CircDCLRE1C Regulated Lipopolysaccharide-Induced Inflammatory Response and Apoptosis by Regulating miR-214b-3p/STAT3 Pathway in Macrophages
Abstract
1. Introduction
2. Results
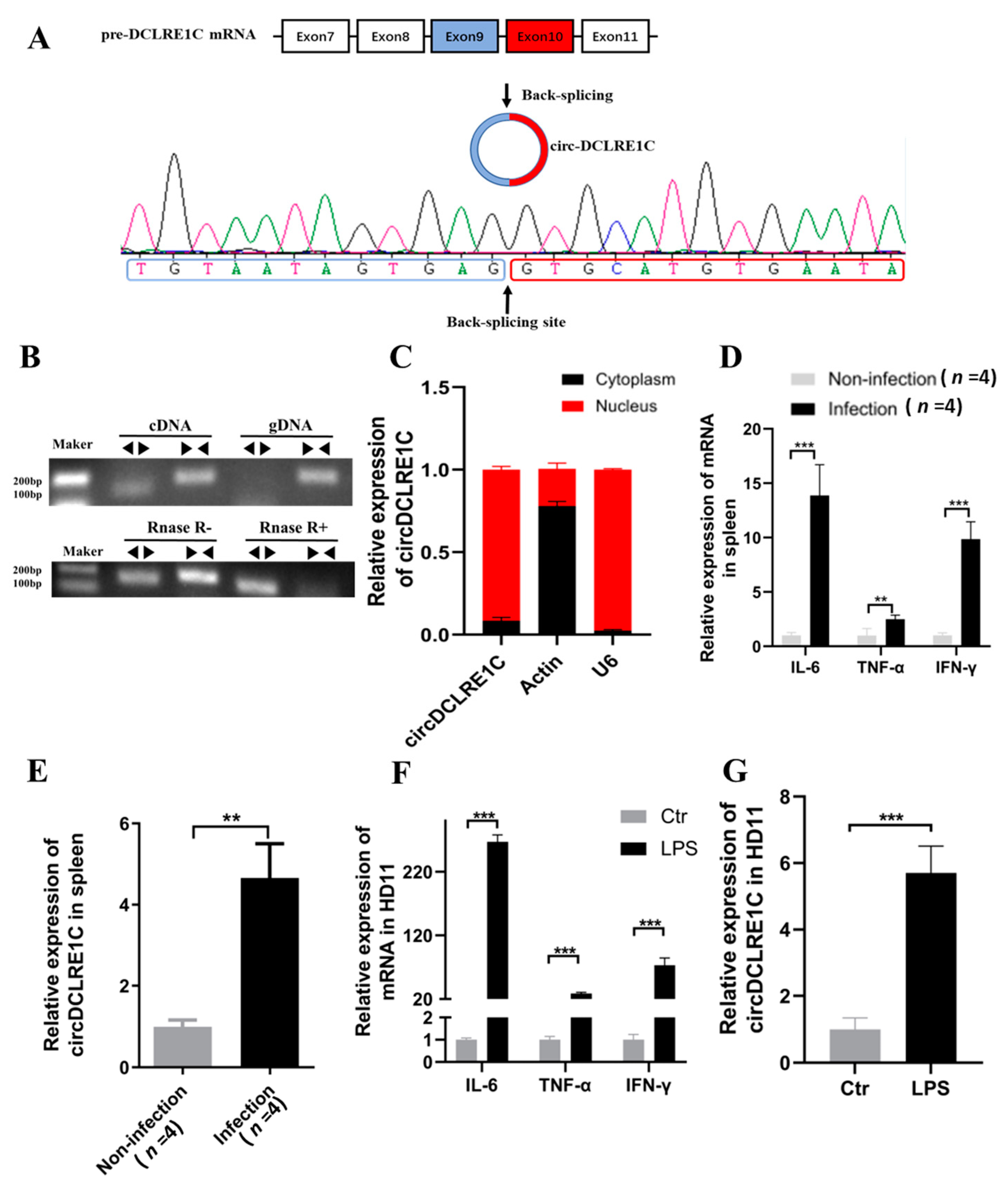
2.1. The Characteristics and Subcellular Localization of circDCLRE1C
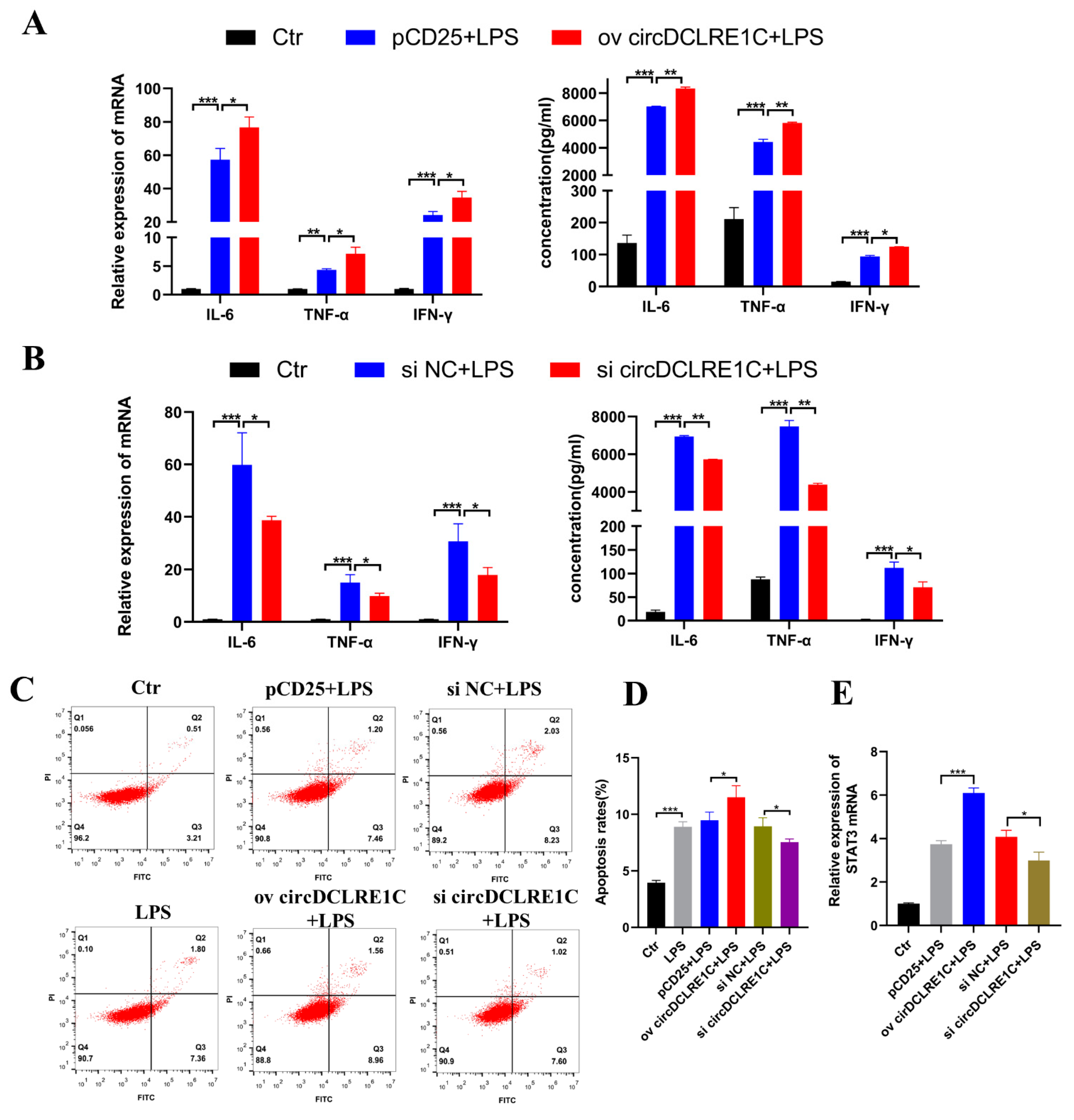
2.2. CircDCLRE1C Aggravated Inflammation and Apoptosis in HD11
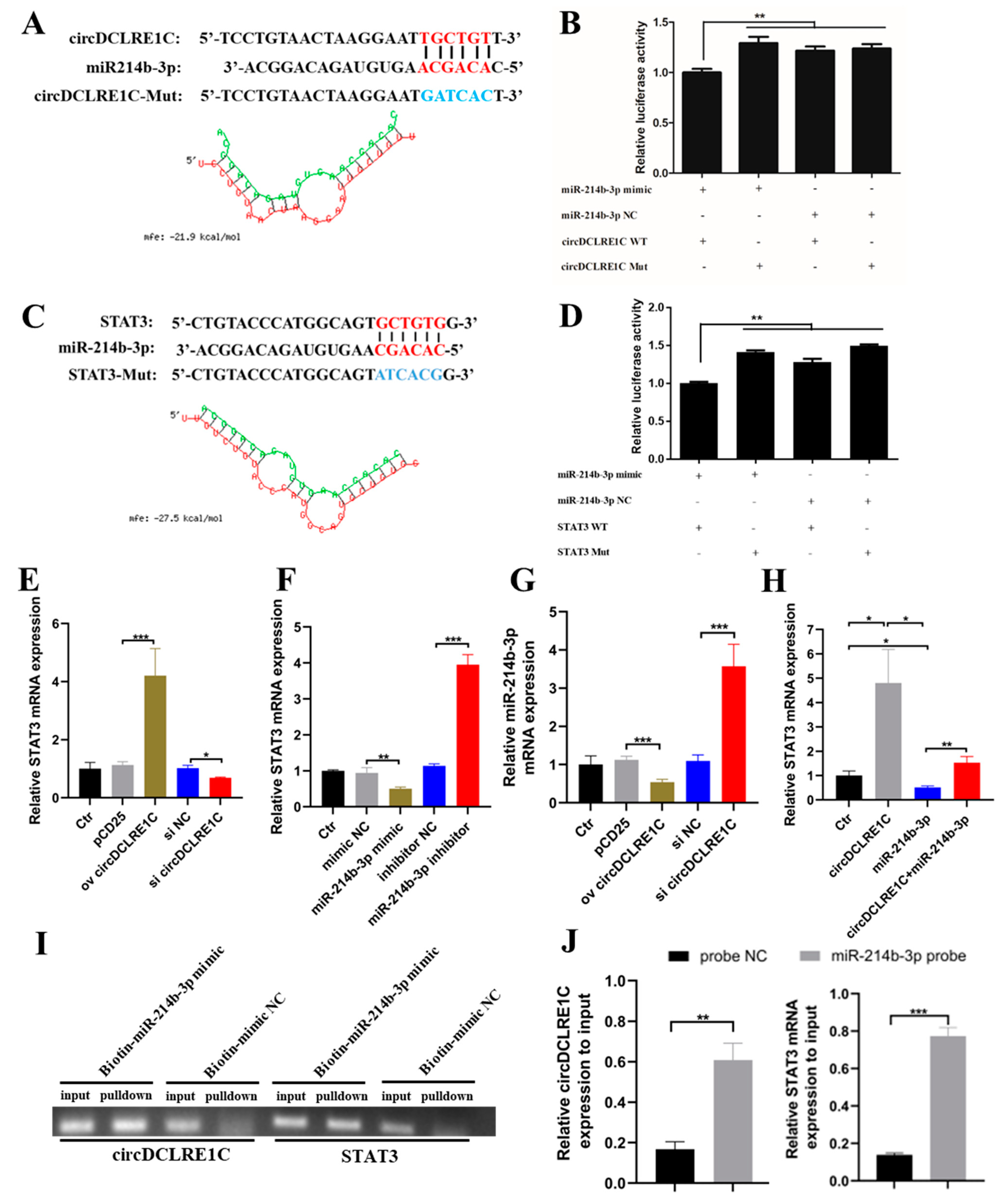
2.3. CircDCLRE1C Could Serve as a Sponge for miR-214b-3p Regulating STAT3 mRNA Expression
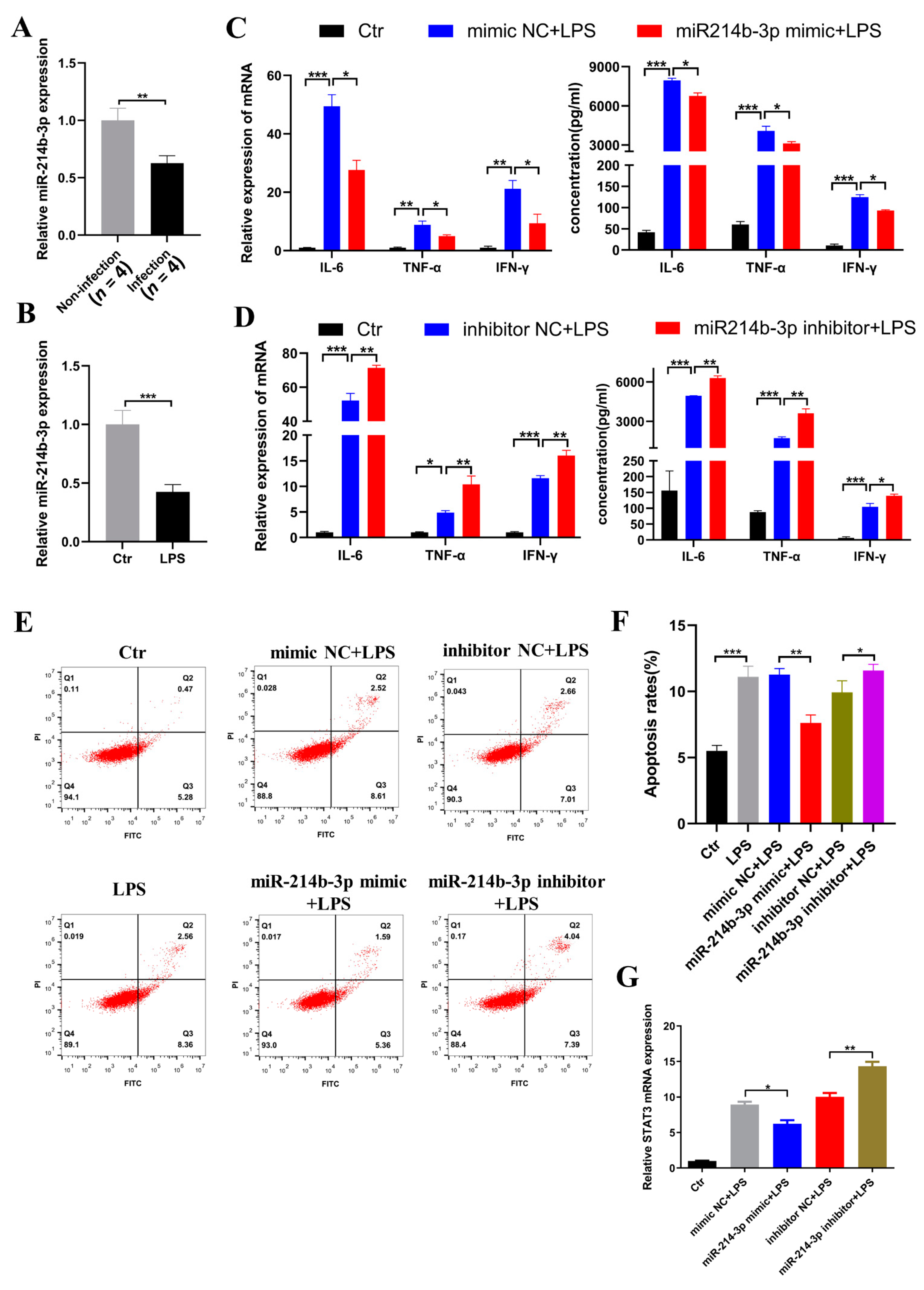
2.4. miR-214b-3p Alleviates Inflammation and Apoptosis in HD11
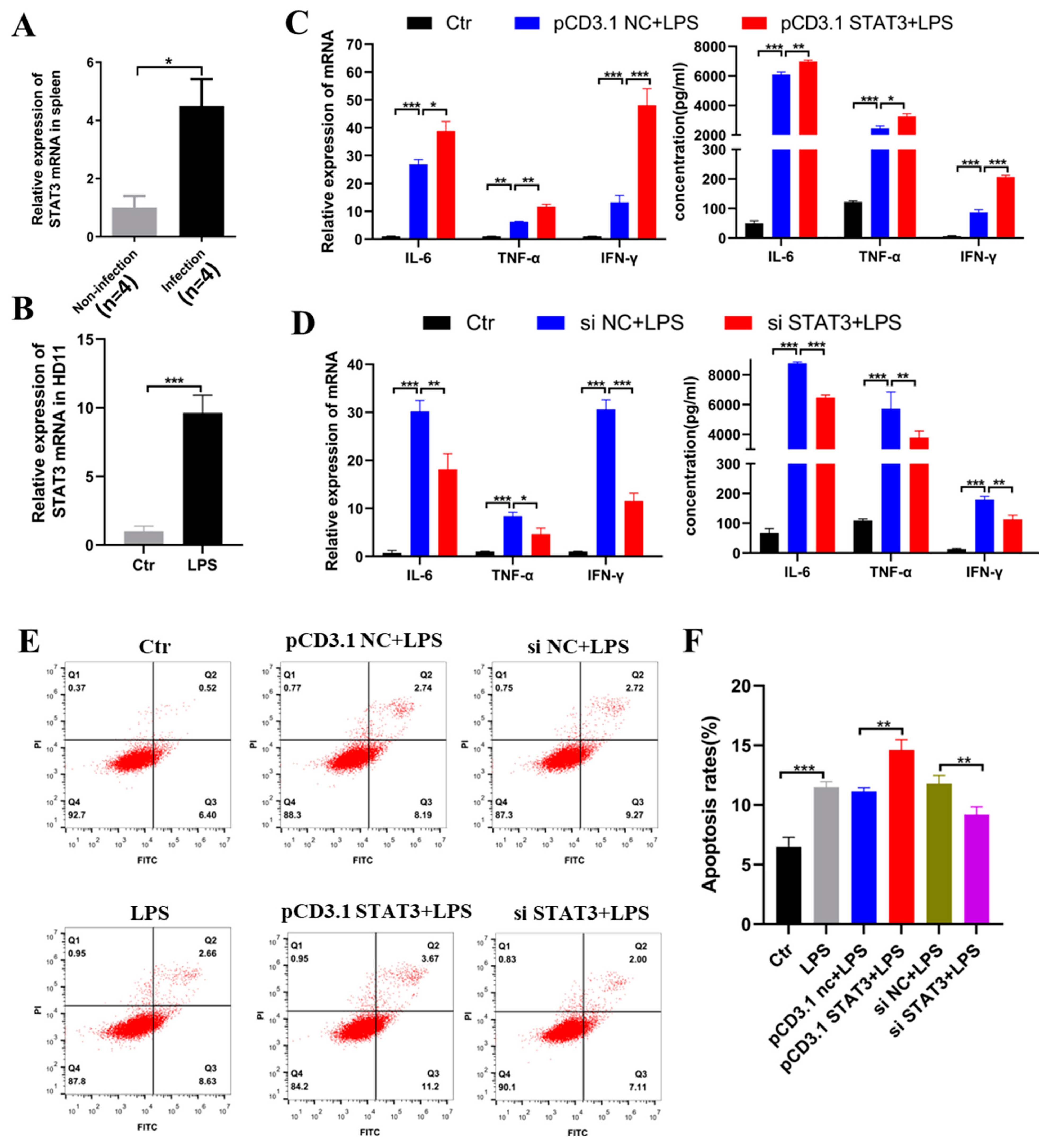
2.5. STAT3 Induced Inflammation and Apoptosis in the HD11
