Molecular Link between DNA Damage Response and Microtubule Dynamics
Abstract
:1. Introduction
2. Microtubule Organization and Stability in Response to DNA Damage
2.1. NEK2
2.2. Centrobin
2.3. Pericentrin (PCNT)
2.4. CEP Family Proteins
2.5. αTAT1
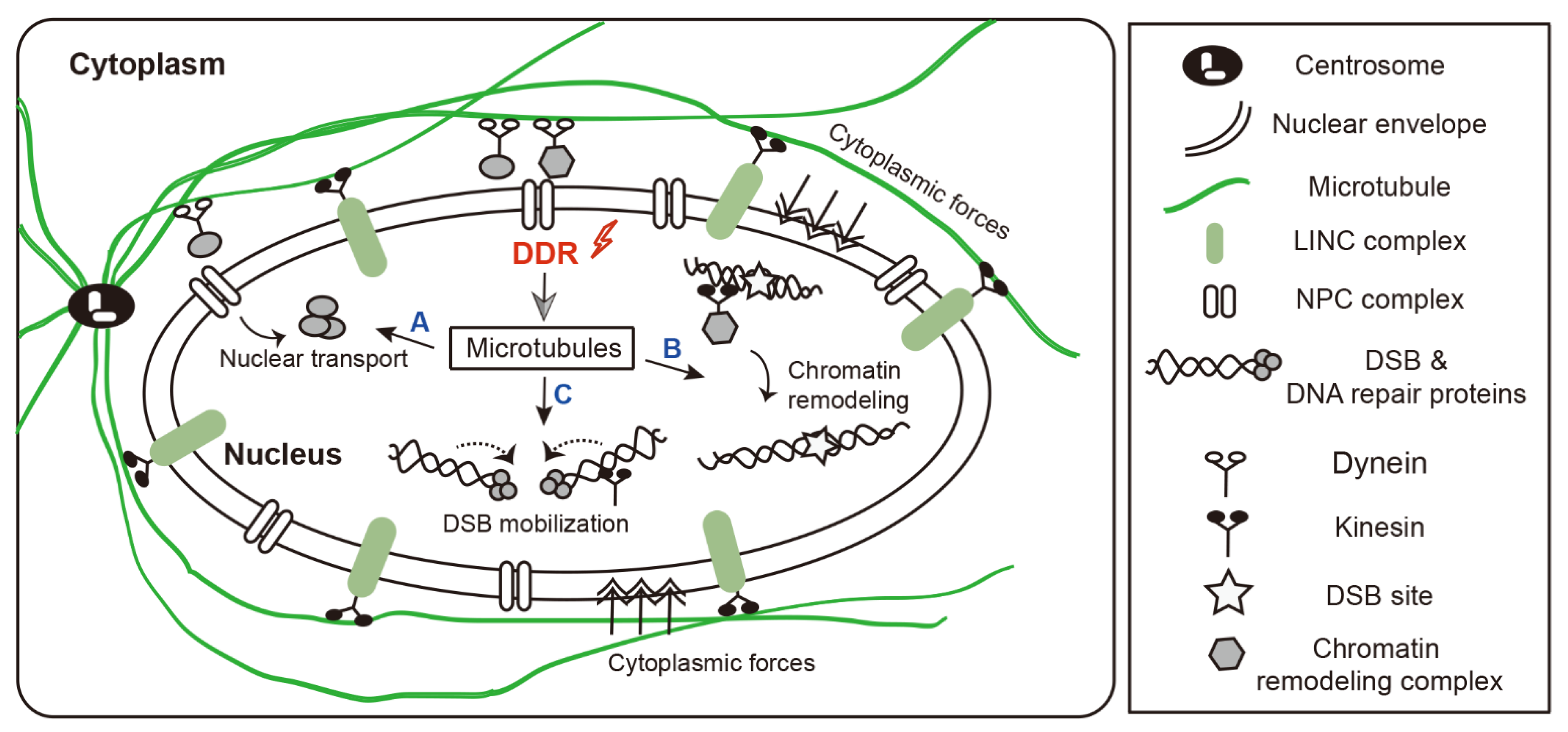
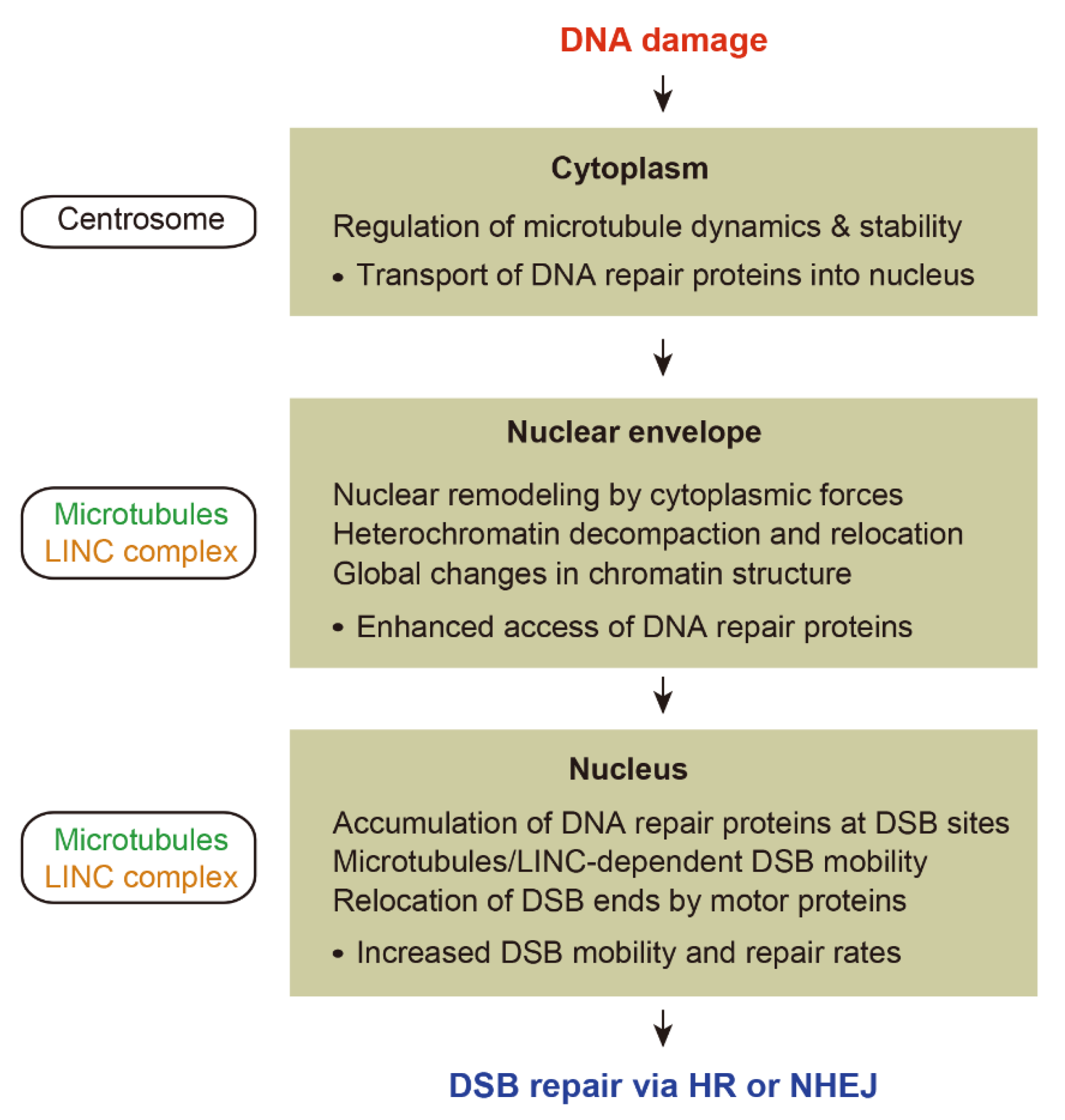
3. Microtubule-Dependent Nuclear Remodeling Following DNA Damage
3.1. Microtubule-Driven Cytoplasmic Forces
3.2. Microtubule and Microtubule-Associated Proteins
3.2.1. γ-Tubulin
3.2.2. KIF4
3.2.3. Actin
4. Microtubule-Dependent Chromatin Mobility Following DNA Damage
4.1. Chromatin Mobility during HR and NHEJ
4.2. KIF2C
4.3. KIF18B
4.4. DNA-PK-AKT
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Conduit, P.T.; Wainman, A.; Raff, J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell. Biol. 2015, 16, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Mitchison, T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kollman, J.M.; Merdes, A.; Mourey, L.; Agard, D.A. Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell. Biol. 2011, 12, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Mandelkow, E.; Mandelkow, E.M. Microtubules and microtubule-associated proteins. Curr. Opin. Cell Biol. 1995, 7, 72–81. [Google Scholar] [CrossRef]
- Gadde, S.; Heald, R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004, 14, R797–R805. [Google Scholar] [CrossRef] [Green Version]
- Petry, S. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 2016, 85, 659–683. [Google Scholar] [CrossRef] [Green Version]
- Gerlitz, G.; Reiner, O.; Bustin, M. Microtubule dynamics alter the interphase nucleus. Cell Mol. Life Sci. 2013, 70, 1255–1268. [Google Scholar] [CrossRef]
- Maizels, Y.; Gerlitz, G. Shaping of interphase chromosomes by the microtubule network. FEBS J. 2015, 282, 3500–3524. [Google Scholar] [CrossRef] [Green Version]
- Shokrollahi, M.; Mekhail, K. Interphase microtubules in nuclear organization and genome maintenance. Trends Cell Biol. 2021, 31, 721–731. [Google Scholar] [CrossRef]
- Graml, V.; Studera, X.; Lawson, J.L.D.; Chessel, A.; Geymonat, M.; Bortfeld-Miller, M.; Walter, T.; Wagstaff, L.; Piddini, E.; Carazo Salas, R.E. A genomic Multiprocess survey of machineries that control and link cell shape, microtubule organization, and cell-cycle progression. Dev. Cell 2014, 31, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Poruchynsky, M.S.; Komlodi-Pasztor, E.; Trostel, S.; Wilkerson, J.; Regairaz, M.; Pommier, Y.; Zhang, X.; Kumar Maity, T.; Robey, R.; Burotto, M.; et al. Microtubule-targeting agents augment the toxicity of DNA-damaging agents by disrupting intracellular trafficking of DNA repair proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 1571–1576. [Google Scholar] [CrossRef] [Green Version]
- Chouinard, G.; Clement, I.; Lafontaine, J.; Rodier, F.; Schmitt, E. Cell cycle-dependent localization of CHK2 at centrosomes during mitosis. Cell Div. 2013, 8, 7. [Google Scholar] [CrossRef] [Green Version]
- Hsu, L.C.; Doan, T.P.; White, R.L. Identification of a gamma-tubulin-binding domain in BRCA1. Cancer Res. 2001, 61, 7713–7718. [Google Scholar]
- Hsu, L.C.; White, R.L. BRCA1 is associated with the centrosome during mitosis. Proc. Natl. Acad. Sci. USA 1998, 95, 12983–12988. [Google Scholar] [CrossRef] [Green Version]
- Kramer, A.; Mailand, N.; Lukas, C.; Syljuasen, R.G.; Wilkinson, C.J.; Nigg, E.A.; Bartek, J.; Lukas, J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat. Cell Biol. 2004, 6, 884–891. [Google Scholar] [CrossRef]
- Shimada, M.; Komatsu, K. Emerging connection between centrosome and DNA repair machinery. J. Radiat. Res. 2009, 50, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Starita, L.M.; Machida, Y.; Sankaran, S.; Elias, J.E.; Griffin, K.; Schlegel, B.P.; Gygi, S.P.; Parvin, J.D. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol. Cell. Biol. 2004, 24, 8457–8466. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Hemmerich, P.; Grosse, F. Centrosomal localization of DNA damage checkpoint proteins. J. Cell. Biochem. 2007, 101, 451–465. [Google Scholar] [CrossRef]
