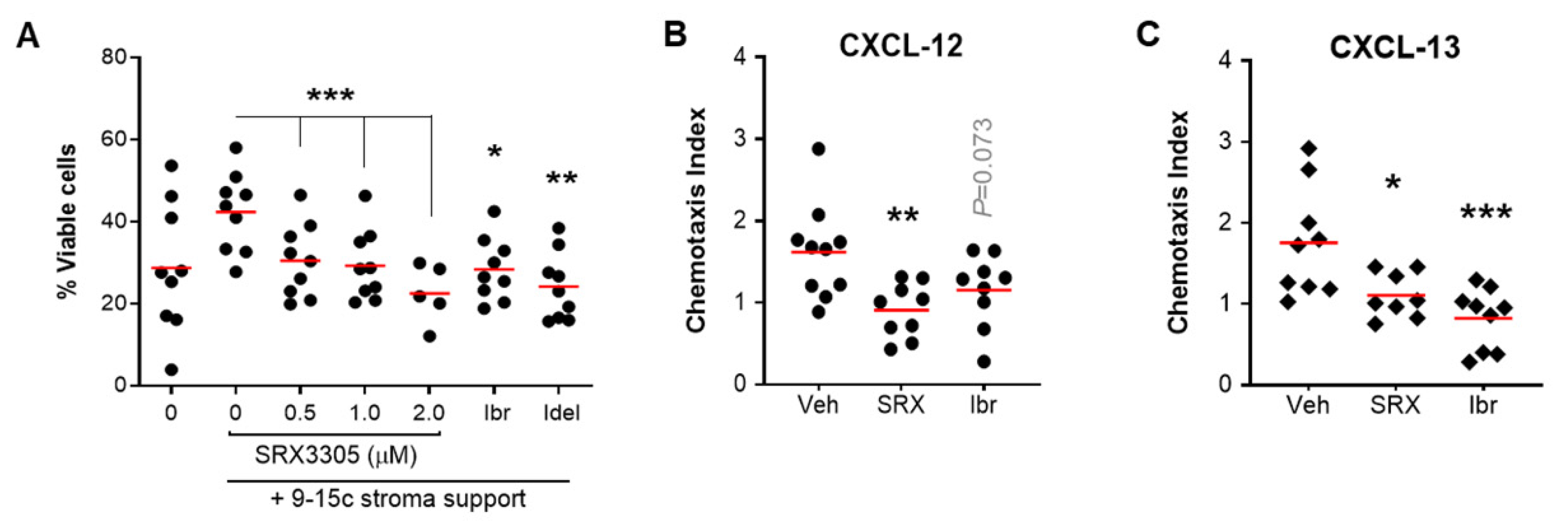
2.4. SRX3305 Disrupts Stroma Survival Support and Chemokine-Induced Migration in CLL
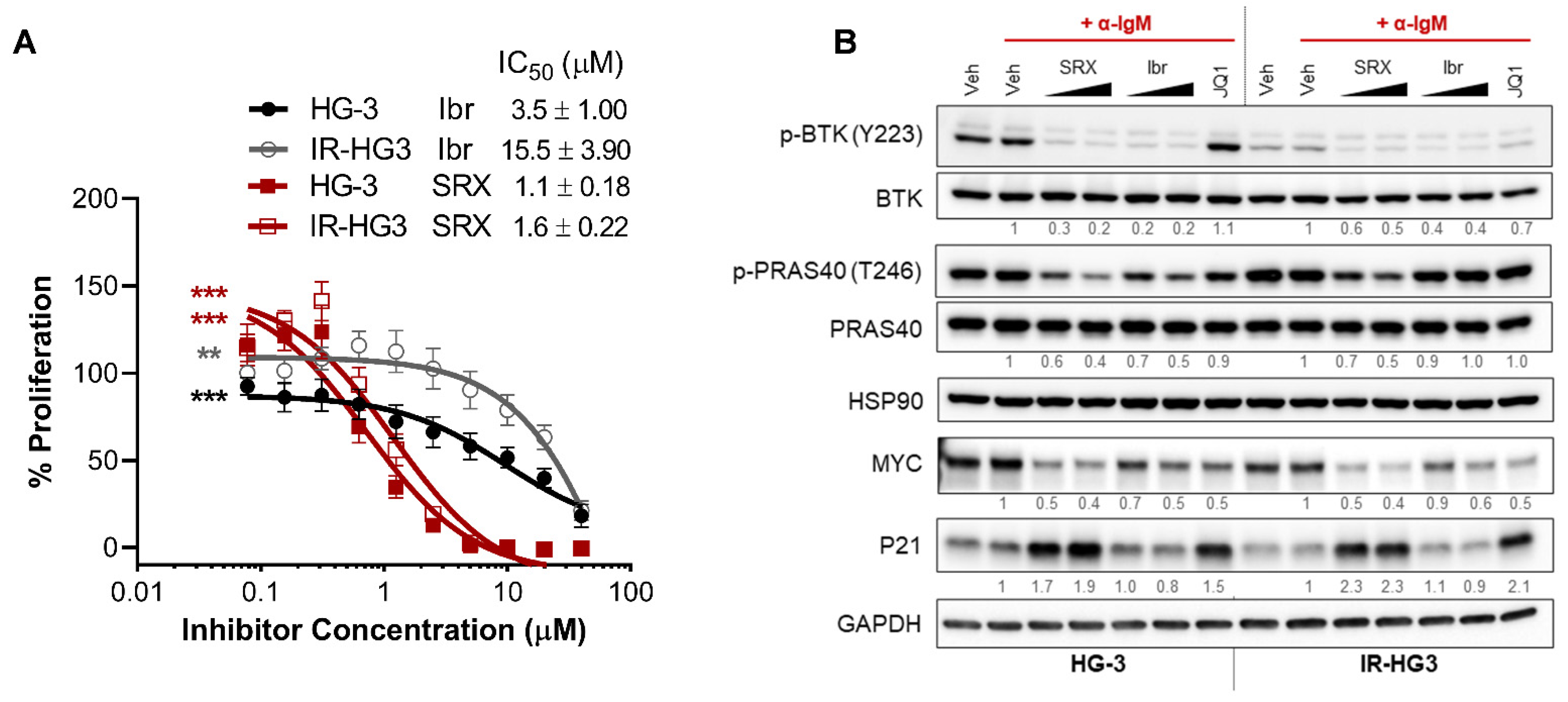
2.5. SRX3305 Is Active in Ibrutinib-Resistant CLL
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Primary Samples, and Inhibitors
4.2. Cell Culture
4.3. Cytotoxicity and Flow Cytometric Studies
4.4. BCR Pathway Activation
4.5. Inhibitor Washout Assay
4.6. Ex Vivo Stimulation of Primary Malignant B-Cells
4.7. Immunoblot Analyses
4.8. Migration Assay
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kipps, T.J.; Stevenson, F.; Wu, C.J.; Croce, C.M.; Packham, G.; Wierda, W.G.; O’Brien, S.; Gribben, J.; Rai, K. Chronic lymphocytic leukaemia. Nat. Rev. Dis. Prim. 2017, 3, 16096. [Google Scholar] [CrossRef]
- Riches, J.C.; Gribben, J.G. Understanding the immunodeficiency in chronic lymphocytic leukemia: Potential clinical implications. Hematol. Oncol. Clin. N. Am. 2013, 27, 207–235. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2019, 94, 1266–1287. [Google Scholar] [CrossRef] [PubMed]
- Herman, S.E.M.; Gordon, A.L.; Hertlein, E.; Ramanunni, A.; Zhang, X.; Jaglowski, S.; Flynn, J.; Jones, J.; Blum, K.A.; Buggy, J.J.; et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood 2011, 117, 6287–6296. [Google Scholar] [CrossRef] [PubMed]
- Herman, S.E.; Jones, J.; Mustafa, R.Z.; Farooqui, M.; Wiestner, A. In Vivo Effects of Ibrutinib on the Migration of Chronic Lymphocytic Leukemia Cells Differ Between Patients and Reduce the Ability of the Bone Marrow Microenvironment to Attract the Tumor Cells. Blood 2013, 122, 604. [Google Scholar] [CrossRef]
- O’Brien, S.; Furman, R.R.; Coutre, S.; Flinn, I.W.; Burger, J.A.; Blum, K.; Sharman, J.; Wierda, W.; Jones, J.; Zhao, W.; et al. Single-agent ibrutinib in treatment-naïve and relapsed/refractory chronic lymphocytic leukemia: A 5-year experience. Blood 2018, 131, 1910–1919. [Google Scholar] [CrossRef]
- Zhao, X.; Lwin, T.; Silva, A.; Shah, B.; Tao, J.; Fang, B.; Zhang, L.; Fu, K.; Bi, C.; Li, J.; et al. Unification of de novo and acquired ibrutinib resistance in mantle cell lymphoma. Nat. Commun. 2017, 8, 14920. [Google Scholar] [CrossRef]
- Rahal, R.; Frick, M.; Romero, R.; Korn, J.M.; Kridel, R.; Chan, F.C.; Meissner, B.; Bhang, H.-E.; Ruddy, D.; Kauffmann, A.; et al. Pharmacological and genomic profiling identifies NF-κB–targeted treatment strategies for mantle cell lymphoma. Nat. Med. 2014, 20, 87–92. [Google Scholar] [CrossRef]
- Moyo, T.K.; Wilson, C.S.; Moore, D.J.; Eischen, C.M. Myc enhances B-cell receptor signaling in precancerous B cells and confers resistance to Btk inhibition. Oncogene 2017, 36, 4653–4661. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L.L.; Wu, W.; Guo, H.; Li, Y.; Sukhanova, M.; Venkataraman, G.; Huang, S.; Zhang, H.; Alikhan, M.; et al. Activation of MYC, a bona fide client of HSP90, contributes to intrinsic ibrutinib resistance in mantle cell lymphoma. Blood Adv. 2018, 2, 2039–2051. [Google Scholar] [CrossRef]
