Human Protein Tyrosine Phosphatase 1B (PTP1B): From Structure to Clinical Inhibitor Perspectives
Abstract
1. Introduction
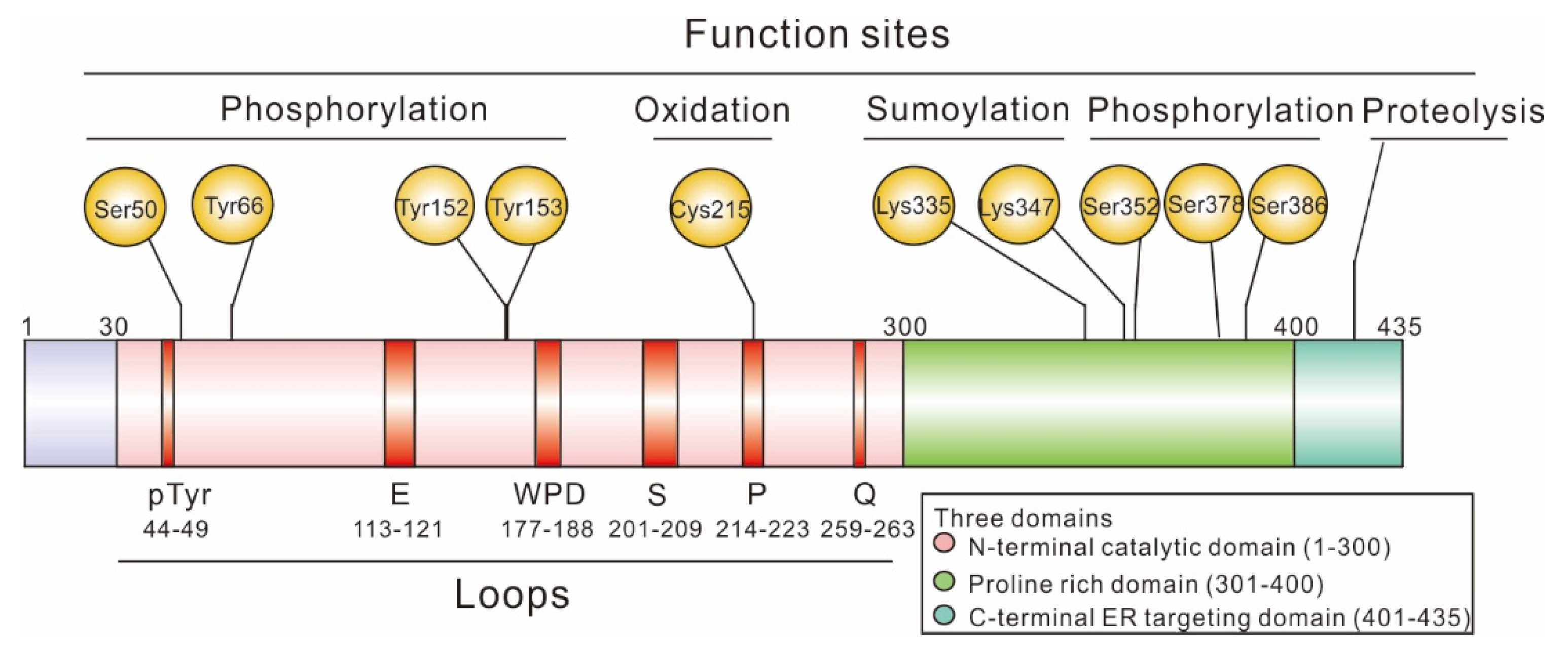
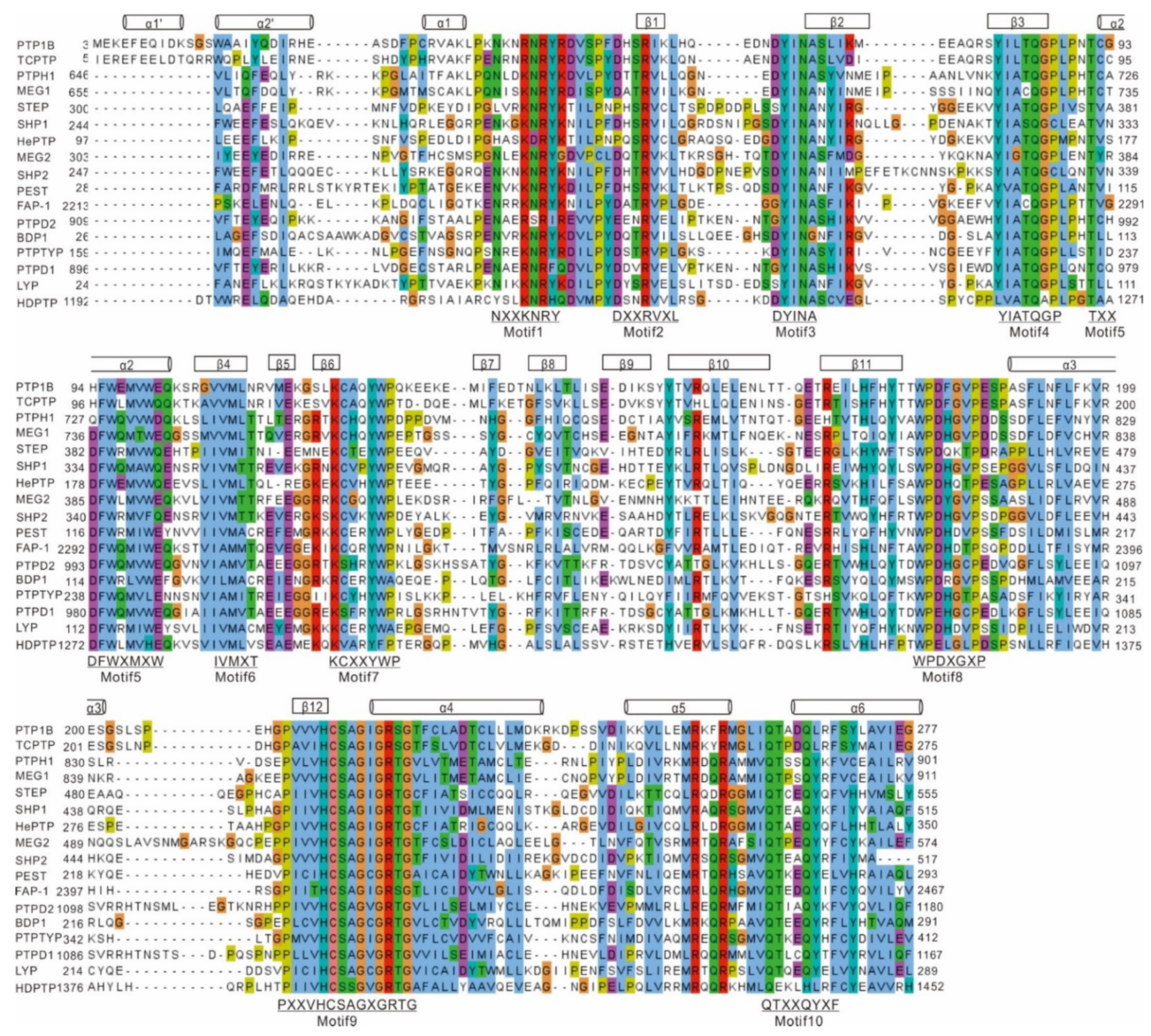
1.1. Structure of PTP1B
1.1.1. N-Terminal Catalytic Domain
1.1.2. Regulatory Domain
1.1.3. C-Terminal Membrane Localization Domain
1.2. Failed PTP1B Inhibitors in Pre-Clinical or Clinical Trials
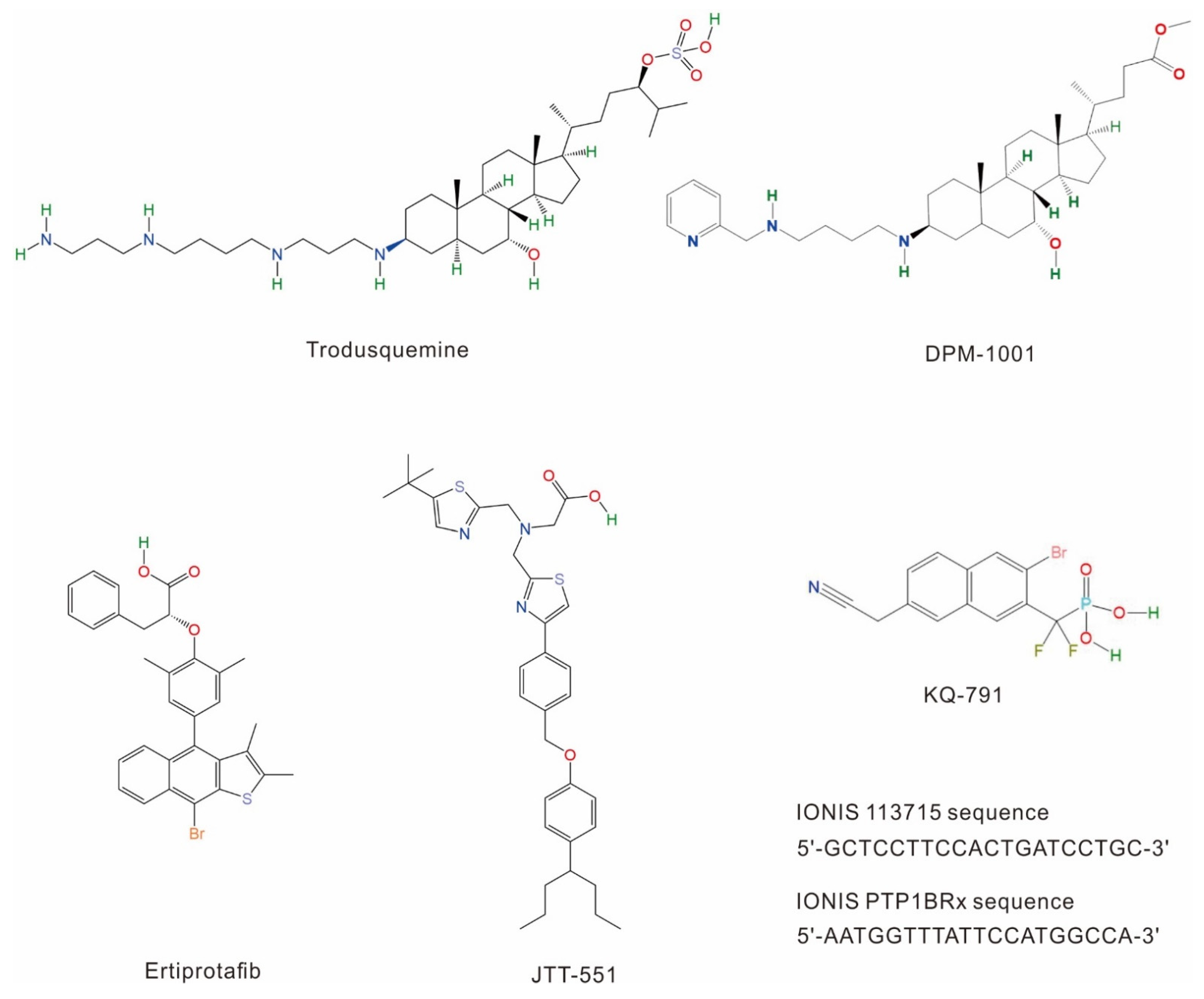
1.2.1. Ertiprotafib
1.2.2. Trodusquemine
1.2.3. JTT-551
1.2.4. Other PTP1B Inhibitors Tested in Clinical Trials
2. Newly Developed Specific PTP1B Inhibitors
2.1. Specific PTP1B Inhibitors Targeting PTP1B A, B, C and D Sites
2.1.1. A Site
2.1.2. B Site
2.1.3. C Site
2.1.4. D Site
2.1.5. Multiple Sites Inhibitors of PTP1B
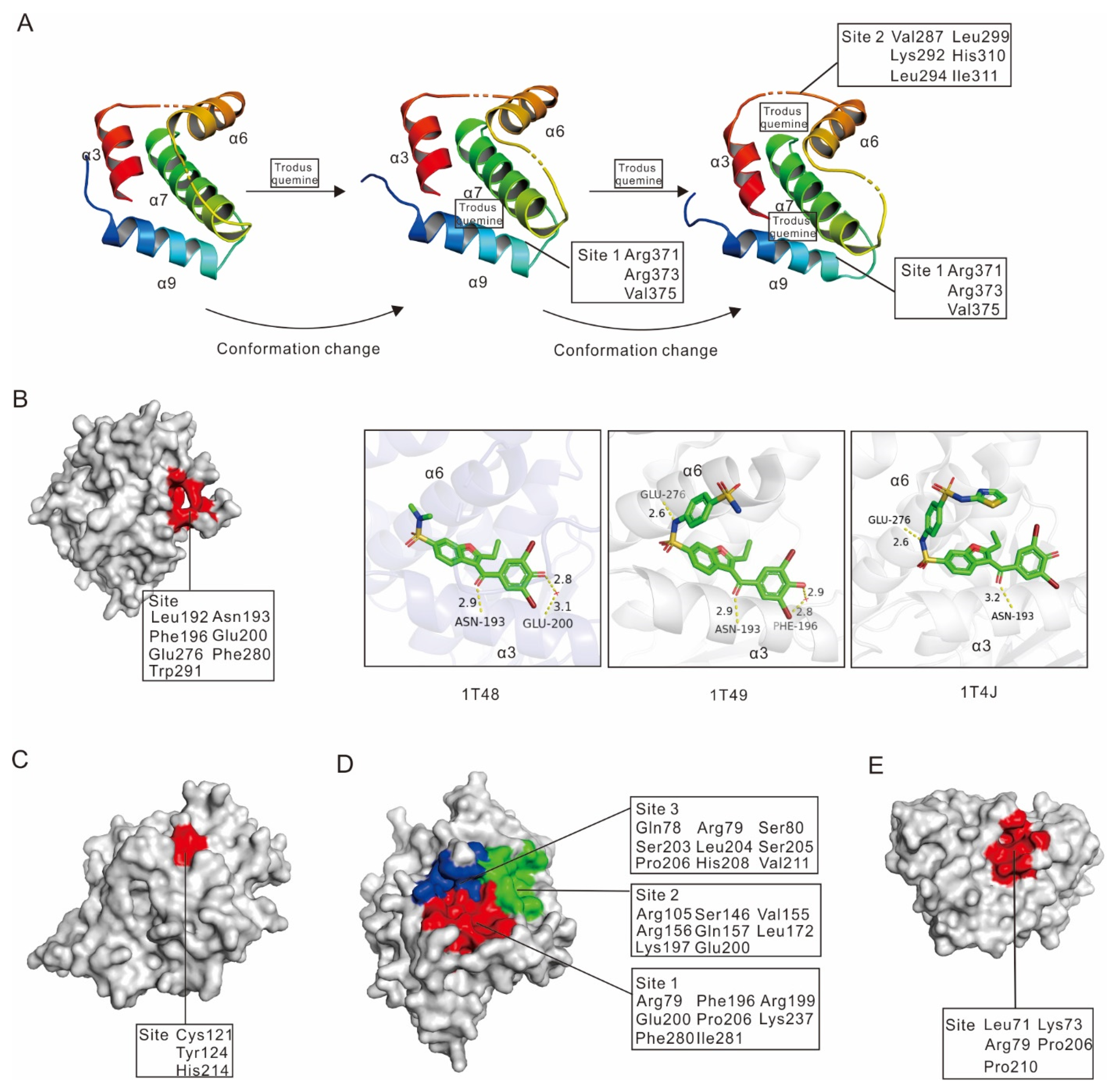
2.2. Allosteric Inhibitors
2.2.1. Allosteric Inhibitors Binding to PTP1B α3-α6-α7 Helices
2.2.2. Allosteric Inhibitors Binding to PTP1B α3–α6–α7–α9 Helices
2.2.3. Allosteric Inhibitors Binding to PTP1B Cys121, Tyr124 and His214 Residues
2.2.4. Allosteric Inhibitors Binding to Three Different Sites on PTP1B
2.2.5. Allosteric Inhibitors Binding to PTP1B Leu71, Lys73 and a Lipophilic Pocket (Arg79, Pro206 and Pro210)
2.3. PTP1B ASOs Inhibitors
3. Further Challenges and Perspectives of PTP1B Inhibitors
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Villamar-Cruz, O.; Loza-Mejía, M.A.; Arias-Romero, L.E.; Camacho-Arroyo, I. Recent advances in PTP1B signaling in metabolism and cancer. Biosci. Rep. 2021, 41, BSR20211994. [Google Scholar] [CrossRef] [PubMed]
- Simoncic, P.D.; McGlade, C.J.; Tremblay, M.L. PTP1B and TC-PTP: Novel roles in immune-cell signalingThis paper is one of a selection of papers published in this Special issue, entitled Second Messengers and Phosphoproteins—12th International Conference. Can. J. Physiol. Pharmacol. 2006, 84, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Pike, K.A.; Tremblay, M.L. TC-PTP and PTP1B: Regulating JAK-STAT signaling, controlling lymphoid malignancies. Cytokine 2016, 82, 52–57. [Google Scholar] [CrossRef]
- Yip, S.-C.; Saha, S.; Chernoff, J. PTP1B: A double agent in metabolism and oncogenesis. Trends Biochem. Sci. 2010, 35, 442–449. [Google Scholar] [CrossRef]
- Haj, F.G.; Zabolotny, J.M.; Kim, Y.B.; Kahn, B.B.; Neel, B.G. Liver-specific protein-tyrosine phosphatase 1B (PTP1B) re-expression alters glucose homeostasis of PTP1B-/-mice. J. Biol. Chem. 2005, 280, 15038–15046. [Google Scholar] [CrossRef]
- Lessard, L.; Stuible, M.; Tremblay, M.L. The two faces of PTP1B in cancer. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 613–619. [Google Scholar] [CrossRef] [PubMed]
- van Montfort, R.L.; Congreve, M.; Tisi, D.; Carr, R.; Jhoti, H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 2003, 423, 773–777. [Google Scholar] [CrossRef]
- Meng, T.-C.; Buckley, D.A.; Galic, S.; Tiganis, T.; Tonks, N.K. Regulation of Insulin Signaling through Reversible Oxidation of the Protein-tyrosine Phosphatases TC45 and PTP1B. J. Biol. Chem. 2004, 279, 37716–37725. [Google Scholar] [CrossRef]
- Dadke, S.; Kusari, A.; Kusari, J. Phosphorylation and activation of protein tyrosine phosphatase (PTP) 1B by insulin receptor. Mol. Cell. Biochem. 2001, 221, 147–154. [Google Scholar] [CrossRef]
- Saha, S.; Chernoff, J. Analysis of PTP1B sumoylation. Methods 2013, 65, 201–206. [Google Scholar] [CrossRef][Green Version]
- Barford, D.; Flint, A.J.; Tonks, N.K. Crystal structure of human protein tyrosine phosphatase 1B. Science 1994, 263, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Tautz, L.; Critton, D.A.; Grotegut, S. Protein Tyrosine Phosphatases: Structure, Function, and Implication in Human Disease. Phosphatase Modul. 2013, 1053, 179–221. [Google Scholar] [CrossRef]