- Fry, A.M.; Meraldi, P.; Nigg, E.A. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998, 17, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Fry, A.M.; O’Regan, L.; Sabir, S.R.; Bayliss, R. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 2012, 125, 4423–4433. [Google Scholar] [CrossRef] [Green Version]
- Moniz, L.; Dutt, P.; Haider, N.; Stambolic, V. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Fry, A.M.; Schultz, S.J.; Bartek, J.; Nigg, E.A. Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J. Biol. Chem. 1995, 270, 12899–12905. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, L.; Cerniglia, G.J.; Nigg, E.A.; Yend, T.J.; Muschel, R.J. Inhibition of centrosome separation after DNA damage: A role for Nek2. Radiat. Res. 2004, 162, 128–135. [Google Scholar] [CrossRef]
- Andreassen, P.R.; Lacroix, F.B.; Villa-Moruzzi, E.; Margolis, R.L. Differential subcellular localization of protein phosphatase-1 alpha, gamma1, and delta isoforms during both interphase and mitosis in mammalian cells. J. Cell Biol. 1998, 141, 1207–1215. [Google Scholar] [CrossRef] [Green Version]
- Mi, J.; Guo, C.; Brautigan, D.L.; Larner, J.M. Protein phosphatase-1alpha regulates centrosome splitting through Nek2. Cancer Res. 2007, 67, 1082–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, Y.; Lee, J.; Kim, K.; Yoo, J.C.; Rhee, K. Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J. Cell Sci. 2007, 120, 2106–2116. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.; Li, J.; Bai, Y.; Gunning, W.T.; Wazer, D.E.; Band, V.; Gao, Q. Centrobin: A novel daughter centriole-associated protein that is required for centriole duplication. J. Cell Biol. 2005, 171, 437–445. [Google Scholar] [CrossRef] [Green Version]
- Gudi, R.; Zou, C.; Li, J.; Gao, Q. Centrobin-tubulin interaction is required for centriole elongation and stability. J. Cell Biol. 2011, 193, 711–725. [Google Scholar] [CrossRef] [Green Version]
- Jeffery, J.M.; Urquhart, A.J.; Subramaniam, V.N.; Parton, R.G.; Khanna, K.K. Centrobin regulates the assembly of functional mitotic spindles. Oncogene 2010, 29, 2649–2658. [Google Scholar] [CrossRef] [Green Version]
- Shin, W.; Yu, N.K.; Kaang, B.K.; Rhee, K. The microtubule nucleation activity of centrobin in both the centrosome and cytoplasm. Cell Cycle 2015, 14, 1925–1931. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Jeong, Y.; Jeong, S.; Rhee, K. Centrobin/NIP2 is a microtubule stabilizer whose activity is enhanced by PLK1 phosphorylation during mitosis. J. Biol. Chem. 2010, 285, 25476–25484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Rhee, K. NEK2 phosphorylation antagonizes the microtubule stabilizing activity of centrobin. Biochem. Biophys. Res. Commun. 2013, 431, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Ryu, N.M.; Kim, J.M. Centrobin plays a role in the cellular response to DNA damage. Cell Cycle 2019, 18, 2660–2671. [Google Scholar] [CrossRef] [PubMed]
- Delaval, B.; Doxsey, S.J. Pericentrin in cellular function and disease. J. Cell Biol. 2010, 188, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Doxsey, S.J.; Stein, P.; Evans, L.; Calarco, P.D.; Kirschner, M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 1994, 76, 639–650. [Google Scholar] [CrossRef]
- Griffith, E.; Walker, S.; Martin, C.A.; Vagnarelli, P.; Stiff, T.; Vernay, B.; Al Sanna, N.; Saggar, A.; Hamel, B.; Earnshaw, W.C.; et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat. Genet. 2008, 40, 232–236. [Google Scholar] [CrossRef] [Green Version]
- Tibelius, A.; Marhold, J.; Zentgraf, H.; Heilig, C.E.; Neitzel, H.; Ducommun, B.; Rauch, A.; Ho, A.D.; Bartek, J.; Kramer, A. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J. Cell Biol. 2009, 185, 1149–1157. [Google Scholar] [CrossRef] [Green Version]
