AB186 Inhibits Migration of Triple-Negative Breast Cancer Cells and Interacts with α-Tubulin
Abstract
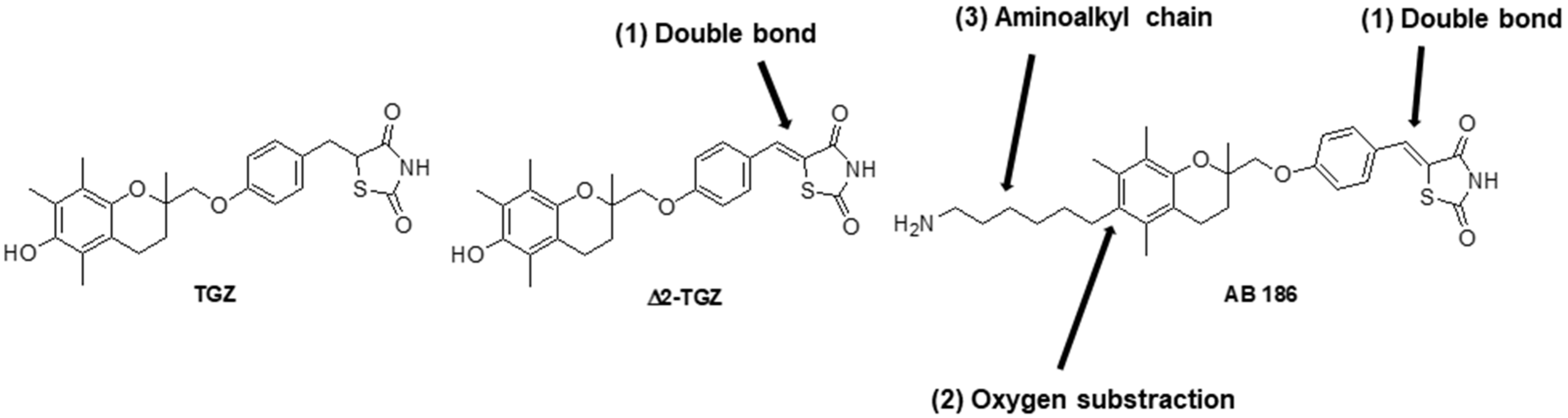
:1. Introduction
2. Results
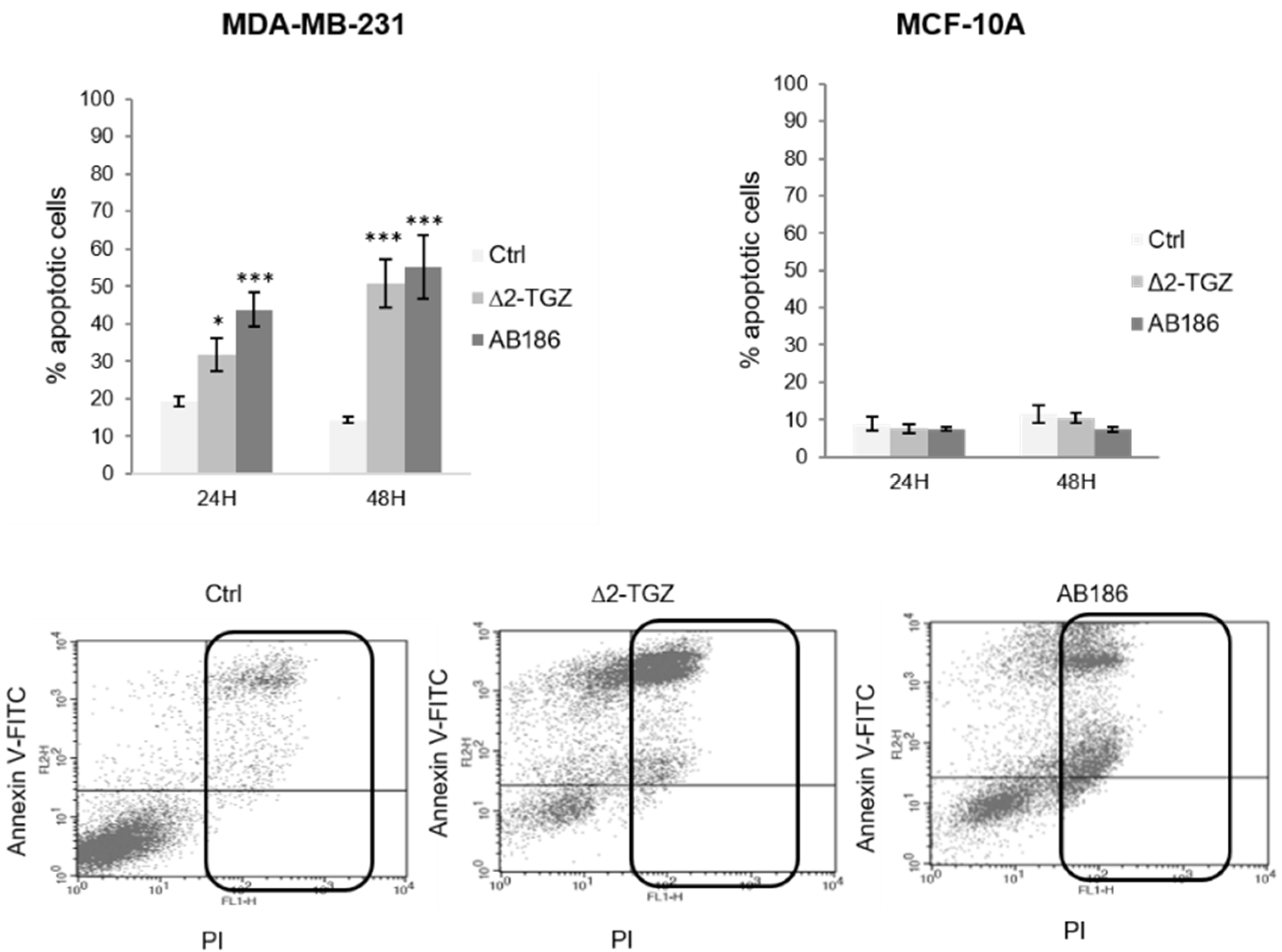
2.1. AB186 Is a More Potent Inducer of Apoptosis Compared to ∆2-TGZ in MDA-MB-231 Breast Cancer Cells and Is Ineffective in the Non-Malignant MCF-10A
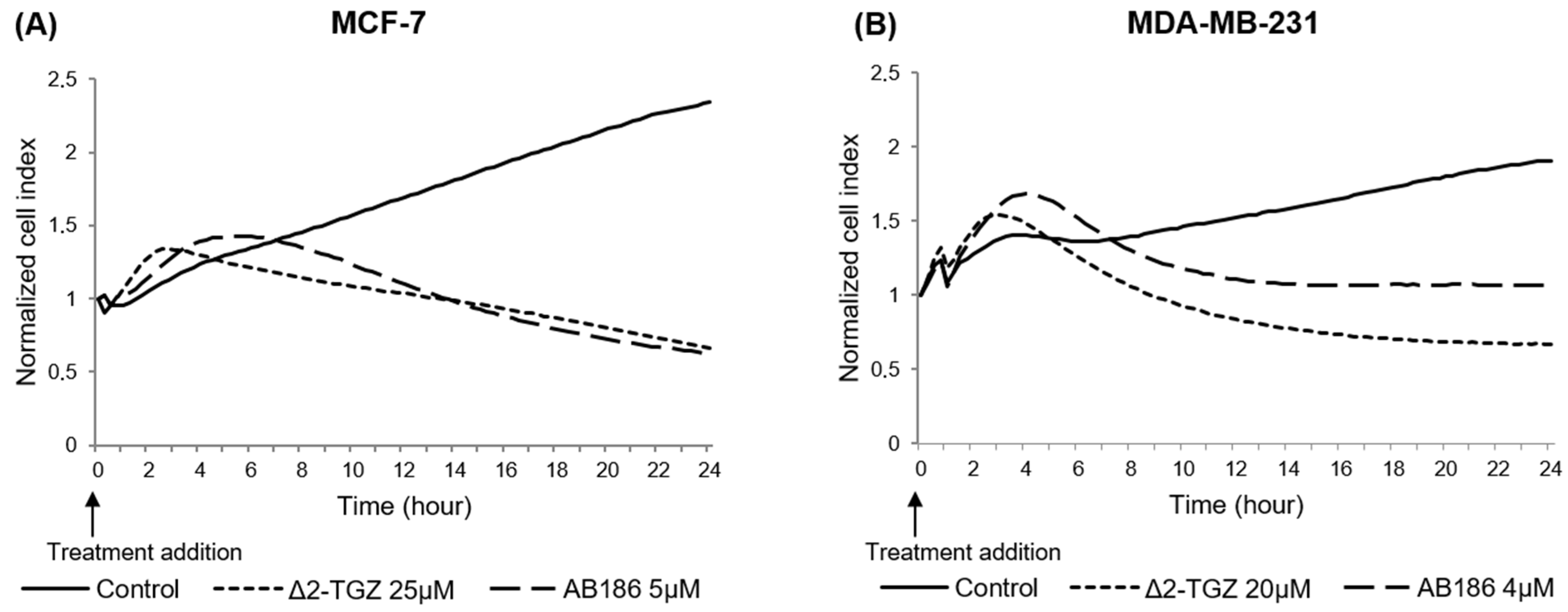
2.2. Variations of Impedance Occur Early after ∆2-TGZ and AB186 Treatment in MCF-7 and MDA-MB-231 Breast Cancer Cells
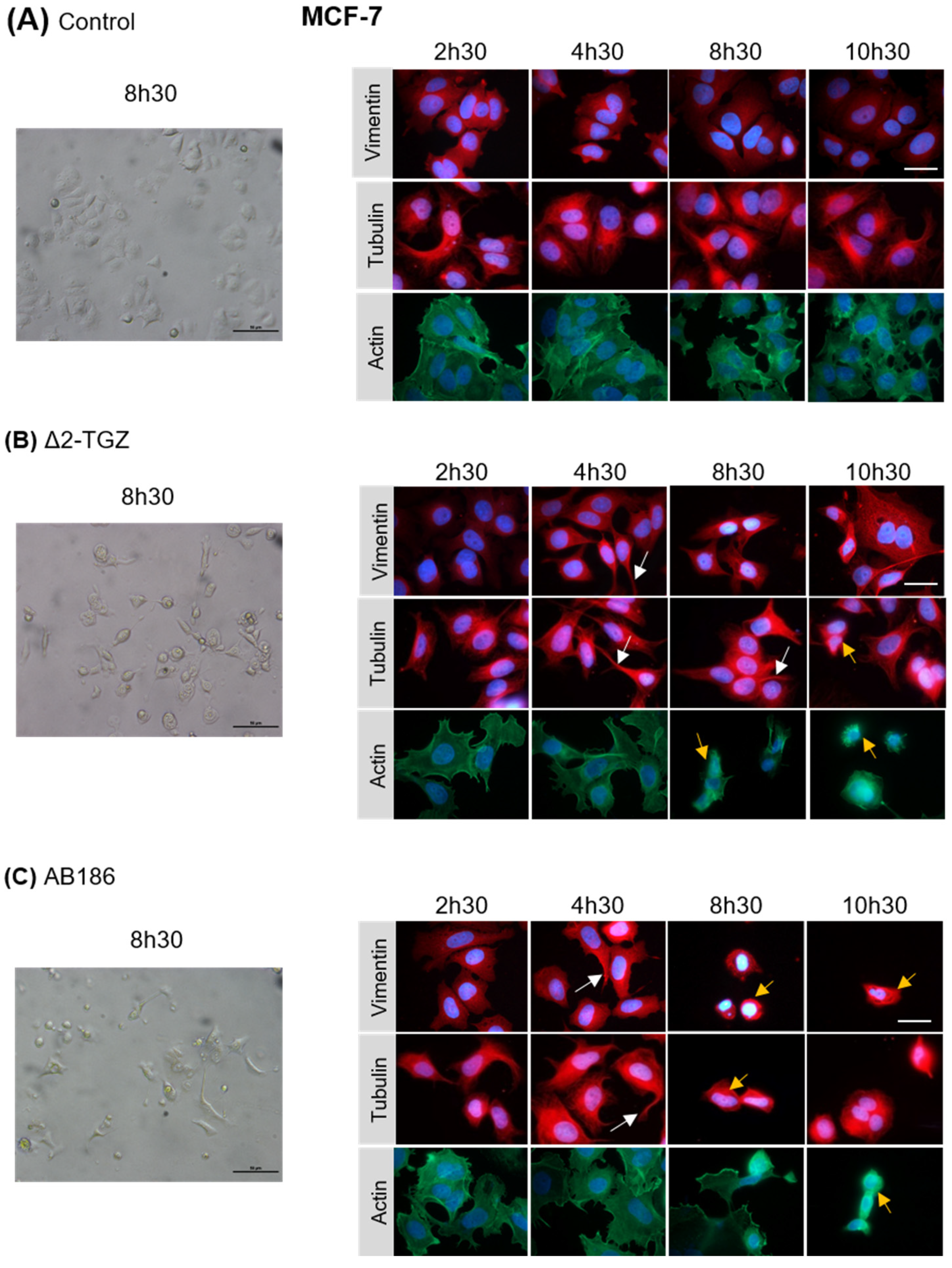
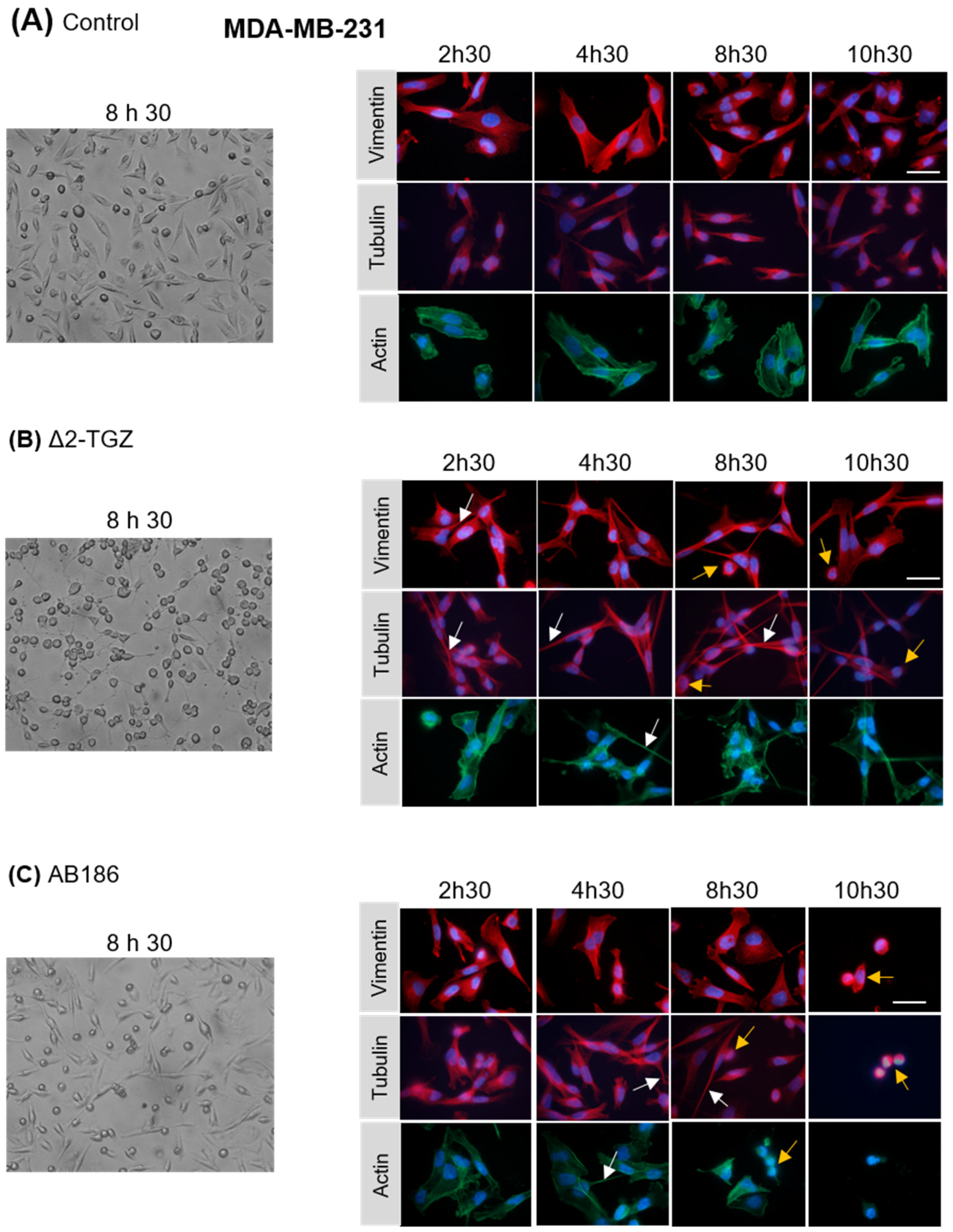
2.3. ∆2-TGZ and AB186 Induce Early Morphological Changes in MCF-7 and MDA-MB-231 Breast Cancer Cells
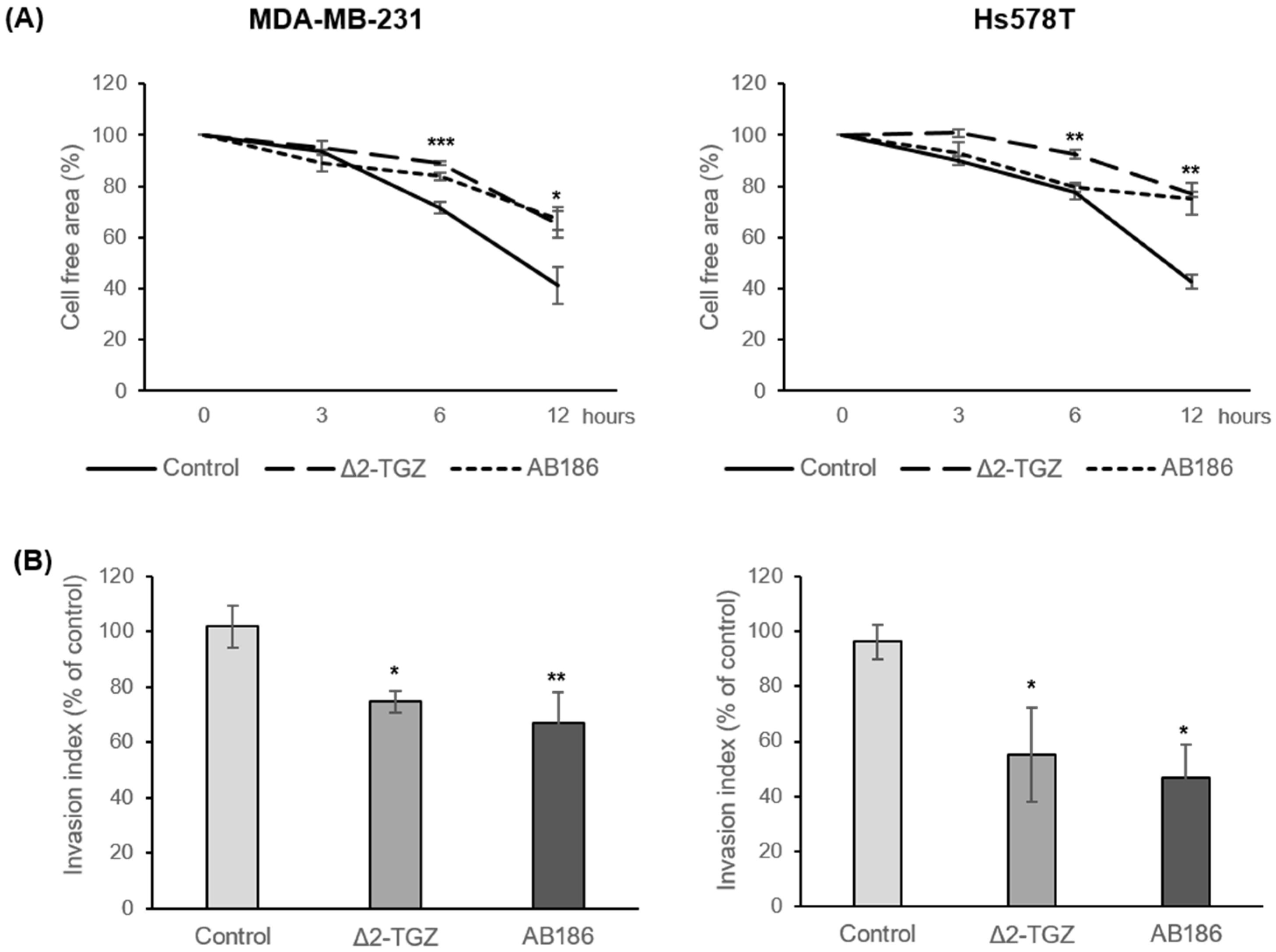
2.4. ∆2-TGZ and AB186 Inhibit Migration Properties of Two Highly Metastatic TNBC Cell Lines: MDA-MB-231 and Hs578T
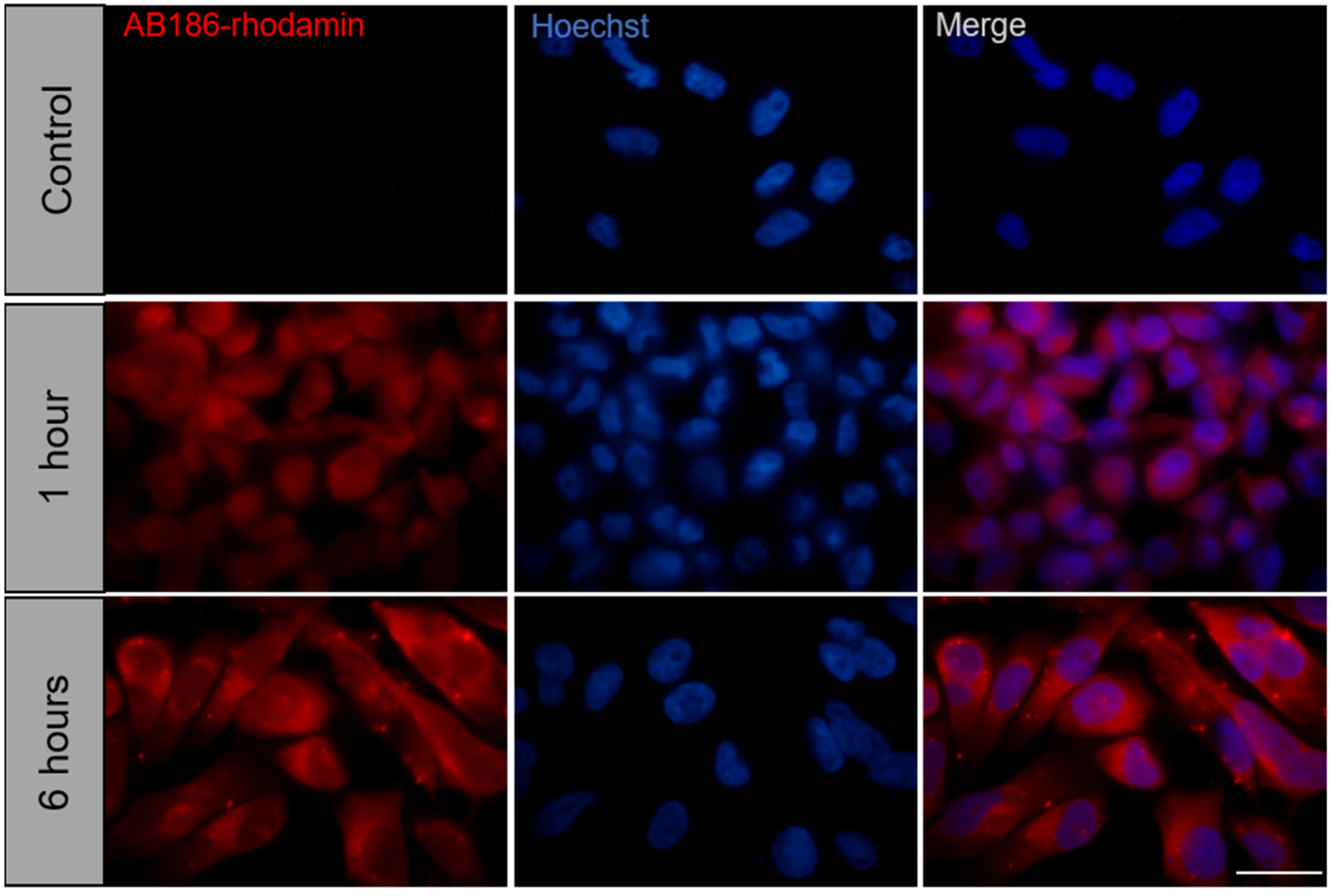
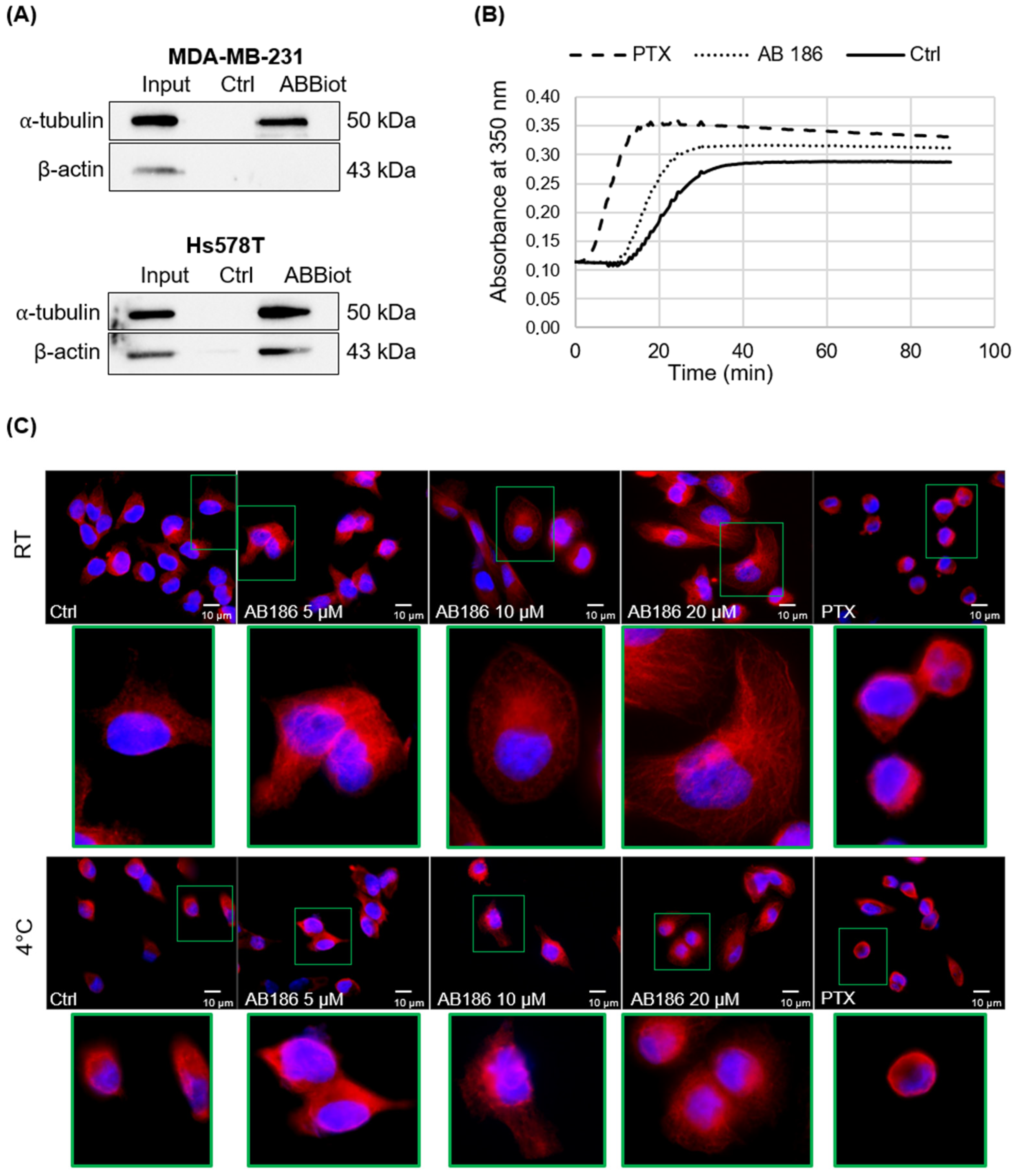
2.5. AB186 Acts through Its Binding to α-Tubulin
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Cell Culture and Treatment
4.2.2. Flow Cytometry
4.2.3. Real-Time Impedance-Based Cell Analysis
4.2.4. Immunofluorescence
4.2.5. Wound Healing Assay
4.2.6. Migration Assay
4.2.7. SPR Binding Analysis
4.2.8. Pull down Assay
4.2.9. Tubulin Polymerization Assay