- Ravichandran, L.V.; Chen, H.; Li, Y.; Quon, M.J. Phosphorylation of PTP1B at Ser(50) by Akt impairs its ability to dephosphorylate the insulin receptor. Mol. Endocrinol. 2001, 15, 1768–1780. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, D.; Kusari, A.; Kenner, K.A.; Liu, F.; Chernoff, J.; Gustafson, T.A.; Kusari, J. Protein-Tyrosine Phosphatase 1B Complexes with the Insulin Receptor In Vivo and Is Tyrosine-phosphorylated in the Presence of Insulin. J. Biol. Chem. 1997, 272, 1639–1645. [Google Scholar] [CrossRef]
- Songyang, Z.; E Shoelson, S.; McGlade, J.; Olivier, P.; Pawson, T.; Bustelo, X.R.; Barbacid, M.; Sabe, H.; Hanafusa, H.; Yi, T. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 1994, 14, 2777–2785. [Google Scholar] [CrossRef]
- Cheatham, B.; Kahn, C.R. Insulin Action and the Insulin Signaling Network*. Endocr. Rev. 1995, 16, 117–142. [Google Scholar] [CrossRef]
- Salmeen, A.; Andersen, J.N.; Myers, M.P.; Meng, T.-C.; Hinks, J.A.; Tonks, N.K.; Barford, D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 2003, 423, 769–773. [Google Scholar] [CrossRef]
- Shinde, R.N.; Kumar, G.S.; Eqbal, S.; Sobhia, M.E. Screening and identification of potential PTP1B allosteric inhibitors using in silico and in vitro approaches. PLoS ONE 2018, 13, e0199020. [Google Scholar] [CrossRef]
- Liu, F.; Hill, D.E.; Chernoff, J. Direct Binding of the Proline-rich Region of Protein Tyrosine Phosphatase 1B to the Src Homology 3 Domain of p130Cas. J. Biol. Chem. 1996, 271, 31290–31295. [Google Scholar] [CrossRef]
- Dadke, S.; Chernoff, J. Protein-tyrosine Phosphatase 1B Mediates the Effects of Insulin on the Actin Cytoskeleton in Immortalized Fibroblasts. J. Biol. Chem. 2003, 278, 40607–40611. [Google Scholar] [CrossRef]
- Liu, F.; Chernoff, J. Protein tyrosine phosphatase 1B interacts with and is tyrosine phosphorylated by the epidermal growth factor receptor. Biochem. J. 1997, 327, 139–145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Flint, A.J.; Gebbink, M.F.; Franza, B.R.; E Hill, D.; Tonks, N.K. Multi-site phosphorylation of the protein tyrosine phosphatase, PTP1B: Identification of cell cycle regulated and phorbol ester stimulated sites of phosphorylation. EMBO J. 1993, 12, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Shifrin, V.I.; Davis, R.J.; Neel, B.G. Phosphorylation of Protein-tyrosine Phosphatase PTP-1B on Identical Sites Suggests Activation of a Common Signaling Pathway during Mitosis and Stress Response in Mammalian Cells. J. Biol. Chem. 1997, 272, 2957–2962. [Google Scholar] [CrossRef] [PubMed]
- Dadke, S.; Cotteret, S.; Yip, S.-C.; Jaffer, Z.M.; Haj, F.; Ivanov, A.; Rauscher, F.; Shuai, K.; Ng, T.; Neel, B.G.; et al. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat. Cell Biol. 2006, 9, 80–85. [Google Scholar] [CrossRef]
- Frangioni, J.V.; Oda, A.; Smith, M.; Salzman, E.W.; Neel, B.G. Calpain-catalyzed cleavage and subcellular relocation of protein phosphotyrosine phosphatase 1B (PTP-1B) in human platelets. EMBO J. 1993, 12, 4843–4856. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.; Andrabi, S.S.; Sahr, K.E.; Kamath, L.; Kuliopulos, A.; Chishti, A.H. Disruption of the mouse mu-calpain gene reveals an essential role in platelet function. Mol. Cell. Biol. 2001, 21, 2213–2220. [Google Scholar] [CrossRef]
- Kuchay, S.M.; Kim, N.; Grunz, E.A.; Fay, W.P.; Chishti, A.H. Double Knockouts Reveal that Protein Tyrosine Phosphatase 1B Is a Physiological Target of Calpain-1 in Platelets. Mol. Cell. Biol. 2007, 27, 6038–6052. [Google Scholar] [CrossRef][Green Version]
- Erbe, D.V.; Wang, S.; Zhang, Y.-L.; Harding, K.; Kung, L.; Tam, M.; Stoltz, L.; Xing, Y.; Furey, S.; Qadri, A.; et al. Ertiprotafib Improves Glycemic Control and Lowers Lipids via Multiple Mechanisms. Mol. Pharmacol. 2004, 67, 69–77. [Google Scholar] [CrossRef]