- Ahn, I.E.; Underbayev, C.; Albitar, A.; Herman, S.E.M.; Tian, X.; Maric, I.; Arthur, D.C.; Wake, L.; Pittaluga, S.; Yuan, C.M.; et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood 2017, 129, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- George, B.; Chowdhury, S.M.; Hart, A.; Sircar, A.; Singh, S.K.; Nath, U.K.; Mamgain, M.; Singhal, N.K.; Sehgal, L.; Jain, N. Ibrutinib Resistance Mechanisms and Treatment Strategies for B-Cell Lymphomas. Cancers 2020, 12, 1328. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Spina, V.; Gaidano, G. Biology and treatment of Richter syndrome. Blood 2018, 131, 2761–2772. [Google Scholar] [CrossRef] [PubMed]
- Skånland, S.S.; Mato, A.R. Overcoming resistance to targeted therapies in chronic lymphocytic leukemia. Blood Adv. 2021, 5, 334–343. [Google Scholar] [CrossRef]
- Timofeeva, N.; Gandhi, V. Ibrutinib combinations in CLL therapy: Scientific rationale and clinical results. Blood Cancer J. 2021, 11, 79. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Mansouri, L.; Wierzbinska, J.A.; Plass, C.; Rosenquist, R. Epigenetic deregulation in chronic lymphocytic leukemia: Clinical and biological impact. Semin. Cancer Biol. 2018, 51, 1–11. [Google Scholar] [CrossRef]
- Xanthopoulos, C.; Kostareli, E. Advances in Epigenetics and Epigenomics in Chronic Lymphocytic Leukemia. Curr. Genet. Med. Rep. 2019, 7, 214–226. [Google Scholar] [CrossRef]
- Jiang, Y.; Redmond, D.; Nie, K.; Eng, K.W.; Clozel, T.; Martin, P.; Tan, L.H.; Melnick, A.M.; Tam, W.; Elemento, O. Deep sequencing reveals clonal evolution patterns and mutation events associated with relapse in B-cell lymphomas. Genome Biol. 2014, 15, 432. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Knapp, S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 2014, 13, 337–356. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-Enhancers in the Control of Cell Identity and Disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Lovén, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef] [PubMed]
- Chapuy, B.; McKeown, M.R.; Lin, C.Y.; Monti, S.; Roemer, M.G.; Qi, J.; Rahl, P.B.; Sun, H.H.; Yeda, K.T.; Doench, J.G.; et al. Discovery and Characterization of Super-Enhancer-Associated Dependencies in Diffuse Large B Cell Lymphoma. Cancer Cell 2013, 24, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.-I.; Robson, S.C.; Chung, C.-W.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef]
- Hogg, S.J.; Newbold, A.; Vervoort, S.J.; Cluse, L.A.; Martin, B.P.; Gregory, G.P.; Lefebure, M.; Vidacs, E.; Tothill, R.W.; Bradner, J.E.; et al. BET Inhibition Induces Apoptosis in Aggressive B-Cell Lymphoma via Epigenetic Regulation of BCL-2 Family Members. Mol. Cancer Ther. 2016, 15, 2030–2041. [Google Scholar] [CrossRef]