4.2.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parise, C.A.; Caggiano, V. Breast Cancer Survival Defined by the ER/PR/HER2 Subtypes and a Surrogate Classification according to Tumor Grade and Immunohistochemical Biomarkers. J. Cancer Epidemiol. 2014, 2014, 469251. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 2005, 365, 1687–1717. [Google Scholar] [CrossRef]
- Nahta, R.; Esteva, F.J. HER2 therapy: Molecular mechanisms of trastuzumab resistance. Breast Cancer Res. 2006, 8, 215. [Google Scholar] [CrossRef]
- Abramson, V.G.; Lehmann, B.D.; Ballinger, T.J.; Pietenpol, J.A. Subtyping of triple-negative breast cancer: Implications for therapy. Cancer 2015, 121, 8–16. [Google Scholar] [CrossRef] [Green Version]
- Mazerbourg, S.; Kuntz, S.; Grillier-Vuissoz, I.; Berthe, A.; Geoffroy, M.; Flament, S.; Bordessa, A.; Boisbrun, M. Reprofiling of Troglitazone Towards More Active and Less Toxic Derivatives: A New Hope for Cancer Treatment? Curr. Top. Med. Chem. 2016, 16, 2115–2124. [Google Scholar] [CrossRef]
- Fröhlich, E.; Wahl, R. Chemotherapy and Chemoprevention by Thiazolidinediones. BioMed Res. Int. 2015, 2015, 845340. [Google Scholar] [CrossRef] [Green Version]
- Prost, S.; Relouzat, F.; Spentchian, M.; Ouzegdouh, Y.; Saliba, J.; Massonnet, G.; Beressi, J.-P.; Verhoeyen, E.; Raggueneau, V.; Maneglier, B.; et al. Erosion of the chronic myeloid leukaemia stem cell pool by PPARγ agonists. Nature 2015, 525, 380–383. [Google Scholar] [CrossRef]
- Rousselot, P.; Prost, S.; Guilhot, J.; Roy, L.; Etienne, G.; Legros, L.; Charbonnier, A.; Coiteux, V.; Cony-Makhoul, P.; Huguet, F.; et al. Pioglitazone together with imatinib in chronic myeloid leukemia: A proof of concept study. Cancer 2017, 123, 1791–1799. [Google Scholar] [CrossRef]