- Zasloff, M.; Williams, J.; Chen, Q.; Anderson, M.; Maeder, T.; Holroyd, K.; Jones, S.; Kinney, W.; Cheshire, K.; McLane, M. A spermine-coupled cholesterol metabolite from the shark with potent appetite suppressant and antidiabetic properties. Int. J. Obes. 2001, 25, 689–697. [Google Scholar] [CrossRef]
- Lantz, K.A.; Hart, S.G.E.; Planey, S.L.; Roitman, M.F.; Ruiz-White, I.A.; Wolfe, H.R.; McLane, M.P. Inhibition of PTP1B by Trodusquemine (MSI-1436) Causes Fat-specific Weight Loss in Diet-induced Obese Mice. Obesity 2010, 18, 1516–1523. [Google Scholar] [CrossRef]
- Fukuda, S.; Ohta, T.; Sakata, S.; Morinaga, H.; Ito, M.; Nakagawa, Y.; Tanaka, M.; Matsushita, M. Pharmacological profiles of a novel protein tyrosine phosphatase 1B inhibitor, JTT-551. Diabetes Obes. Metab. 2010, 12, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Fukuda, S.; Sakata, S.; Morinaga, H.; Ohta, T. Pharmacological effects of JTT-551, a novel protein tyrosine phosphatase 1B inhibitor, in diet-induced obesity mice. J. Diabetes Res. 2014, 2014, 680348. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zheng, M.; Jiang, B.; Li, C.; Guo, S.; Wang, L.; Li, X.; Yu, R.; Shi, D. Antidiabetic activity in vitro and in vivo of BDB, a selective inhibitor of protein tyrosine phosphatase 1B, from Rhodomela confervoides. Br. J. Pharmacol. 2020, 177, 4464–4480. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Magid, A.F. The Inhibitors of Protein Tyrosine Phosphatase Nonreceptor Type 2 (PTPN2) as Potential Enhancers of Cancer Immunotherapy and Type 1 (PTPN1) as Treatment of Metabolic Diseases. ACS Med. Chem. Lett. 2021, 13, 19–21. [Google Scholar] [CrossRef]
- Qian, S.; Zhang, M.; He, Y.; Wang, W.; Liu, S. Recent advances in the development of protein tyrosine phosphatase 1B inhibitors for Type 2 diabetes. Futur. Med. Chem. 2016, 8, 1239–1258. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Lee, S.Y. PTP1B inhibitors as potential therapeutics in the treatment of type 2 diabetes and obesity. Expert Opin. Investig. Drugs 2003, 12, 223–233. [Google Scholar] [CrossRef]
- Matulis, D.; Kranz, J.K.; Salemme, F.R.; Todd, M.J. Thermodynamic Stability of Carbonic Anhydrase: Measurements of Binding Affinity and Stoichiometry Using ThermoFluor. Biochemistry 2005, 44, 5258–5266. [Google Scholar] [CrossRef]
- Weber, P.C.; Salemme, F.R. Applications of calorimetric methods to drug discovery and the study of protein interactions. Curr. Opin. Struct. Biol. 2003, 13, 115–121. [Google Scholar] [CrossRef]
- Shrestha, S.; Bhattarai, B.R.; Cho, H.; Choi, J.K.; Cho, H. PTP1B inhibitor Ertiprotafib is also a potent inhibitor of IkappaB kinase beta (IKK-beta). Bioorganic Med. Chem. Lett. 2007, 17, 2728–2730. [Google Scholar] [CrossRef]
- Krishnan, N.; Koveal, D.; Miller, D.H.; Xue, B.; Akshinthala, S.D.; Kragelj, J.; Jensen, M.R.; Gauss, C.-M.; Page, R.; Blackledge, M.; et al. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat. Chem. Biol. 2014, 10, 558–566. [Google Scholar] [CrossRef]
- Liu, G.; Xin, Z.; Liang, H.; Abad-Zapatero, C.; Hajduk, P.J.; Janowick, D.A.; Szczepankiewicz, B.G.; Pei, Z.; Hutchins, C.W.; Ballaron, S.J.; et al. Selective Protein Tyrosine Phosphatase 1B Inhibitors: Targeting the Second Phosphotyrosine Binding Site with Non-Carboxylic Acid-Containing Ligands. J. Med. Chem. 2003, 46, 3437–3440. [Google Scholar] [CrossRef] [PubMed]
- Bourdeau, A.; Dubé, N.; Tremblay, M.L. Cytoplasmic protein tyrosine phosphatases, regulation and function: The roles of PTP1B and TC-PTP. Curr. Opin. Cell Biol. 2005, 17, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Tamrakar, A.K.; Maurya, C.K.; Rai, A.K. PTP1B inhibitors for type 2 diabetes treatment: A patent review (2011–2014). Expert Opin. Ther. Patents 2014, 24, 1101–1115. [Google Scholar] [CrossRef] [PubMed]