- Purohit, A.; Tynan, S.H.; Vallee, R.; Doxsey, S.J. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 1999, 147, 481–492. [Google Scholar] [CrossRef]
- Zimmerman, W.C.; Sillibourne, J.; Rosa, J.; Doxsey, S.J. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 2004, 15, 3642–3657. [Google Scholar] [CrossRef]
- Alderton, G.K.; Joenje, H.; Varon, R.; Borglum, A.D.; Jeggo, P.A.; O’Driscoll, M. Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum. Mol. Genet. 2004, 13, 3127–3138. [Google Scholar] [CrossRef]
- O’Driscoll, M.; Ruiz-Perez, V.L.; Woods, C.G.; Jeggo, P.A.; Goodship, J.A. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003, 33, 497–501. [Google Scholar] [CrossRef] [Green Version]
- Rauch, A.; Thiel, C.T.; Schindler, D.; Wick, U.; Crow, Y.J.; Ekici, A.B.; van Essen, A.J.; Goecke, T.O.; Al-Gazali, L.; Chrzanowska, K.H.; et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 2008, 319, 816–819. [Google Scholar] [CrossRef]
- Sillibourne, J.E.; Delaval, B.; Redick, S.; Sinha, M.; Doxsey, S.J. Chromatin remodeling proteins interact with pericentrin to regulate centrosome integrity. Mol. Biol. Cell 2007, 18, 3667–3680. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.; Dejsuphong, D.; Balestrini, A.; Hampel, M.; Lenz, C.; Takeda, S.; Vindigni, A.; Costanzo, V. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat. Cell Biol. 2009, 11, 278–285. [Google Scholar] [CrossRef] [Green Version]
- Sir, J.H.; Barr, A.R.; Nicholas, A.K.; Carvalho, O.P.; Khurshid, M.; Sossick, A.; Reichelt, S.; D’Santos, C.; Woods, C.G.; Gergely, F. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 2011, 43, 1147–1153. [Google Scholar] [CrossRef] [Green Version]
- Kalay, E.; Yigit, G.; Aslan, Y.; Brown, K.E.; Pohl, E.; Bicknell, L.S.; Kayserili, H.; Li, Y.; Tuysuz, B.; Nurnberg, G.; et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat. Genet. 2011, 43, 23–26. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.J.; Marjanovic, M.; Luders, J.; Stracker, T.H.; Costanzo, V. Cep63 and cep152 cooperate to ensure centriole duplication. PLoS ONE 2013, 8, e69986. [Google Scholar] [CrossRef] [Green Version]
- Sivasubramaniam, S.; Sun, X.; Pan, Y.R.; Wang, S.; Lee, E.Y. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 2008, 22, 587–600. [Google Scholar] [CrossRef] [Green Version]
- Akella, J.S.; Wloga, D.; Kim, J.; Starostina, N.G.; Lyons-Abbott, S.; Morrissette, N.S.; Dougan, S.T.; Kipreos, E.T.; Gaertig, J. MEC-17 is an alpha-tubulin acetyltransferase. Nature 2010, 467, 218–222. [Google Scholar] [CrossRef] [Green Version]
- Kalebic, N.; Sorrentino, S.; Perlas, E.; Bolasco, G.; Martinez, C.; Heppenstall, P.A. alphaTAT1 is the major alpha-tubulin acetyltransferase in mice. Nat. Commun. 2013, 4, 1962. [Google Scholar] [CrossRef] [Green Version]
- Janke, C.; Bulinski, J.C. Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat. Rev. Mol. Cell. Biol. 2011, 12, 773–786. [Google Scholar] [CrossRef]
- Janke, C.; Montagnac, G. Causes and Consequences of Microtubule Acetylation. Curr. Biol. 2017, 27, R1287–R1292. [Google Scholar] [CrossRef] [Green Version]
- Ryu, N.M.; Kim, J.M. The role of the alpha-tubulin acetyltransferase alphaTAT1 in the DNA damage response. J. Cell Sci. 2020, 133, jcs246702. [Google Scholar] [CrossRef]
- Magiera, M.M.; Janke, C. Post-translational modifications of tubulin. Curr. Biol. 2014, 24, R351–R354. [Google Scholar] [CrossRef] [Green Version]
- Reed, N.A.; Cai, D.; Blasius, T.L.; Jih, G.T.; Meyhofer, E.; Gaertig, J.; Verhey, K.J. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 2006, 16, 2166–2172. [Google Scholar] [CrossRef] [Green Version]
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.F.; Yao, T.P. HDAC6 is a microtubule-associated deacetylase. Nature 2002, 417, 455–458. [Google Scholar] [CrossRef]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef]