- Smallridge, R.C.; Copland, J.A.; Brose, M.S.; Wadsworth, J.T.; Houvras, Y.; Menefee, M.E.; Bible, K.C.; Shah, M.H.; Gramza, A.W.; Klopper, J.P.; et al. Efatutazone, an oral PPAR-γ agonist, in combination with paclitaxel in anaplastic thyroid cancer: Results of a multicenter phase 1 trial. J. Clin. Endocrinol. Metab. 2013, 98, 2392–2400. [Google Scholar] [CrossRef] [Green Version]
- Ishay-Ronen, D.; Diepenbruck, M.; Kalathur, R.K.R.; Sugiyama, N.; Tiede, S.; Ivanek, R.; Bantug, G.; Morini, M.F.; Wang, J.; Hess, C.; et al. Gain Fat—Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis. Cancer Cell 2019, 35, 17–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chojkier, M. Troglitazone and liver injury: In search of answers. Hepatology 2005, 41, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Salamone, S.; Colin, C.; Grillier-Vuissoz, I.; Kuntz, S.; Mazerbourg, S.; Flament, S.; Martin, H.; Richert, L.; Chapleur, Y.; Boisbrun, M. Synthesis of new troglitazone derivatives: Anti-proliferative activity in breast cancer cell lines and preliminary toxicological study. Eur. J. Med. Chem. 2012, 51, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Bordessa, A.; Colin, C.; Grillier-Vuissoz, I.; Kuntz, S.; Mazerbourg, S.; Husson, G.; Vo, M.; Flament, S.; Martin, H.; Chapleur, Y.; et al. Optimization of troglitazone derivatives as potent anti-proliferative agents: Towards more active and less toxic compounds. Eur. J. Med. Chem. 2014, 83, 129–140. [Google Scholar] [CrossRef]