- Huang, B.; Yang, X.-D.; Zhou, M.-M.; Ozato, K.; Chen, L.-F. Brd4 Coactivates Transcriptional Activation of NF-κB via Specific Binding to Acetylated RelA. Mol. Cell. Biol. 2009, 29, 1375–1387. [Google Scholar] [CrossRef]
- Mansouri, L.; Papakonstantinou, N.; Ntoufa, S.; Stamatopoulos, K.; Rosenquist, R. NF-κB activation in chronic lymphocytic leukemia: A point of convergence of external triggers and intrinsic lesions. Semin. Cancer Biol. 2016, 39, 40–48. [Google Scholar] [CrossRef]
- Chaidos, A.; Caputo, V.; Karadimitris, A. Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: Emerging preclinical and clinical evidence. Ther. Adv. Hematol. 2015, 6, 128–141. [Google Scholar] [CrossRef]
- Stathis, A.; Bertoni, F. BET Proteins as Targets for Anticancer Treatment. Cancer Discov. 2018, 8, 24–36. [Google Scholar] [CrossRef]
- Ozer, H.G.; El-Gamal, D.; Powell, B.; Hing, Z.A.; Blachly, J.S.; Harrington, B.; Mitchell, S.; Grieselhuber, N.R.; Williams, K.; Lai, T.-H.; et al. BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor. Cancer Discov. 2018, 8, 458–477. [Google Scholar] [CrossRef]
- Spriano, F.; Stathis, A.; Bertoni, F. Targeting BET bromodomain proteins in cancer: The example of lymphomas. Pharmacol. Ther. 2020, 215, 107631. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Hacken, E.T.; Sivina, M.; Clarke, A.; Thompson, P.A.; Jain, N.; Ferrajoli, A.; Estrov, Z.; Keating, M.J.; Wierda, W.G.; et al. The BET inhibitor GS-5829 targets chronic lymphocytic leukemia cells and their supportive microenvironment. Leukemia 2019, 34, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Carrà, G.; Nicoli, P.; Lingua, M.F.; Maffeo, B.; Cartellà, A.; Circosta, P.; Brancaccio, M.; Parvis, G.; Gaidano, V.; Guerrasio, A.; et al. Inhibition of bromodomain and extra-terminal proteins increases sensitivity to venetoclax in chronic lymphocytic leukaemia. J. Cell. Mol. Med. 2019, 24, 1650–1657. [Google Scholar] [CrossRef] [PubMed]
- Tarantelli, C.; Lange, M.; Gaudio, E.; Cascione, L.; Spriano, F.; Kwee, I.; Arribas, A.J.; Rinaldi, A.; Jourdan, T.; Berthold, M.; et al. Copanlisib synergizes with conventional and targeted agents including venetoclax in B- and T-cell lymphoma models. Blood Adv. 2020, 4, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Fiskus, W.; Mill, C.P.; Perera, D.; Birdwell, C.; Deng, Q.; Yang, H.; Lara, B.H.; Jain, N.; Burger, J.; Ferrajoli, A.; et al. BET proteolysis targeted chimera-based therapy of novel models of Richter Transformation-diffuse large B-cell lymphoma. Leukemia 2021, 35, 2621–2634. [Google Scholar] [CrossRef] [PubMed]
- Morales, G.A.; Garlich, J.R.; Durden, D.L. Single Molecule Compounds Providing Multi-Target Inhibition of Btk and Other Proteins and Methods of Use Thereof. U.S. Patent Application 17/262,358, 30 September 2021. [Google Scholar]