- Zhang, H.; Head, P.E.; Daddacha, W.; Park, S.H.; Li, X.; Pan, Y.; Madden, M.Z.; Duong, D.M.; Xie, M.; Yu, B.; et al. ATRIP Deacetylation by SIRT2 Drives ATR Checkpoint Activation by Promoting Binding to RPA-ssDNA. Cell Rep. 2016, 14, 1435–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Hu, C.; Moses, N.; Haakenson, J.; Xiang, S.; Quan, D.; Fang, B.; Yang, Z.; Bai, W.; Bepler, G.; et al. HDAC6 regulates DNA damage response via deacetylating MLH1. J. Biol. Chem. 2019, 294, 5813–5826. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.; Toseland, C.P. Regulation of Nuclear Mechanics and the Impact on DNA Damage. Int. J. Mol. Sci. 2021, 22, 3178. [Google Scholar] [CrossRef]
- Hauer, M.H.; Gasser, S.M. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017, 31, 2204–2221. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, A.; Cook, A.W.; Gough, R.E.; Schilling, M.; Olszok, N.A.; Brown, I.; Wang, L.; Aaron, J.; Martin-Fernandez, M.L.; Rehfeldt, F.; et al. DNA damage alters nuclear mechanics through chromatin reorganization. Nucleic Acids Res. 2021, 49, 340–353. [Google Scholar] [CrossRef]
- Spichal, M.; Fabre, E. The Emerging Role of the Cytoskeleton in Chromosome Dynamics. Front. Genet. 2017, 8, 60. [Google Scholar] [CrossRef] [Green Version]
- Bolhy, S.; Bouhlel, I.; Dultz, E.; Nayak, T.; Zuccolo, M.; Gatti, X.; Vallee, R.; Ellenberg, J.; Doye, V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J. Cell Biol. 2011, 192, 855–871. [Google Scholar] [CrossRef] [Green Version]
- Zuleger, N.; Robson, M.I.; Schirmer, E.C. The nuclear envelope as a chromatin organizer. Nucleus 2011, 2, 339–349. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.; Worman, H.J.; Gundersen, G.G. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 2015, 208, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Rothballer, A.; Kutay, U. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma 2013, 122, 415–429. [Google Scholar] [CrossRef] [Green Version]
- Starr, D.A.; Fridolfsson, H.N. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 2010, 26, 421–444. [Google Scholar] [CrossRef] [Green Version]
- Tajik, A.; Zhang, Y.; Wei, F.; Sun, J.; Jia, Q.; Zhou, W.; Singh, R.; Khanna, N.; Belmont, A.S.; Wang, N. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 2016, 15, 1287–1296. [Google Scholar] [CrossRef] [Green Version]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef] [Green Version]
- van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef] [Green Version]
- Furusawa, T.; Rochman, M.; Taher, L.; Dimitriadis, E.K.; Nagashima, K.; Anderson, S.; Bustin, M. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 2015, 6, 6138. [Google Scholar] [CrossRef] [Green Version]
- Schreiner, S.M.; Koo, P.K.; Zhao, Y.; Mochrie, S.G.; King, M.C. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 2015, 6, 7159. [Google Scholar] [CrossRef] [Green Version]
- Amaral, N.; Ryu, T.; Li, X.; Chiolo, I. Nuclear Dynamics of Heterochromatin Repair. Trends Genet. 2017, 33, 86–100. [Google Scholar] [CrossRef] [Green Version]
- Chiolo, I.; Minoda, A.; Colmenares, S.U.; Polyzos, A.; Costes, S.V.; Karpen, G.H. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 2011, 144, 732–744. [Google Scholar] [CrossRef] [Green Version]
- Fortuny, A.; Polo, S.E. The response to DNA damage in heterochromatin domains. Chromosoma 2018, 127, 291–300. [Google Scholar] [CrossRef]
- Jakob, B.; Splinter, J.; Conrad, S.; Voss, K.O.; Zink, D.; Durante, M.; Lobrich, M.; Taucher-Scholz, G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011, 39, 6489–6499. [Google Scholar] [CrossRef]
- Janssen, A.; Breuer, G.A.; Brinkman, E.K.; van der Meulen, A.I.; Borden, S.V.; van Steensel, B.; Bindra, R.S.; LaRocque, J.R.; Karpen, G.H. A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes Dev. 2016, 30, 1645–1657. [Google Scholar] [CrossRef] [Green Version]