- Huang, J.-W.; Shiau, C.-W.; Yang, J.; Wang, D.-S.; Chiu, H.-C.; Chen, C.-S.; Chen, A.C.-Y. Development of Small-Molecule Cyclin D1-Ablative Agents. J. Med. Chem. 2006, 49, 4684–4689. [Google Scholar] [CrossRef]
- Shiau, C.-W.; Yang, C.-C.; Kulp, S.K.; Chen, K.-F.; Chen, C.-S.; Huang, J.-W.; Chen, C.-S. Thiazolidenediones Mediate Apoptosis in Prostate Cancer Cells in Part through Inhibition of Bcl-xL/Bcl-2 Functions Independently of PPARgamma. Cancer Res. 2005, 65, 1561–1569. [Google Scholar] [CrossRef] [Green Version]
- Lecomte, J.; Flament, S.; Salamone, S.; Boisbrun, M.; Mazerbourg, S.; Chapleur, Y.; Grillier-Vuissoz, I. Disruption of ERalpha signalling pathway by PPARgamma agonists: Evidences of PPARgamma-independent events in two hormone-dependent breast cancer cell lines. Breast Cancer Res. Treat. 2008, 112, 437–451. [Google Scholar] [CrossRef]
- Yang, C.-C.; Ku, C.-Y.; Wei, S.; Shiau, C.-W.; Chen, C.-S.; Pinzone, J.J.; Ringel, M.D.; Chen, C.-S. Peroxisome Proliferator-Activated Receptor gamma-Independent Repression of Prostate-Specific Antigen Expression by Thiazolidinediones in Prostate Cancer Cells. Mol. Pharmacol. 2006, 69, 1564–1570. [Google Scholar] [CrossRef] [Green Version]