- Cho, H. Protein Tyrosine Phosphatase 1B (PTP1B) and Obesity. Vitam. Horm. 2013, 91, 405–424. [Google Scholar] [CrossRef]
- Tsou, R.C.; Bence, K.K. The Genetics of PTPN1 and Obesity: Insights from Mouse Models of Tissue-Specific PTP1B Deficiency. J. Obes. 2012, 2012, 1–8. [Google Scholar] [CrossRef]
- Dubé, N.; Bourdeau, A.; Heinonen, K.M.; Cheng, A.; Loy, A.L.; Tremblay, M.L. Genetic Ablation of Protein Tyrosine Phosphatase 1B Accelerates Lymphomagenesis of p53-Null Mice through the Regulation of B-Cell Development. Cancer Res. 2005, 65, 10088–10095. [Google Scholar] [CrossRef]
- Le Sommer, S.; Morrice, N.; Pesaresi, M.; Thompson, D.; Vickers, M.A.; Murray, G.I.; Mody, N.; Neel, B.G.; Bence, K.K.; Wilson, H.M.; et al. Deficiency in Protein Tyrosine Phosphatase PTP1B Shortens Lifespan and Leads to Development of Acute Leukemia. Cancer Res. 2018, 78, 75–87. [Google Scholar] [CrossRef]
- Yu, M.; Liu, Z.; Liu, Y.; Zhou, X.; Sun, F.; Liu, Y.; Li, L.; Hua, S.; Zhao, Y.; Gao, H.; et al. PTP 1B markedly promotes breast cancer progression and is regulated by miR-193a-3p. FEBS J. 2018, 286, 1136–1153. [Google Scholar] [CrossRef]
- Franco, M.M.R.; Rodriguez, E.L.; Benitez, B.M.; Rodriguez, L.G.V.; Gonzalez, M.D.L.L.S.; Alonso, A.A. Association of PTP1B with Outcomes of Breast Cancer Patients who Underwent Neoadjuvant Chemotherapy. Breast Cancer Basic Clin. Res. 2016, 10, 177–184. [Google Scholar] [CrossRef]
- You-Ten, K.E.; Muise, E.; Itié, A.; Michaliszyn, E.; Wagner, J.; Jothy, S.; Lapp, W.S.; Tremblay, M.L. Impaired Bone Marrow Microenvironment and Immune Function in T Cell Protein Tyrosine Phosphatase–deficient Mice. J. Exp. Med. 1997, 186, 683–693. [Google Scholar] [CrossRef]
- Galic, S.; Klingler-Hoffmann, M.; Fodero-Tavoletti, M.T.; Puryer, M.A.; Meng, T.-C.; Tonks, N.K.; Tiganis, T. Regulation of Insulin Receptor Signaling by the Protein Tyrosine Phosphatase TCPTP. Mol. Cell. Biol. 2003, 23, 2096–2108. [Google Scholar] [CrossRef] [PubMed]
- Ala, P.J.; Gonneville, L.; Hillman, M.; Becker-Pasha, M.; Yue, E.W.; Douty, B.; Wayland, B.; Polam, P.; Crawley, M.L.; McLaughlin, E.; et al. Structural Insights into the Design of Nonpeptidic Isothiazolidinone-containing Inhibitors of Protein-tyrosine Phosphatase 1B. J. Biol. Chem. 2006, 281, 38013–38021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, Y. The development of protein tyrosine phosphatase1B inhibitors defined by binding sites in crystalline complexes. Futur. Med. Chem. 2018, 10, 2345–2367. [Google Scholar] [CrossRef]
- Koller, E.; A Gaarde, W.; Monia, B.P. Elucidating cell signaling mechanisms using antisense technology. Trends Pharmacol. Sci. 2000, 21, 142–148. [Google Scholar] [CrossRef]
- Liu, G. Protein tyrosine phosphatase 1B inhibition: Opportunities and challenges. Curr. Med. Chem. 2003, 10, 1407–1421. [Google Scholar] [CrossRef]
- Andersen, J.N.; Mortensen, O.H.; Peters, G.H.; Drake, P.G.; Iversen, L.F.; Olsen, O.H.; Jansen, P.G.; Andersen, H.S.; Tonks, N.K.; Møller, N.P.H. Structural and Evolutionary Relationships among Protein Tyrosine Phosphatase Domains. Mol. Cell. Biol. 2001, 21, 7117–7136. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Barford, D.; Flint, A.J.; Tonks, N.K. Structural Basis for Phosphotyrosine Peptide Recognition by Protein Tyrosine Phosphatase 1B. Science 1995, 268, 1754–1758. [Google Scholar] [CrossRef] [PubMed]
- Brandão, T.A.; Hengge, A.C.; Johnson, S.J. Insights into the Reaction of Protein-tyrosine Phosphatase 1B: Crystal Structures for transition state analogs of both catalytic steps. J. Biol. Chem. 2010, 285, 15874–15883. [Google Scholar] [CrossRef]