- Pal, D.; Vann, K.R.; Joshi, S.; Sahar, N.E.; Morales, G.A.; El-Gamal, D.; Kutateladze, T.G.; Durden, D.L. The BTK/PI3K/BRD4 axis inhibitor SRX3262 overcomes Ibrutinib resistance in mantle cell lymphoma. iScience 2021, 24, 102931. [Google Scholar] [CrossRef]
- Vann, K.R.; Pal, D.; Smith, A.L.; Sahar, N.-E.; Krishnaiah, M.; El-Gamal, D.; Kutateladze, T.G. Combinatorial inhibition of BTK, PI3K-AKT and BRD4-MYC as a strategy for treatment of mantle cell lymphoma. Mol. Biomed. 2022, 3, 2. [Google Scholar] [CrossRef]
- Rasul, E.; Salamon, D.; Nagy, N.; Leveau, B.; Banati, F.; Szenthe, K.; Koroknai, A.; Minarovits, J.; Klein, G.; Klein, E. The MEC1 and MEC2 lines represent two CLL subclones in different stages of progression towards prolymphocytic leukemia. PLoS ONE 2014, 9, e106008. [Google Scholar] [CrossRef][Green Version]
- Stacchini, A.; Aragno, M.; Vallario, A.; Alfarano, A.; Circosta, P.; Gottardi, D.; Faldella, A.; Rege-Cambrin, G.; Thunberg, U.; Nilsson, K.; et al. MEC1 and MEC2: Two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk. Res. 1999, 23, 127–136. [Google Scholar] [CrossRef]
- Rosén, A.; Bergh, A.C.; Gogok, P.; Evaldsson, C.; Myhrinder, A.L.; Hellqvist, E.; Rasul, A.; Björkholm, M.; Jansson, M.; Mansouri, L.; et al. Lymphoblastoid cell line with B1 cell characteristics established from a chronic lymphocytic leukemia clone by in vitro EBV infection. Oncoimmunology 2012, 1, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Mehra, S.; Messner, H.; Minden, M.; Chaganti, R.S. Molecular cytogenetic characterization of non-Hodgkin lymphoma cell lines. Genes Chromosomes Cancer 2002, 33, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Quentmeier, H.; Pommerenke, C.; Dirks, W.G.; Eberth, S.; Koeppel, M.; MacLeod, R.A.; Nagel, S.; Steube, K.; Uphoff, C.C.; Drexler, H.G. The LL-100 panel: 100 cell lines for blood cancer studies. Sci. Rep. 2019, 9, 8218. [Google Scholar] [CrossRef] [PubMed]
- Drexler, H.G.; Quentmeier, H. The LL-100 Cell Lines Panel: Tool for Molecular Leukemia-Lymphoma Research. Int. J. Mol. Sci. 2020, 21, 5800. [Google Scholar] [CrossRef]
- Davis, R.E.; Ngo, V.N.; Lenz, G.; Tolar, P.; Young, R.M.; Romesser, P.B.; Kohlhammer, H.; Lamy, L.; Zhao, H.; Yang, Y.; et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010, 463, 88–92. [Google Scholar] [CrossRef]
- Ngo, V.N.; Young, R.M.; Schmitz, R.; Jhavar, S.; Xiao, W.; Lim, K.H.; Kohlhammer, H.; Xu, W.; Yang, Y.; Zhao, H.; et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011, 470, 115–119. [Google Scholar] [CrossRef]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef]
- Seda, V.; Mraz, M. B-cell receptor signalling and its crosstalk with other pathways in normal and malignant cells. Eur. J. Haematol. 2014, 94, 193–205. [Google Scholar] [CrossRef]