- Colin, C.; Salamone, S.; Grillier-Vuissoz, I.; Boisbrun, M.; Kuntz, S.; Lecomte, J.; Chapleur, Y.; Flament, S. New troglitazone derivatives devoid of PPARgamma agonist activity display an increased antiproliferative effect in both hormone-dependent and hormone-independent breast cancer cell lines. Breast Cancer Res. Treat. 2010, 124, 101–110. [Google Scholar] [CrossRef]
- Colin-Cassin, C.; Yao, X.; Cerella, C.; Chbicheb, S.; Kuntz, S.; Mazerbourg, S.; Boisbrun, M.; Chapleur, Y.; Diederich, M.; Flament, S.; et al. PPARγ-inactive Δ2-troglitazone independently triggers ER stress and apoptosis in breast cancer cells. Mol. Carcinog. 2015, 54, 393–404. [Google Scholar] [CrossRef]
- Geoffroy, M.; Kleinclauss, A.; Grandemange, S.; Hupont, S.; Boisbrun, M.; Flament, S.; Kuntz, S. Pro-apoptotic effect of Δ2-TGZ in “claudin-1-low” triple-negative breast cancer cells: Involvement of claudin-1. Breast Cancer Res. Treat. 2017, 165, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Fife, C.M.; McCarroll, J.A.; Kavallaris, M. Movers and shakers: Cells cytoskeleton in cancer metastasis. Br. J. Pharmacol. 2014, 171, 5507–5523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouleau, A.; El Osta, M.; Lucchi, G.; Ducoroy, P.; Boireau, W. Immuno-MALDI-MS in Human Plasma and On-Chip Biomarker Characterizations at the Femtomole Level. Sensors 2012, 12, 15119–15132. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zeng, R.R.; Guo, Q.; Xu, J.; Sun, X.; Wang, L. Identifying a novel anticancer agent with microtubule-stabilizing effects through computational cell-based bioactivity prediction models and bioassays. Org. Biomol. Chem. 2019, 17, 1519–1530. [Google Scholar] [CrossRef]