- Choy, M.S.; Li, Y.; Machado, L.E.; Kunze, M.; Connors, C.R.; Wei, X.; Lindorff-Larsen, K.; Page, R.; Peti, W. Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery. Mol. Cell 2017, 65, 644–658.e5. [Google Scholar] [CrossRef]
- Ruddraraju, K.V.; Zhang, Z.-Y. Covalent inhibition of protein tyrosine phosphatases. Mol. BioSyst. 2017, 13, 1257–1279. [Google Scholar] [CrossRef]
- Pannifer, A.D.B.; Flint, A.J.; Tonks, N.K.; Barford, D. Visualization of the Cysteinyl-phosphate Intermediate of a Protein-tyrosine Phosphatase by X-ray Crystallography. J. Biol. Chem. 1998, 273, 10454–10462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Wang, Y.; Dixon, J.E. Dissecting the catalytic mechanism of protein-tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 1994, 91, 1624–1627. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, L.; Noh, S.J.; Guan, K.-L.; Zhang, Z.-Y. Altering the Nucleophile Specificity of a Protein-tyrosine Phosphatase-catalyzed Reaction. J. Biol. Chem. 1998, 273, 5484–5492. [Google Scholar] [CrossRef] [PubMed]
- Tonks, N.K. PTP1B: From the sidelines to the front lines! FEBS Lett. 2003, 546, 140–148. [Google Scholar] [CrossRef]
- Puius, Y.A.; Zhao, Y.; Sullivan, M.; Lawrence, D.S.; Almo, S.C.; Zhang, Z.-Y. Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: A paradigm for inhibitor design. Proc. Natl. Acad. Sci. USA 1997, 94, 13420–13425. [Google Scholar] [CrossRef] [PubMed]
- Salmeen, A.; Andersen, J.; Myers, M.P.; Tonks, N.K.; Barford, D. Molecular Basis for the Dephosphorylation of the Activation Segment of the Insulin Receptor by Protein Tyrosine Phosphatase 1B. Mol. Cell 2000, 6, 1401–1412. [Google Scholar] [CrossRef]
- Low, J.L.; Chai, C.L.; Yao, S.Q. Bidentate Inhibitors of Protein Tyrosine Phosphatases. Antioxid. Redox Signal. 2014, 20, 2225–2250. [Google Scholar] [CrossRef]
- Chen, X.; Gan, Q.; Feng, C.; Liu, X.; Zhang, Q. Virtual Screening of Novel and Selective Inhibitors of Protein Tyrosine Phosphatase 1B over T-Cell Protein Tyrosine Phosphatase Using a Bidentate Inhibition Strategy. J. Chem. Inf. Model. 2018, 58, 837–847. [Google Scholar] [CrossRef]
- Asante-Appiah, E.; Ball, K.; Bateman, K.; Skorey, K.; Friesen, R.; Desponts, C.; Payette, P.; Bayly, C.; Zamboni, R.; Scapin, G.; et al. The YRD Motif Is a Major Determinant of Substrate and Inhibitor Specificity in T-cell Protein-tyrosine Phosphatase. J. Biol. Chem. 2001, 276, 26036–26043. [Google Scholar] [CrossRef]
- Iversen, L.F.; Andersen, H.S.; Branner, S.; Mortensen, S.B.; Peters, G.H.; Norris, K.; Olsen, O.H.; Jeppesen, C.B.; Lundt, B.F.; Ripka, W.; et al. Structure-based Design of a Low Molecular Weight, Nonphosphorus, Nonpeptide, and Highly Selective Inhibitor of Protein-tyrosine Phosphatase 1B. J. Biol. Chem. 2000, 275, 10300–10307. [Google Scholar] [CrossRef]
- Szczepankiewicz, B.G.; Liu, G.; Hajduk, P.J.; Abad-Zapatero, C.; Pei, Z.; Xin, Z.; Lubben, T.H.; Trevillyan, J.M.; Stashko, M.A.; Ballaron, S.J.; et al. Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Using a Linked-Fragment Strategy. J. Am. Chem. Soc. 2003, 125, 4087–4096. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, L.; Shi, D. The design strategy of selective PTP1B inhibitors over TCPTP. Bioorganic Med. Chem. 2016, 24, 3343–3352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Yu, R.; Xu, Q.; Li, X.; Luo, J.; Jiang, B.; Wang, L.; Guo, S.; Wu, N.; Shi, D. Discovery and evaluation of the hybrid of bromophenol and saccharide as potent and selective protein tyrosine phosphatase 1B inhibitors. Eur. J. Med. Chem. 2017, 134, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Asante-Appiah, E.; Patel, S.; Dufresne, C.; Roy, P.; Wang, Q.; Patel, V.; Friesen, R.W.; Ramachandran, C.; Becker, J.W.; Leblanc, Y.; et al. The Structure of PTP-1B in Complex with a Peptide Inhibitor Reveals an Alternative Binding Mode for Bisphosphonates. Biochemistry 2002, 41, 9043–9051. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Maclean, D.; McNamara, D.J.; Sawyer, T.K.; Dixon, J.E. Protein Tyrosine Phosphatase Substrate Specificity: Size and Phosphotyrosine Positioning Requirements in Peptide Substrates. Biochemistry 1994, 33, 2285–2290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Thieme-Sefler, A.M.; Maclean, D.; McNamara, D.J.; Dobrusin, E.M.; Sawyer, T.K.; E Dixon, J. Substrate specificity of the protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 1993, 90, 4446–4450. [Google Scholar] [CrossRef]
- Bilwes, A.M.; Hertog, J.D.; Hunter, T.; Noel, J.P. Structural basis for inhibition of receptor protein-tyrosine phosphatase-α by dimerization. Nature 1996, 382, 555–559. [Google Scholar] [CrossRef]
- Peters, G.H.; Iversen, L.F.; Branner, S.; Andersen, H.S.; Mortensen, S.B.; Olsen, O.H.; Møller, K.B.; Møller, N.P.H. Residue 259 Is a Key Determinant of Substrate Specificity of Protein-tyrosine Phosphatases 1B and α. J. Biol. Chem. 2000, 275, 18201–18209. [Google Scholar] [CrossRef]
- Buist, A.; Zhang, Y.-L.; Keng, Y.-F.; Wu, L.; Zhang, Z.-Y.; Hertog, J.D. Restoration of Potent Protein−Tyrosine Phosphatase Activity into the Membrane-Distal Domain of Receptor Protein−Tyrosine Phosphatase α. Biochemistry 1998, 38, 914–922. [Google Scholar] [CrossRef]
- Adams, D.R.; Abraham, A.; Asano, J.; Breslin, C.; Dick, C.A.; Ixkes, U.; Johnston, B.F.; Johnston, D.; Kewnay, J.; Takano, Y.; et al. 2-Aryl-3,3,3-trifluoro-2-hydroxypropionic acids: A new class of protein tyrosine phosphatase 1B inhibitors. Bioorganic Med. Chem. Lett. 2007, 17, 6579–6583. [Google Scholar] [CrossRef]
- Xin, Z.; Oost, T.K.; Abad-Zapatero, C.; Hajduk, P.J.; Pei, Z.; Szczepankiewicz, B.G.; Hutchins, C.W.; Ballaron, S.J.; A Stashko, M.; Lubben, T.; et al. Potent, selective inhibitors of protein tyrosine phosphatase 1B. Bioorganic Med. Chem. Lett. 2003, 13, 1887–1890. [Google Scholar] [CrossRef]
- Lund, I.K.; A Hansen, J.; Andersen, H.S.; Møller, N.P.H.; Billestrup, N. Mechanism of protein tyrosine phosphatase 1B-mediated inhibition of leptin signalling. J. Mol. Endocrinol. 2005, 34, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhu, L.; Cao, Y.; Wu, G.; Liu, X.; Chen, Y.; Wang, Q.; Shi, T.; Zhao, Y.; Wang, Y.; et al. ASD: A comprehensive database of allosteric proteins and modulators. Nucleic Acids Res. 2010, 39, D663–D669. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Mou, L.; Shen, Q.; Lu, S.; Li, C.; Liu, X.; Wang, G.; Li, S.; Geng, L.; Liu, Y.; et al. ASD v2.0: Updated content and novel features focusing on allosteric regulation. Nucleic Acids Res. 2013, 42, D510–D516. [Google Scholar] [CrossRef] [PubMed]
- Wiesmann, C.; Barr, K.J.; Kung, J.; Zhu, J.; A Erlanson, D.; Shen, W.; Fahr, B.J.; Zhong, M.; Taylor, L.; Randal, M.; et al. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat. Struct. Mol. Biol. 2004, 11, 730–737. [Google Scholar] [CrossRef]
- Li, S.; Zhang, J.; Lu, S.; Huang, W.; Geng, L.; Shen, Q.; Zhang, J. The Mechanism of Allosteric Inhibition of Protein Tyrosine Phosphatase 1B. PLoS ONE 2014, 9, e97668. [Google Scholar] [CrossRef]
- Kamerlin, S.C.L.; Rucker, R.; Boresch, S. A molecular dynamics study of WPD-loop flexibility in PTP1B. Biochem. Biophys. Res. Commun. 2007, 356, 1011–1016. [Google Scholar] [CrossRef]
- Kamerlin, S.C.L.; Rucker, R.; Boresch, S. A targeted molecular dynamics study of WPD loop movement in PTP1B. Biochem. Biophys. Res. Commun. 2006, 345, 1161–1166. [Google Scholar] [CrossRef]