- Tsouroula, K.; Furst, A.; Rogier, M.; Heyer, V.; Maglott-Roth, A.; Ferrand, A.; Reina-San-Martin, B.; Soutoglou, E. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol. Cell 2016, 63, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Goodarzi, A.A.; Kurka, T.; Jeggo, P.A. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 2011, 18, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Noon, A.T.; Deckbar, D.; Ziv, Y.; Shiloh, Y.; Lobrich, M.; Jeggo, P.A. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 2008, 31, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Papamichos-Chronakis, M.; Watanabe, S.; Rando, O.J.; Peterson, C.L. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 2011, 144, 200–213. [Google Scholar] [CrossRef] [Green Version]
- Park, E.J.; Hur, S.K.; Lee, H.S.; Lee, S.A.; Kwon, J. The human Ino80 binds to microtubule via the E-hook of tubulin: Implications for the role in spindle assembly. Biochem. Biophys. Res. Commun. 2011, 416, 416–420. [Google Scholar] [CrossRef]
- Yokoyama, H.; Gruss, O.J. New mitotic regulators released from chromatin. Front. Oncol. 2013, 3, 308. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, H.; Rybina, S.; Santarella-Mellwig, R.; Mattaj, I.W.; Karsenti, E. ISWI is a RanGTP-dependent MAP required for chromosome segregation. J. Cell Biol. 2009, 187, 813–829. [Google Scholar] [CrossRef] [Green Version]
- Dutcher, S.K. The tubulin fraternity: Alpha to eta. Curr. Opin. Cell Biol. 2001, 13, 49–54. [Google Scholar] [CrossRef]
- Stathatos, G.G.; Dunleavy, J.E.M.; Zenker, J.; O’Bryan, M.K. Delta and epsilon tubulin in mammalian development. Trends Cell Biol. 2021, 31, 774–787. [Google Scholar] [CrossRef]
- Chang, P.; Stearns, T. Delta-tubulin and epsilon-tubulin: Two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2000, 2, 30–35. [Google Scholar] [CrossRef]
- Alvarado-Kristensson, M. Gamma-tubulin as a signal-transducing molecule and meshwork with therapeutic potential. Signal. Transduct. Target. Ther. 2018, 3, 24. [Google Scholar] [CrossRef]
- Vazquez, M.; Cooper, M.T.; Zurita, M.; Kennison, J.A. GammaTub23C interacts genetically with brahma chromatin-remodeling complexes in Drosophila melanogaster. Genetics 2008, 180, 835–843. [Google Scholar] [CrossRef] [Green Version]
- Hoog, G.; Zarrizi, R.; von Stedingk, K.; Jonsson, K.; Alvarado-Kristensson, M. Nuclear localization of gamma-tubulin affects E2F transcriptional activity and S-phase progression. FASEB J. 2011, 25, 3815–3827. [Google Scholar] [CrossRef] [Green Version]
- Harbour, J.W.; Dean, D.C. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000, 14, 2393–2409. [Google Scholar] [CrossRef] [Green Version]
- Brehm, A.; Miska, E.A.; McCance, D.J.; Reid, J.L.; Bannister, A.J.; Kouzarides, T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 1998, 391, 597–601. [Google Scholar] [CrossRef]
- Dunaief, J.L.; Strober, B.E.; Guha, S.; Khavari, P.A.; Alin, K.; Luban, J.; Begemann, M.; Crabtree, G.R.; Goff, S.P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 1994, 79, 119–130. [Google Scholar] [CrossRef]
- Robertson, K.D.; Ait-Si-Ali, S.; Yokochi, T.; Wade, P.A.; Jones, P.L.; Wolffe, A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000, 25, 338–342. [Google Scholar] [CrossRef]
- Lesca, C.; Germanier, M.; Raynaud-Messina, B.; Pichereaux, C.; Etievant, C.; Emond, S.; Burlet-Schiltz, O.; Monsarrat, B.; Wright, M.; Defais, M. DNA damage induce gamma-tubulin-RAD51 nuclear complexes in mammalian cells. Oncogene 2005, 24, 5165–5172. [Google Scholar] [CrossRef] [Green Version]
- Hirokawa, N.; Noda, Y.; Tanaka, Y.; Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell. Biol. 2009, 10, 682–696. [Google Scholar] [CrossRef]
- Miki, H.; Setou, M.; Kaneshiro, K.; Hirokawa, N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA 2001, 98, 7004–7011. [Google Scholar] [CrossRef] [Green Version]
- Mazumdar, M.; Sundareshan, S.; Misteli, T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J. Cell Biol. 2004, 166, 613–620. [Google Scholar] [CrossRef]
- Lee, Y.M.; Lee, S.; Lee, E.; Shin, H.; Hahn, H.; Choi, W.; Kim, W. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem. J. 2001, 360, 549–556. [Google Scholar] [CrossRef]