- Colin, C.; Meyer, M.; Cerella, C.; Kleinclauss, A.; Monard, G.; Boisbrun, M.; Diederich, M.; Flament, S.; Grillier-Vuissoz, I.; Kuntz, S. Biotinylation enhances the anticancer effects of 15d-PGJ2 against breast cancer cells. Int. J. Oncol. 2018, 52, 1991–2000. [Google Scholar] [CrossRef]
- Blanquicett, C.; Roman, J.; Hart, C.M. Thiazolidinediones as anti-cancer agents. Cancer Ther. 2008, 6, 25–34. [Google Scholar]
- Okumura, T.; Motomura, W.; Nagamine, M.; Tanno, S.; Sawamukai, M.; Takahashi, N.; Kohgo, Y. Inhibition of cell invasion and morphological change by troglitazone in human pancreatic cancer cells. J. Gastroenterol. 2004, 39, 461–468. [Google Scholar] [CrossRef]
- Wang, P.; Chou, F.; Porchia, L.; Saji, M.; Pinzone, J.J. Troglitazone inhibits cell migration, adhesion, and spreading by modulating cytoskeletal rearrangement in human breast cancer cells. Mol. Carcinog. 2008, 47, 905–915. [Google Scholar] [CrossRef]
- Yang, Y.-C.; Ho, T.-C.; Chen, S.-L.; Lai, H.-Y.; Wu, J.-Y.; Tsao, Y.-P. Inhibition of cell motility by troglitazone in human ovarian carcinoma cell line. BMC Cancer 2007, 7, 216. [Google Scholar] [CrossRef] [Green Version]
- Folkman, J.; Moscona, A. Role of cell shape in growth control. Nature 1978, 273, 345–349. [Google Scholar] [CrossRef]
- Sun, X.; Shi, X.; Liu, M.; Li, D.; Zhang, L.; Liu, X.; Zhou, J. Mdp3 is a novel microtubule-binding protein that regulates microtubule assembly and stability. Cell Cycle 2011, 10, 3929–3937. [Google Scholar] [CrossRef] [Green Version]