- Kumar, R.; Shinde, R.N.; Ajay, D.; Sobhia, M.E. Probing Interaction Requirements in PTP1B Inhibitors: A Comparative Molecular Dynamics Study. J. Chem. Inf. Model. 2010, 50, 1147–1158. [Google Scholar] [CrossRef]
- Olmez, E.O.; Alakent, B. Alpha7 Helix Plays an Important Role in the Conformational Stability of PTP1B. J. Biomol. Struct. Dyn. 2011, 28, 675–693. [Google Scholar] [CrossRef]
- Cui, W.; Cheng, Y.-H.; Geng, L.-L.; Liang, D.-S.; Hou, T.-J.; Ji, M.-J. Unraveling the Allosteric Inhibition Mechanism of PTP1B by Free Energy Calculation Based on Umbrella Sampling. J. Chem. Inf. Model. 2013, 53, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Hassan, I.; Aijaz, A.; Ahmad, F. Structural and functional analysis of human prostatic acid phosphatase. Expert Rev. Anticancer Ther. 2010, 10, 1055–1068. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, L.; Tian, J.; Ye, F.; Xiao, Z. Integrated Approach to Identify Selective PTP1B Inhibitors Targeting the Allosteric Site. J. Chem. Inf. Model. 2021, 61, 4720–4732. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.; Konidaris, K.F.; Gasser, G.; Tonks, N.K. A potent, selective, and orally bioavailable inhibitor of the protein-tyrosine phosphatase PTP1B improves insulin and leptin signaling in animal models. J. Biol. Chem. 2018, 293, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.; Felice, C.; Rivera, K.; Pappin, D.J.; Tonks, N.K. DPM-1001 decreased copper levels and ameliorated deficits in a mouse model of Wilson’s disease. Genes Dev. 2018, 32, 944–952. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, L.; Yuan, C.; Pei, K.; Liu, Z.; Guo, M.; Zhu, M. Potent inhibition of protein tyrosine phosphatase 1B by copper complexes: Implications for copper toxicity in biological systems. Chem. Commun. 2010, 46, 3547–3549. [Google Scholar] [CrossRef]
- Hongdusit, A.; Zwart, P.H.; Sankaran, B.; Fox, J.M. Minimally disruptive optical control of protein tyrosine phosphatase 1B. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Hansen, S.K.; Cancilla, M.T.; Shiau, T.P.; Kung, J.; Chen, T.; Erlanson, D.A. Allosteric Inhibition of PTP1B Activity by Selective Modification of a Non-Active Site Cysteine Residue. Biochemistry 2005, 44, 7704–7712. [Google Scholar] [CrossRef]
- Punthasee, P.; Laciak, A.R.; Cummings, A.H.; Ruddraraju, K.V.; Lewis, S.M.; Hillebrand, R.; Singh, H.; Tanner, J.J.; Gates, K.S. Covalent Allosteric Inactivation of Protein Tyrosine Phosphatase 1B (PTP1B) by an Inhibitor–Electrophile Conjugate. Biochemistry 2017, 56, 2051–2060. [Google Scholar] [CrossRef]
- Khan, S.; Bjij, I.; Soliman, M.E.S. Selective Covalent Inhibition of “Allosteric Cys121” Distort the Binding of PTP1B Enzyme: A Novel Therapeutic Approach for Cancer Treatment. Cell Biophys. 2019, 77, 203–211. [Google Scholar] [CrossRef]
- Kumar, A.P.; Nguyen, M.N.; Verma, C.; Lukman, S. Structural analysis of protein tyrosine phosphatase 1B reveals potentially druggable allosteric binding sites. Proteins Struct. Funct. Bioinform. 2018, 86, 301–321. [Google Scholar] [CrossRef]
- Maccari, R.; Paoli, P.; Ottanà, R.; Jacomelli, M.; Ciurleo, R.; Manao, G.; Steindl, T.; Langer, T.; Vigorita, M.G.; Camici, G. 5-Arylidene-2,4-thiazolidinediones as inhibitors of protein tyrosine phosphatases. Bioorganic Med. Chem. 2007, 15, 5137–5149. [Google Scholar] [CrossRef] [PubMed]