- Mazumdar, M.; Sung, M.H.; Misteli, T. Chromatin maintenance by a molecular motor protein. Nucleus 2011, 2, 591–600. [Google Scholar] [CrossRef] [Green Version]
- Krishnakumar, R.; Kraus, W.L. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 2010, 39, 8–24. [Google Scholar] [CrossRef] [Green Version]
- Midorikawa, R.; Takei, Y.; Hirokawa, N. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell 2006, 125, 371–383. [Google Scholar] [CrossRef] [Green Version]
- Geiman, T.M.; Sankpal, U.T.; Robertson, A.K.; Chen, Y.; Mazumdar, M.; Heale, J.T.; Schmiesing, J.A.; Kim, W.; Yokomori, K.; Zhao, Y.; et al. Isolation and characterization of a novel DNA methyltransferase complex linking DNMT3B with components of the mitotic chromosome condensation machinery. Nucleic Acids Res. 2004, 32, 2716–2729. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.M.; Kim, W. Association of human kinesin superfamily protein member 4 with BRCA2-associated factor 35. Biochem. J. 2003, 374, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhou, L.; Khidr, L.; Guo, X.E.; Kim, W.; Lee, Y.M.; Krasieva, T.; Chen, P.L. A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle 2008, 7, 2013–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caridi, C.P.; D’Agostino, C.; Ryu, T.; Zapotoczny, G.; Delabaere, L.; Li, X.; Khodaverdian, V.Y.; Amaral, N.; Lin, E.; Rau, A.R.; et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Farrants, A.K. Chromatin remodelling and actin organisation. FEBS Lett. 2008, 582, 2041–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, P.; Shen, X. Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol. 2014, 24, 238–246. [Google Scholar] [CrossRef] [Green Version]
- Treisman, R. Shedding light on nuclear actin dynamics and function. Trends Biochem. Sci. 2013, 38, 376–377. [Google Scholar] [CrossRef]
- Dion, V.; Gasser, S.M. Chromatin movement in the maintenance of genome stability. Cell 2013, 152, 1355–1364. [Google Scholar] [CrossRef] [Green Version]
- Hauer, M.H.; Seeber, A.; Singh, V.; Thierry, R.; Sack, R.; Amitai, A.; Kryzhanovska, M.; Eglinger, J.; Holcman, D.; Owen-Hughes, T.; et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol. 2017, 24, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Herbert, S.; Brion, A.; Arbona, J.M.; Lelek, M.; Veillet, A.; Lelandais, B.; Parmar, J.; Fernandez, F.G.; Almayrac, E.; Khalil, Y.; et al. Chromatin stiffening underlies enhanced locus mobility after DNA damage in budding yeast. EMBO J. 2017, 36, 2595–2608. [Google Scholar] [CrossRef]
- Mine-Hattab, J.; Rothstein, R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 2012, 14, 510–517. [Google Scholar] [CrossRef]
- Bermejo, R.; Capra, T.; Jossen, R.; Colosio, A.; Frattini, C.; Carotenuto, W.; Cocito, A.; Doksani, Y.; Klein, H.; Gomez-Gonzalez, B.; et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 2011, 146, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Neumann, F.R.; Dion, V.; Gehlen, L.R.; Tsai-Pflugfelder, M.; Schmid, R.; Taddei, A.; Gasser, S.M. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev. 2012, 26, 369–383. [Google Scholar] [CrossRef] [Green Version]
- Brandsma, I.; Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Burby, P.E.; Simmons, L.A. Regulation of Cell Division in Bacteria by Monitoring Genome Integrity and DNA Replication Status. J. Bacteriol. 2020, 202, e00408-19. [Google Scholar] [CrossRef]
- Lenhart, J.S.; Schroeder, J.W.; Walsh, B.W.; Simmons, L.A. DNA repair and genome maintenance in Bacillus subtilis. Microbiol Mol. Biol. Rev. 2012, 76, 530–564. [Google Scholar] [CrossRef] [Green Version]
- Butala, M.; Zgur-Bertok, D.; Busby, S.J. The bacterial LexA transcriptional repressor. Cell Mol. Life Sci. 2009, 66, 82–93. [Google Scholar] [CrossRef]
- Puig, J.; Knodlseder, N.; Quera, J.; Algara, M.; Guell, M. DNA Damage Protection for Enhanced Bacterial Survival Under Simulated Low Earth Orbit Environmental Conditions in Escherichia coli. Front. Microbiol. 2021, 12, 789668. [Google Scholar] [CrossRef]