- Russu, W.A. Thiazolidinedione anti-cancer activity: Is inhibition of microtubule assembly implicated? Med. Hypotheses 2007, 68, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Matson, D.R.; Stukenberg, T. Spindle Poisons and Cell Fate: A Tale of Two Pathways. Mol. Interv. 2011, 11, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Lehtimäki, J.; Hakala, M.; Lappalainen, P. Actin Filament Structures in Migrating Cells. Handb. Exp. Pharmacol. 2017, 235, 123–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geoffroy, M.; Kleinclauss, A.; Kuntz, S.; Grillier-Vuissoz, I. Claudin 1 inhibits cell migration and increases intercellular adhesion in triple-negative breast cancer cell line. Mol. Biol. Rep. 2020, 47, 7643–7653. [Google Scholar] [CrossRef]
| Time of CI Maximal Difference | Time of CI below Control | ||||
|---|---|---|---|---|---|
| Cell Line | Experiment | ∆2-TGZ/Control | AB186/Control | ∆2-TGZ | AB186 |
| MCF-7 | 1 | 2 h | 2 h 45 min | 6 h | 4 h 30 min |
| 2 | 1 h 45 min | 4 h | 3 h 30 min | 6 h 45 min | |
| 3 | 2h | 5 h 15 min | 3 h 45 min | 9 h 30 min | |
| Mean ± SD | 1 h 55 min ± 8 min | 4 h ± 75 min | 4 h 25 min ± 82 min | 6 h 55 min ± 150 min | |
| MDA-MB-231 | 1 | 1 h 45 min | 3 h 30 min | 5 h | 7 h 15 min |
| 2 | 1 h 45 min | 4 h | 3 h 45 min | 7 h | |
| 3 | 2 h | 4 h 15 min | 5 h 45 min | 8 h 15 min | |
| Mean ± SD | 1 h 50 min ± 5min | 3 h 55 min ± 13 min | 4 h 50 min ± 60 min | 7 h 30 min ± 40 min | |