2.6. CircDCLRE1C Aggravated LPS-Induced Inflammation and Apoptosis via miR-214b-3p/STAT3 Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Sample Sources
4.2. Transfection
4.3. Flow Cytometry Assay
4.4. Construction of RNA Oligonucleotides and Plasmids
4.5. Circular Structure Confirmation
4.6. MiRNA Targets Prediction and RNAhybrid Detection
4.7. RNA Pulldown
4.8. Reverse-Transcription Polymerase Chain Reaction (RT-PCR) and Quantitative PCRQ-PCR)
4.9. Enzyme-Linked Immunosorbent assay (ELISA)
4.10. Dual-Luciferase Reporter Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blake, D.P.; Tomley, F.M. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014, 30, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zou, W.; Mi, C.; Wang, Q.; Shi, H.J.P.S. Expression of T cell-related cytokines in chicken cecal and spleen tissues following Eimeria tenella infection in vivo. Poult. Sci. 2021, 100, 101161. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Brownlie, R.; Allan, B. Avian toll-like receptors. Cell Tissue Res. 2011, 343, 121–130. [Google Scholar] [CrossRef]
- Sandholt, A.K.S.; Wattrang, E.; Lilja, T.; Ahola, H.; Lundén, A.; Troell, K.; Svärd, S.G.; Söderlund, R. Dual RNA-seq transcriptome analysis of caecal tissue during primary Eimeria tenella infection in chickens. BMC Genom. 2021, 22, 660. [Google Scholar] [CrossRef]
- Swaggerty, C.L.; Pevzner, I.Y.; Kogut, M.H. Selection for pro-inflammatory mediators produces chickens more resistant to Eimeria tenella. Poult. Sci. 2015, 94, 37–42. [Google Scholar] [CrossRef]