- Almiron, M.; Link, A.J.; Furlong, D.; Kolter, R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992, 6, 2646–2654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, A.R.; Barton, J.K. DNA protection by the bacterial ferritin Dps via DNA charge transport. J. Am. Chem. Soc. 2013, 135, 15726–15729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Holowka, J.; Zakrzewska-Czerwinska, J. Nucleoid Associated Proteins: The Small Organizers That Help to Cope With Stress. Front. Microbiol. 2020, 11, 590. [Google Scholar] [CrossRef] [PubMed]
- Karas, V.O.; Westerlaken, I.; Meyer, A.S. The DNA-Binding Protein from Starved Cells (Dps) Utilizes Dual Functions To Defend Cells against Multiple Stresses. J. Bacteriol. 2015, 197, 3206–3215. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.S.; Bumbaca, D.; Kosman, J.; Setlow, P.; Jedrzejas, M.J. Structure of a protein-DNA complex essential for DNA protection in spores of Bacillus species. Proc. Natl. Acad. Sci. USA 2008, 105, 2806–2811. [Google Scholar] [CrossRef] [Green Version]
- Molan, K.; Zgur Bertok, D. Small Prokaryotic DNA-Binding Proteins Protect Genome Integrity throughout the Life Cycle. Int. J. Mol. Sci. 2022, 23, 4008. [Google Scholar] [CrossRef]
- Pallares, M.C.; Marcuello, C.; Botello-Morte, L.; Gonzalez, A.; Fillat, M.F.; Lostao, A. Sequential binding of FurA from Anabaena sp. PCC 7120 to iron boxes: Exploring regulation at the nanoscale. Biochim. Biophys. Acta 2014, 1844, 623–631. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus subtilis: Their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef]
- Shereda, R.D.; Kozlov, A.G.; Lohman, T.M.; Cox, M.M.; Keck, J.L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 289–318. [Google Scholar] [CrossRef] [Green Version]
- Rudin, N.; Haber, J.E. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol. Cell. Biol. 1988, 8, 3918–3928. [Google Scholar] [CrossRef]
- Weiner, A.; Zauberman, N.; Minsky, A. Recombinational DNA repair in a cellular context: A search for the homology search. Nat. Rev. Microbiol. 2009, 7, 748–755. [Google Scholar] [CrossRef]
- Forget, A.L.; Kowalczykowski, S.C. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature 2012, 482, 423–427. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, K.S.; Tapley, E.C.; Cruz, V.E.; Li, Q.; Aung, K.; Hart, K.C.; Schwartz, T.U.; Starr, D.A.; Engebrecht, J. LINC complexes promote homologous recombination in part through inhibition of nonhomologous end joining. J. Cell Biol. 2016, 215, 801–821. [Google Scholar] [CrossRef] [Green Version]
- Kruhlak, M.J.; Celeste, A.; Dellaire, G.; Fernandez-Capetillo, O.; Muller, W.G.; McNally, J.G.; Bazett-Jones, D.P.; Nussenzweig, A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006, 172, 823–834. [Google Scholar] [CrossRef] [Green Version]
- Nelms, B.E.; Maser, R.S.; MacKay, J.F.; Lagally, M.G.; Petrini, J.H. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 1998, 280, 590–592. [Google Scholar] [CrossRef] [Green Version]
- Soutoglou, E.; Dorn, J.F.; Sengupta, K.; Jasin, M.; Nussenzweig, A.; Ried, T.; Danuser, G.; Misteli, T. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 2007, 9, 675–682. [Google Scholar] [CrossRef] [Green Version]
- Dimitrova, N.; Chen, Y.C.; Spector, D.L.; de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 2008, 456, 524–528. [Google Scholar] [CrossRef]
- Lottersberger, F.; Karssemeijer, R.A.; Dimitrova, N.; de Lange, T. 53BP1 and the LINC Complex Promote Microtubule-Dependent DSB Mobility and DNA Repair. Cell 2015, 163, 880–893. [Google Scholar] [CrossRef] [Green Version]
- Lawrimore, J.; Barry, T.M.; Barry, R.M.; York, A.C.; Friedman, B.; Cook, D.M.; Akialis, K.; Tyler, J.; Vasquez, P.; Yeh, E.; et al. Microtubule dynamics drive enhanced chromatin motion and mobilize telomeres in response to DNA damage. Mol. Biol. Cell 2017, 28, 1701–1711. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Thompson, S.L.; Manning, A.L.; Compton, D.A. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 2009, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Gwon, M.R.; Cho, J.H.; Kim, J.R. Mitotic centromere-associated kinase (MCAK/Kif2C) regulates cellular senescence in human primary cells through a p53-dependent pathway. FEBS Lett. 2012, 586, 4148–4156. [Google Scholar] [CrossRef] [Green Version]