- Zhuang, J.; Hawkins, S.F.; Glenn, M.A.; Lin, K.; Johnson, G.G.; Carter, A.; Cawley, J.C.; Pettitt, A.R. Akt is activated in chronic lymphocytic leukemia cells and delivers a pro-survival signal: The therapeutic potential of Akt inhibition. Haematologica 2009, 95, 110–118. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Murray, L.J.; Habeshaw, J.A.; Stansfeld, A.G. Lymphocytic lymphoma/B-chronic lymphocytic leukaemia—An immunohistopathological study of peripheral B lymphocyte neoplasia. Br. J. Cancer 1984, 50, 587–599. [Google Scholar] [CrossRef][Green Version]
- Decker, T.; Schneller, F.; Sparwasser, T.; Tretter, T.; Lipford, G.B.; Wagner, H.; Peschel, C. Immunostimulatory CpG-oligonucleotides cause proliferation, cytokine production, and an immunogenic phenotype in chronic lymphocytic leukemia B cells. Blood 2000, 95, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Bichi, R.; Shinton, S.A.; Martin, E.S.; Koval, A.; Calin, G.A.; Cesari, R.; Russo, G.; Hardy, R.R.; Croce, C.M. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6955–6960. [Google Scholar] [CrossRef]
- Johnson, A.J.; Lucas, D.M.; Muthusamy, N.; Smith, L.L.; Edwards, R.B.; De Lay, M.D.; Croce, C.M.; Grever, M.R.; Byrd, J.C. Characterization of the TCL-1 transgenic mouse as a preclinical drug development tool for human chronic lymphocytic leukemia. Blood 2006, 108, 1334–1338. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.A.; El-Gamal, D.; Bonnie, H.K.; Zachary, H.A.; Virginia, G.M.; Rose, M.; Smith, L.L.; Yu, L.; Johnson, A.J.; Byrd, J.C.; et al. The Eµ-Myc/TCL1 Transgenic Mouse as a New Aggressive B-Cell Malignancy Model Suitable for Preclinical Therapeutics Testing. Blood 2015, 126, 2752. [Google Scholar] [CrossRef]
- Lucas, F.; Rogers, K.A.; Harrington, B.K.; Pan, A.; Yu, L.; Breitbach, J.; Bundschuh, R.; Goettl, V.M.; Hing, Z.A.; Kanga, P.; et al. Eμ-TCL1xMyc: A Novel Mouse Model for Concurrent CLL and B-Cell Lymphoma. Clin. Cancer Res. 2019, 25, 6260–6273. [Google Scholar] [CrossRef]
- Herishanu, Y.; Katz, B.-Z.; Lipsky, A.; Wiestner, A. Biology of Chronic Lymphocytic Leukemia in Different Microenvironments. Hematol. Clin. N. Am. 2013, 27, 173–206. [Google Scholar] [CrossRef]
- Ramsay, A.D.; Rodriguez-Justo, M. Chronic lymphocytic leukaemia—The role of the microenvironment pathogenesis and therapy. Br. J. Haematol. 2013, 162, 15–24. [Google Scholar] [CrossRef]
- Kurtova, A.V.; Balakrishnan, K.; Chen, R.; Ding, W.; Schnabl, S.; Quiroga, M.P.; Sivina, M.; Wierda, W.G.; Estrov, Z.; Keating, M.J.; et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: Development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood 2009, 114, 4441–4450. [Google Scholar] [CrossRef]
- Burger, J.A.; Wiestner, A. Targeting B cell receptor signalling in cancer: Preclinical and clinical advances. Nat. Cancer 2018, 18, 148–167. [Google Scholar] [CrossRef]
- Hartmann, T.N.; Grabovsky, V.; Wang, W.; Desch, P.; Rubenzer, G.; Wollner, S.; Binsky, I.; Vallon-Eberhard, A.; Sapoznikov, A.; Burger, M.; et al. Circulating B-Cell Chronic Lymphocytic Leukemia Cells Display Impaired Migration to Lymph Nodes and Bone Marrow. Cancer Res. 2009, 69, 3121–3130. [Google Scholar] [CrossRef] [PubMed]