- Yu, H.; Zou, W.; Xin, S.; Wang, X.; Mi, C.; Dai, G.; Zhang, T.; Zhang, G.; Xie, K.; Wang, J.; et al. Association Analysis of Single Nucleotide Polymorphisms in the 5’ Regulatory Region of the IL-6 Gene with Eimeria tenella Resistance in Jinghai Yellow Chickens. Genes 2019, 10, 890. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.H.; Chaudhari, A.A.; Lillehoj, H.S. Involvement of T Cell Immunity in Avian Coccidiosis. Front. Immunol. 2019, 10, 2732. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Tang, X.; Wang, S. Roles of CircRNAs in Autoimmune Diseases. Front. Immunol. 2019, 10, 639. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yang, T.; Ding, Z.; Cao, Y. Circ_0026579 alleviates LPS-induced WI-38 cells inflammation injury in infantile pneumonia. Innate Immun. 2022, 28, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880. [Google Scholar] [CrossRef]
- Salas, A.; Hernandez-Rocha, C.; Duijvestein, M.; Faubion, W.; McGovern, D.; Vermeire, S.; Vetrano, S.; Vande Casteele, N. JAK-STAT pathway targeting for the treatment of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Hernández, A.; Forbes, L.R. JAK/STAT proteins and their biological impact on NK cell development and function. Mol. Immunol. 2019, 115, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M. Activating mutations of STAT3: Impact on human growth. Mol. Cell Endocrinol. 2020, 518, 110979. [Google Scholar] [CrossRef]
- Yoshimura, A.; Ito, M.; Chikuma, S.; Akanuma, T.; Nakatsukasa, H. Negative Regulation of Cytokine Signaling in Immunity. Cold Spring Harb. Perspect. Biol. 2018, 10, a028571. [Google Scholar] [CrossRef]
- Lokau, J.; Schoeder, V.; Haybaeck, J.; Garbers, C. Jak-Stat Signaling Induced by Interleukin-6 Family Cytokines in Hepatocellular Carcinoma. Cancers 2019, 11, 1704. [Google Scholar] [CrossRef]
- Szilveszter, K.P.; Németh, T.; Mócsai, A. Tyrosine Kinases in Autoimmune and Inflammatory Skin Diseases. Front. Immunol. 2019, 10, 1862. [Google Scholar] [CrossRef]
- He, G.; Karin, M. NF-κB and STAT3—key players in liver inflammation and cancer. Cell Res. 2011, 21, 159–168. [Google Scholar] [CrossRef]
- Chen, X.L.; Wang, Z.J.; Chen, Y.F.; Akinci, I.; Luo, W.; Xu, Y.B.; Jebessa, E.; Blake, D.; Sparks, N.; Hanotte, O.H.; et al. Whole transcriptome sequencing analysis of circRNAs, miRNAs, and mRNAs in Sasso chickens during the challenge of coccidiosis. Front. Immunol. submitted.