| Protein | UniProt ID | # Peptides | Mascot Score | |
|---|---|---|---|---|
| Actin | ACTG1 | P63261 | 8 | 316.4 |
| Tubulin beta chain | TUBB | P07437 | 8 | 144.9 |
| Filamin-A | FLNA | P21333 | 7 | 82.8 |
| Tubulin alpha chain | TUBA1C | Q9BQE3 | 4 | 75.6 |
| Moesin | MSN | P26038 | 4 | 65.9 |
| Talin-1 | TLN1 | Q9Y490 | 8 | 55.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geoffroy, M.; Lemesle, M.; Kleinclauss, A.; Mazerbourg, S.; Batista, L.; Barberi-Heyob, M.; Bastogne, T.; Boireau, W.; Rouleau, A.; Dupommier, D.; et al. AB186 Inhibits Migration of Triple-Negative Breast Cancer Cells and Interacts with α-Tubulin. Int. J. Mol. Sci. 2022, 23, 6859. https://doi.org/10.3390/ijms23126859
Geoffroy M, Lemesle M, Kleinclauss A, Mazerbourg S, Batista L, Barberi-Heyob M, Bastogne T, Boireau W, Rouleau A, Dupommier D, et al. AB186 Inhibits Migration of Triple-Negative Breast Cancer Cells and Interacts with α-Tubulin. International Journal of Molecular Sciences. 2022; 23(12):6859. https://doi.org/10.3390/ijms23126859
Chicago/Turabian StyleGeoffroy, Marine, Marine Lemesle, Alexandra Kleinclauss, Sabine Mazerbourg, Levy Batista, Muriel Barberi-Heyob, Thierry Bastogne, Wilfrid Boireau, Alain Rouleau, Dorian Dupommier, and et al. 2022. "AB186 Inhibits Migration of Triple-Negative Breast Cancer Cells and Interacts with α-Tubulin" International Journal of Molecular Sciences 23, no. 12: 6859. https://doi.org/10.3390/ijms23126859