- Scielzo, C.; Bertilaccio, M.T.S.; Simonetti, G.; Dagklis, A.; Hacken, E.T.; Fazi, C.; Muzio, M.; Caiolfa, V.; Kitamura, D.; Restuccia, U.; et al. HS1 has a central role in the trafficking and homing of leukemic B cells. Blood 2010, 116, 3537–3546. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Burger, M.; Kipps, T.J. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood 1999, 94, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Jones, D.; Springer, T.A. The Chemokine Receptor CXCR4 Is Required for the Retention of B Lineage and Granulocytic Precursors within the Bone Marrow Microenvironment. Immunity 1999, 10, 463–471. [Google Scholar] [CrossRef]
- Ansel, K.; Harris, R.B.; Cyster, J.G. CXCL13 Is Required for B1 Cell Homing, Natural Antibody Production, and Body Cavity Immunity. Immunity 2002, 16, 67–76. [Google Scholar] [CrossRef]
- Ponader, S.; Chen, S.-S.; Buggy, J.J.; Balakrishnan, K.; Gandhi, V.; Wierda, W.G.; Keating, M.J.; O’Brien, S.; Chiorazzi, N.; Burger, J.A. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood 2012, 119, 1182–1189. [Google Scholar] [CrossRef]
- Spina, V.; Forestieri, G.; Zucchetto, A.; Bruscaggin, A.; Bittolo, T.; Di Bergamo, L.T.; Szenes, E.; Condoluci, A.; Tissino, E.; De Paoli, L.; et al. Mechanisms of Adaptation to Ibrutinib in High Risk Chronic Lymphocytic Leukemia. Blood 2018, 132, 585. [Google Scholar] [CrossRef]
- Dubois, N.; Crompot, E.; Meuleman, N.; Bron, D.; Lagneaux, L.; Stamatopoulos, B. Importance of Crosstalk Between Chronic Lymphocytic Leukemia Cells and the Stromal Microenvironment: Direct Contact, Soluble Factors, and Extracellular Vesicles. Front. Oncol. 2020, 10, 1422. [Google Scholar] [CrossRef]
- Shain, K.H.; Dalton, W.S.; Tao, J. The tumor microenvironment shapes hallmarks of mature B-cell malignancies. Oncogene 2015, 34, 4673–4682. [Google Scholar] [CrossRef]
- De Rooij, M.F.M.; Kuil, A.; Geest, C.R.; Eldering, E.; Chang, B.Y.; Buggy, J.J.; Pals, S.T.; Spaargaren, M. The clinically active BTK inhibitor PCI-32765 targets B-cell receptor– and chemokine-controlled adhesion and migration in chronic lymphocytic leukemia. Blood 2012, 119, 2590–2594. [Google Scholar] [CrossRef]
- Hoellenriegel, J.; Meadows, S.A.; Sivina, M.; Wierda, W.G.; Kantarjian, H.; Keating, M.J.; Giese, N.; O’Brien, S.; Yu, A.; Miller, L.L.; et al. The phosphoinositide 3′-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood 2011, 118, 3603–3612. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-Q.; Cao, Z.-R.; Wang, Y.; Zhang, X.; Xu, L.; Wang, Y.-X.; Chen, Y.; Yang, C.-H.; Ding, J.; Meng, L.