- Lee, Y.; Lu, M.; Lillehoj, H.S. Coccidiosis: Recent Progress in Host Immunity and Alternatives to Antibiotic Strategies. Vaccines 2022, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Mesa-Pineda, C.; Navarro-Ruíz, J.L.; López-Osorio, S.; Chaparro-Gutiérrez, J.J.; Gómez-Osorio, L.M. Chicken Coccidiosis: From the Parasite Lifecycle to Control of the Disease. Front. Vet. Sci. 2021, 8, 787653. [Google Scholar] [CrossRef] [PubMed]
- Salem, H.M.; Salem, M.A.; Soliman, M.M.; Althobaiti, S.A.; Khafaga, A.K.; El-Tahan, A.M.; El-Saadony, M.T.; Attia, M.M. Parasitological and histopathological examination of Cocktail lovebirds infected with Eimeria aratinga (Apicomplexa: Eimeriidae). Poult. Sci. 2022, 101, 101781. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Podinovskaia, M.; Descoteaux, A. Leishmania and the macrophage: A multifaceted interaction. Future Microbiol. 2015, 10, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of Human Macrophage Polarization in Inflammation during Infectious Diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef]
- Beug, H.; von Kirchbach, A.; Döderlein, G.; Conscience, J.F.; Graf, T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 1979, 18, 375–390. [Google Scholar] [CrossRef]
- Zheng, S.; Zhang, X.; Odame, E.; Xu, X.; Chen, Y.; Ye, J.; Zhou, H.; Dai, D.; Kyei, B.; Zhan, S.; et al. CircRNA-Protein Interactions in Muscle Development and Diseases. Int. J. Mol. Sci. 2021, 22, 3262. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.D.; Jiang, L.H.; Sun, D.W.; Hou, J.C.; Ji, Z.L. CircRNA: A novel type of biomarker for cancer. Breast Cancer 2018, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Prats, A.C.; David, F.; Diallo, L.H.; Roussel, E.; Tatin, F.; Garmy-Susini, B.; Lacazette, E. Circular RNA, the Key for Translation. Int. J. Mol. Sci. 2020, 21, 8591. [Google Scholar] [CrossRef] [PubMed]
- Humblet-Baron, S.; Schönefeldt, S.; Garcia-Perez, J.E.; Baron, F.; Pasciuto, E.; Liston, A. Cytotoxic T-lymphocyte-associated protein 4-Ig effectively controls immune activation and inflammatory disease in a novel murine model of leaky severe combined immunodeficiency. J. Allergy Clin. Immunol. 2017, 140, 1394–1403. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hillmer, E.J.; Zhang, H.; Li, H.S.; Watowich, S.S. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 2016, 31, 1–15. [Google Scholar] [CrossRef]
- An, M.; Zheng, H.; Huang, J.; Lin, Y.; Luo, Y.; Kong, Y.; Pang, M.; Zhang, D.; Yang, J.; Chen, J.; et al. Aberrant Nuclear Export of circNCOR1 Underlies SMAD7-Mediated Lymph Node Metastasis of Bladder Cancer. Cancer Res. 2022, 82, 2239–2253. [Google Scholar] [CrossRef]
- Chen, R.X.; Chen, X.; Xia, L.P.; Zhang, J.X.; Pan, Z.Z.; Ma, X.D.; Han, K.; Chen, J.W.; Judde, J.G.; Deas, O.; et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef]
- Cao, Y.; Tang, S.; Nie, X.; Zhou, Z.; Ruan, G.; Han, W.; Zhu, Z.; Ding, C. Decreased miR-214-3p activates NF-κB pathway and aggravates osteoarthritis progression. EBioMedicine 2021, 65, 103283. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.N.; Xu, S.B.; Wu, H.; Dai, B.; Jian, D.D.; Yang, M.; Wu, Y.T.; Feng, Q.; Zhu, J.H.; et al. MicroRNA-214-3p: A link between autophagy and endothelial cell dysfunction in atherosclerosis. Acta Physiol. 2018, 222, e12973. [Google Scholar] [CrossRef]
- Yan, Z.; Zang, B.; Gong, X.; Ren, J.; Wang, R. MiR-214-3p exacerbates kidney damages and inflammation induced by hyperlipidemic pancreatitis complicated with acute renal injury. Life Sci. 2020, 241, 117118. [Google Scholar] [CrossRef] [PubMed]