- Manning, A.L.; Ganem, N.J.; Bakhoum, S.F.; Wagenbach, M.; Wordeman, L.; Compton, D.A. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell 2007, 18, 2970–2979. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Paydar, M.; Wang, F.; Li, Y.; Wang, L.; Barrette, B.; Bessho, T.; Kwok, B.H.; Peng, A. Kinesin Kif2C in regulation of DNA double strand break dynamics and repair. eLife 2020, 9, e53402. [Google Scholar] [CrossRef]
- Lee, Y.M.; Kim, E.; Park, M.; Moon, E.; Ahn, S.M.; Kim, W.; Hwang, K.B.; Kim, Y.K.; Choi, W.; Kim, W. Cell cycle-regulated expression and subcellular localization of a kinesin-8 member human KIF18B. Gene 2010, 466, 16–25. [Google Scholar] [CrossRef]
- Luessing, J.; Sakhteh, M.; Sarai, N.; Frizzell, L.; Tsanov, N.; Ramberg, K.O.; Maretto, S.; Crowley, P.B.; Lowndes, N.F. The nuclear kinesin KIF18B promotes 53BP1-mediated DNA double-strand break repair. Cell Rep. 2021, 35, 109306. [Google Scholar] [CrossRef]
- Ma, S.; Rong, Z.; Liu, C.; Qin, X.; Zhang, X.; Chen, Q. DNA damage promotes microtubule dynamics through a DNA-PK-AKT axis for enhanced repair. J. Cell Biol. 2021, 220, e201911025. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.N.; Zheng, L.L.; Wang, D.; Liang, X.X.; Gao, F.; Zhou, X.L. Recent advances in microtubule-stabilizing agents. Eur. J. Med. Chem. 2018, 143, 806–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinmetz, M.O.; Prota, A.E. Microtubule-Targeting Agents: Strategies To Hijack the Cytoskeleton. Trends Cell Biol. 2018, 28, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Haschka, M.; Karbon, G.; Fava, L.L.; Villunger, A. Perturbing mitosis for anti-cancer therapy: Is cell death the only answer? EMBO Rep. 2018, 19, e45440. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.J.; Pineda, J.; Shi, J.; Florian, S. Is inflammatory micronucleation the key to a successful anti-mitotic cancer drug? Open Biol. 2017, 7, 170182. [Google Scholar] [CrossRef] [Green Version]
- Korman, A.J.; Peggs, K.S.; Allison, J.P. Checkpoint blockade in cancer immunotherapy. Adv. Immunol. 2006, 90, 297–339. [Google Scholar] [CrossRef] [Green Version]
- Zitvogel, L.; Galluzzi, L.; Smyth, M.J.; Kroemer, G. Mechanism of action of conventional and targeted anticancer therapies: Reinstating immunosurveillance. Immunity 2013, 39, 74–88. [Google Scholar] [CrossRef] [Green Version]
- Paz-Ares, L.; Luft, A.; Vicente, D.; Tafreshi, A.; Gumus, M.; Mazieres, J.; Hermes, B.; Cay Senler, F.; Csoszi, T.; Fulop, A.; et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2040–2051. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Socinski, M.A.; Jotte, R.M.; Cappuzzo, F.; Orlandi, F.; Stroyakovskiy, D.; Nogami, N.; Rodriguez-Abreu, D.; Moro-Sibilot, D.; Thomas, C.A.; Barlesi, F.; et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N. Engl. J. Med. 2018, 378, 2288–2301. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.M. Molecular Link between DNA Damage Response and Microtubule Dynamics. Int. J. Mol. Sci. 2022, 23, 6986. https://doi.org/10.3390/ijms23136986
Kim JM. Molecular Link between DNA Damage Response and Microtubule Dynamics. International Journal of Molecular Sciences. 2022; 23(13):6986. https://doi.org/10.3390/ijms23136986
Chicago/Turabian StyleKim, Jung Min. 2022. "Molecular Link between DNA Damage Response and Microtubule Dynamics" International Journal of Molecular Sciences 23, no. 13: 6986. https://doi.org/10.3390/ijms23136986
APA StyleKim, J. M. (2022). Molecular Link between DNA Damage Response and Microtubule Dynamics. International Journal of Molecular Sciences, 23(13), 6986. https://doi.org/10.3390/ijms23136986