-H. Repressing MYC by targeting BET synergizes with selective inhibition of PI3Kα against B cell lymphoma. Cancer Lett. 2021, 524, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, E.; Mondello, P.; Erazo, T.; Portelinha, A.; Liu, Y.; Scallion, M.; Asgari, Z.; Philip, J.; Hilden, P.; Valli, D.; et al. BET Inhibition-Induced GSK3β Feedback Enhances Lymphoma Vulnerability to PI3K Inhibitors. Cell Rep. 2018, 24, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- Sears, R.; Nuckolls, F.; Haura, E.; Taya, Y.; Tamai, K.; Nevins, J.R. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000, 14, 2501–2514. [Google Scholar] [CrossRef]
- Zhu, J.; Blenis, J.; Yuan, J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. USA 2008, 105, 6584–6589. [Google Scholar] [CrossRef]
- Schaffer, M.; Chaturvedi, S.; Davis, C.; Aquino, R.; Stepanchick, E.; Versele, M.; Liu, Y.; Yang, J.; Lu, R.; Balasubramanian, S. Identification of potential ibrutinib combinations in hematological malignancies using a combination high-throughput screen. Leuk. Lymphoma 2017, 59, 931–940. [Google Scholar] [CrossRef]
- Niemann, C.U.; Mora-Jensen, H.I.; Dadashian, E.L.; Krantz, F.; Covey, T.; Chen, S.-S.; Chiorazzi, N.; Izumi, R.; Ulrich, R.; Lannutti, B.J.; et al. Combined BTK and PI3Kδ Inhibition with Acalabrutinib and ACP-319 Improves Survival and Tumor Control in CLL Mouse Model. Clin. Cancer Res. 2017, 23, 5814–5823. [Google Scholar] [CrossRef]
- Andrews, F.H.; Singh, A.R.; Joshi, S.; Smith, C.A.; Morales, G.A.; Garlich, J.R.; Durden, D.L.; Kutateladze, T.G. Dual-activity PI3K–BRD4 inhibitor for the orthogonal inhibition of MYC to block tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2017, 114, E1072–E1080. [Google Scholar] [CrossRef]
- Sestier, M.; Hillis, C.; Fraser, G.; Leong, D. Bruton’s tyrosine kinase Inhibitors and Cardiotoxicity: More Than Just Atrial Fibrillation. Curr. Oncol. Rep. 2021, 23, 113. [Google Scholar] [CrossRef]
- Lampson, B.L.; Brown, J.R. The Evolving Use of Phosphatidylinositol 3-Kinase Inhibitors for the Treatment of Chronic Lymphocytic Leukemia. Hematol. Clin. N. Am. 2021, 35, 807–826. [Google Scholar] [CrossRef]
- Falchook, G.; Rosen, S.; LoRusso, P.; Watts, J.; Gupta, S.; Coombs, C.C.; Talpaz, M.; Kurzrock, R.; Mita, M.; Cassaday, R.; et al. Development of 2 Bromodomain and Extraterminal Inhibitors with Distinct Pharmacokinetic and Pharmacodynamic Profiles for the Treatment of Advanced Malignancies. Clin. Cancer Res. 2020, 26, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Piha-Paul, S.A.; Sachdev, J.C.; Barve, M.; Lorusso, P.; Szmulewitz, R.; Patel, S.P.; Lara, P.N., Jr.; Chen, X.; Hu, B.; Freise, K.J.; et al. First-in-Human Study of Mivebresib (ABBV-075), an Oral Pan-Inhibitor of Bromodomain and Extra Terminal Proteins, in Patients with Relapsed/Refractory Solid Tumors. Clin. Cancer Res. 2019, 25, 6309–6319. [Google Scholar] [CrossRef] [PubMed]