- Li, J.A.; Wang, Y.D.; Wang, K.; Wang, Z.L.; Jia, D.Y.; Yang, B.Y.; Xiong, C.B. Downregulation of miR-214-3p May Contribute to Pathogenesis of Ulcerative Colitis via Targeting STAT6. Biomed. Res. Int. 2017, 2017, 8524972. [Google Scholar] [CrossRef] [PubMed]
- Marques-Rocha, J.L.; Samblas, M.; Milagro, F.I.; Bressan, J.; Martínez, J.A.; Marti, A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015, 29, 3595–3611. [Google Scholar] [CrossRef] [PubMed]
- Vergadi, E.; Vaporidi, K.; Tsatsanis, C. Regulation of Endotoxin Tolerance and Compensatory Anti-inflammatory Response Syndrome by Non-coding RNAs. Front. Immunol. 2018, 9, 2705. [Google Scholar] [CrossRef]
- Severgnini, M.; Takahashi, S.; Rozo, L.M.; Homer, R.J.; Kuhn, C.; Jhung, J.W.; Perides, G.; Steer, M.; Hassoun, P.M.; Fanburg, B.L.; et al. Activation of the STAT pathway in acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2004, 286, L1282–L1292. [Google Scholar] [CrossRef]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Degboé, Y.; Rauwel, B.; Baron, M.; Boyer, J.F.; Ruyssen-Witrand, A.; Constantin, A.; Davignon, J.L. Polarization of Rheumatoid Macrophages by TNF Targeting Through an IL-10/STAT3 Mechanism. Front. Immunol. 2019, 10, 3. [Google Scholar] [CrossRef]
- Kong, H.; You, N.; Chen, H.; Teng, Y.S.; Liu, Y.G.; Lv, Y.P.; Mao, F.Y.; Cheng, P.; Chen, W.; Zhao, Z.; et al. Helicobacter pylori-induced adrenomedullin modulates IFN-γ-producing T-cell responses and contributes to gastritis. Cell Death Dis. 2020, 11, 189. [Google Scholar] [CrossRef]
- Boyle, M.J.; Jagannathan, P.; Bowen, K.; McIntyre, T.I.; Vance, H.M.; Farrington, L.A.; Greenhouse, B.; Nankya, F.; Rek, J.; Katureebe, A.; et al. Effector Phenotype of Plasmodium falciparum-Specific CD4+ T Cells Is Influenced by Both Age and Transmission Intensity in Naturally Exposed Populations. J. Infect. Dis. 2015, 212, 416–425. [Google Scholar] [CrossRef]
- Gbedande, K.; Carpio, V.H.; Stephens, R. Using two phases of the CD4 T cell response to blood-stage murine malaria to understand regulation of systemic immunity and placental pathology in Plasmodium falciparum infection. Immunol. Rev. 2020, 293, 88–114. [Google Scholar] [CrossRef]
- Leão, L.; Puty, B.; Dolabela, M.F.; Povoa, M.M.; Né, Y.G.S.; Eiró, L.G.; Fagundes, N.C.F.; Maia, L.C.; Lima, R.R. Association of cerebral malaria and TNF-α levels: A systematic review. BMC Infect. Dis. 2020, 20, 442. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Huang, Y.; Zhang, S.; Guo, L.; Wu, R.; Fang, X.; Chen, X.; Xu, H.; Nie, Q. CircDCLRE1C Regulated Lipopolysaccharide-Induced Inflammatory Response and Apoptosis by Regulating miR-214b-3p/STAT3 Pathway in Macrophages. Int. J. Mol. Sci. 2022, 23, 6822. https://doi.org/10.3390/ijms23126822
Xu Y, Huang Y, Zhang S, Guo L, Wu R, Fang X, Chen X, Xu H, Nie Q. CircDCLRE1C Regulated Lipopolysaccharide-Induced Inflammatory Response and Apoptosis by Regulating miR-214b-3p/STAT3 Pathway in Macrophages. International Journal of Molecular Sciences. 2022; 23(12):6822. https://doi.org/10.3390/ijms23126822
Chicago/Turabian StyleXu, Yibin, Yulin Huang, Siyu Zhang, Lijin Guo, Ruiquan Wu, Xiang Fang, Xiaolan Chen, Haiping Xu, and Qinghua Nie. 2022. "CircDCLRE1C Regulated Lipopolysaccharide-Induced Inflammatory Response and Apoptosis by Regulating miR-214b-3p/STAT3 Pathway in Macrophages" International Journal of Molecular Sciences 23, no. 12: 6822. https://doi.org/10.3390/ijms23126822
APA StyleXu, Y., Huang, Y., Zhang, S., Guo, L., Wu, R., Fang, X., Chen, X., Xu, H., & Nie, Q. (2022). CircDCLRE1C Regulated Lipopolysaccharide-Induced Inflammatory Response and Apoptosis by Regulating miR-214b-3p/STAT3 Pathway in Macrophages. International Journal of Molecular Sciences, 23(12), 6822. https://doi.org/10.3390/ijms23126822