- Davids, M.S.; Kim, H.T.; Nicotra, A.; Savell, A.; Francoeur, K.; Hellman, J.M.; Bazemore, J.; Miskin, H.P.; Sportelli, P.; Stampleman, L.; et al. Umbralisib in combination with ibrutinib in patients with relapsed or refractory chronic lymphocytic leukaemia or mantle cell lymphoma: A multicentre phase 1–1b study. Lancet Haematol. 2018, 6, e38–e47. [Google Scholar] [CrossRef]
- Matulonis, U.A.; Wulf, G.M.; Barry, W.T.; Birrer, M.; Westin, S.N.; Farooq, S.; Bell-McGuinn, K.M.; Obermayer, E.; Whalen, C.; Spagnoletti, T.; et al. Phase I dose escalation study of the PI3kinase pathway inhibitor BKM120 and the oral poly (ADP ribose) polymerase (PARP) inhibitor olaparib for the treatment of high-grade serous ovarian and breast cancer. Ann. Oncol. 2017, 28, 512–518. [Google Scholar] [CrossRef]
- Hertlein, E.; Beckwith, K.A.; Lozanski, G.; Chen, T.L.; Towns, W.H.; Johnson, A.J.; Lehman, A.; Ruppert, A.S.; Bolon, B.; Andritsos, L.; et al. Characterization of a New Chronic Lymphocytic Leukemia Cell Line for Mechanistic In Vitro and In Vivo Studies Relevant to Disease. PLoS ONE 2013, 8, e76607. [Google Scholar] [CrossRef]
- Yamada, Y.; Sakurada, K.; Takeda, Y.; Gojo, S.; Umezawa, A. Single-cell-derived mesenchymal stem cells overexpressing Csx/Nkx2.5 and GATA4 undergo the stochastic cardiomyogenic fate and behave like transient amplifying cells. Exp. Cell Res. 2007, 313, 698–706. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, A.L.; Eiken, A.P.; Skupa, S.A.; Moore, D.Y.; Umeta, L.T.; Smith, L.M.; Lyden, E.R.; D’Angelo, C.R.; Kallam, A.; Vose, J.M.; et al. A Novel Triple-Action Inhibitor Targeting B-Cell Receptor Signaling and BRD4 Demonstrates Preclinical Activity in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2022, 23, 6712. https://doi.org/10.3390/ijms23126712
Smith AL, Eiken AP, Skupa SA, Moore DY, Umeta LT, Smith LM, Lyden ER, D’Angelo CR, Kallam A, Vose JM, et al. A Novel Triple-Action Inhibitor Targeting B-Cell Receptor Signaling and BRD4 Demonstrates Preclinical Activity in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences. 2022; 23(12):6712. https://doi.org/10.3390/ijms23126712
Chicago/Turabian StyleSmith, Audrey L., Alexandria P. Eiken, Sydney A. Skupa, Dalia Y. Moore, Lelisse T. Umeta, Lynette M. Smith, Elizabeth R. Lyden, Christopher R. D’Angelo, Avyakta Kallam, Julie M. Vose, and et al. 2022. "A Novel Triple-Action Inhibitor Targeting B-Cell Receptor Signaling and BRD4 Demonstrates Preclinical Activity in Chronic Lymphocytic Leukemia" International Journal of Molecular Sciences 23, no. 12: 6712. https://doi.org/10.3390/ijms23126712
APA StyleSmith, A. L., Eiken, A. P., Skupa, S. A., Moore, D. Y., Umeta, L. T., Smith, L. M., Lyden, E. R., D’Angelo, C. R., Kallam, A., Vose, J. M., Kutateladze, T. G., & El-Gamal, D. (2022). A Novel Triple-Action Inhibitor Targeting B-Cell Receptor Signaling and BRD4 Demonstrates Preclinical Activity in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences, 23(12), 6712. https://doi.org/10.3390/ijms23126712