- Ottanà, R.; Maccari, R.; Ciurleo, R.; Paoli, P.; Jacomelli, M.; Manao, G.; Camici, G.; Laggner, C.; Langer, T. 5-Arylidene-2-phenylimino-4-thiazolidinones as PTP1B and LMW-PTP inhibitors. Bioorganic Med. Chem. 2009, 17, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Ottanà, R.; Maccari, R.; Amuso, S.; Wolber, G.; Schuster, D.; Herdlinger, S.; Manao, G.; Camici, G.; Paoli, P. New 4-[(5-arylidene-2-arylimino-4-oxo-3-thiazolidinyl)methyl]benzoic acids active as protein tyrosine phosphatase inhibitors endowed with insulinomimetic effect on mouse C2C12 skeletal muscle cells. Eur. J. Med. Chem. 2012, 50, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Ottanà, R.; Maccari, R.; Mortier, J.; Caselli, A.; Amuso, S.; Camici, G.; Rotondo, A.; Wolber, G.; Paoli, P. Synthesis, biological activity and structure–activity relationships of new benzoic acid-based protein tyrosine phosphatase inhibitors endowed with insulinomimetic effects in mouse C2C12 skeletal muscle cells. Eur. J. Med. Chem. 2014, 71, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Ottanà, R.; Paoli, P.; Naß, A.; Lori, G.; Cardile, V.; Adornato, I.; Rotondo, A.; Graziano, A.C.E.; Wolber, G.; Maccari, R. Discovery of 4-[(5-arylidene-4-oxothiazolidin-3-yl)methyl]benzoic acid derivatives active as novel potent allosteric inhibitors of protein tyrosine phosphatase 1B: In silico studies and in vitro evaluation as insulinomimetic and anti-inflammatory agents. Eur. J. Med. Chem. 2016, 127, 840–858. [Google Scholar] [CrossRef]
- Zinker, B.A.; Rondinone, C.M.; Trevillyan, J.M.; Gum, R.J.; Clampit, J.E.; Waring, J.F.; Xie, N.; Wilcox, D.; Jacobson, P.; Frost, L.; et al. PTP1B antisense oligonucleotide lowers PTP1B protein, normalizes blood glucose, and improves insulin sensitivity in diabetic mice. Proc. Natl. Acad. Sci. USA 2002, 99, 11357–11362. [Google Scholar] [CrossRef]
- Monia, B.P.; A Lesnik, E.; Gonzalez, C.; Lima, W.F.; McGee, D.; Guinosso, C.J.; Kawasaki, A.M.; Cook, P.D.; Freier, S.M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993, 268, 14514–14522. [Google Scholar] [CrossRef]
- Swarbrick, M.M.; Havel, P.J.; Levin, A.A.; Bremer, A.A.; Stanhope, K.L.; Butler, M.; Booten, S.L.; Graham, J.L.; McKay, R.A.; Murray, S.F.; et al. Inhibition of Protein Tyrosine Phosphatase-1B with Antisense Oligonucleotides Improves Insulin Sensitivity and Increases Adiponectin Concentrations in Monkeys. Endocrinology 2009, 150, 1670–1679. [Google Scholar] [CrossRef]
- Koizumi, M.; Takagi-Sato, M.; Okuyama, R.; Araki, K.; Sun, W.; Nakai, D. In vivo antisense activity of ENA(R) oligonucleotides targeting PTP1B mRNA in comparison of that of 2′-MOE-modified oligonucleotides. Nucleic Acids Symp. Ser. 2007, 51, 111–112. [Google Scholar] [CrossRef][Green Version]
- Digenio, A.; Pham, N.C.; Watts, L.M.; Morgan, E.S.; Jung, S.W.; Baker, B.F.; Geary, R.S.; Bhanot, S. Antisense Inhibition of Protein Tyrosine Phosphatase 1B With IONIS-PTP-1BRx Improves Insulin Sensitivity and Reduces Weight in Overweight Patients With Type 2 Diabetes. Diabetes Care 2018, 41, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Chen, Z.; Pang, L.; Wang, L.; Jiang, H.; Chen, Y.; Zhang, Z.; Fu, C.; Ren, B.; Zhang, J. Oral Nano Drug Delivery Systems for the Treatment of Type 2 Diabetes Mellitus: An Available Administration Strategy for Antidiabetic Phytocompounds. Int. J. Nanomed. 2020, 15, 10215–10240. [Google Scholar] [CrossRef] [PubMed]
- Kostrzewa, T.; Styszko, J.; Gorska-Ponikowska, M.; Sledzinski, T.; Kuban-Jankowska, A. Inhibitors of Protein Tyrosine Phosphatase PTP1B With Anticancer Potential. Anticancer Res. 2019, 39, 3379–3384. [Google Scholar] [CrossRef]
- Bellomo, E.; Singh, K.B.; Massarotti, A.; Hogstrand, C.; Maret, W. The metal face of protein tyrosine phosphatase 1B. Coord. Chem. Rev. 2016, 327, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Bellomo, E.; Abro, A.; Hogstrand, C.; Maret, W.; Domene, C. Role of Zinc and Magnesium Ions in the Modulation of Phosphoryl Transfer in Protein Tyrosine Phosphatase 1B. J. Am. Chem. Soc. 2018, 140, 4446–4454. [Google Scholar] [CrossRef] [PubMed]
- Gorgulla, C.; Boeszoermenyi, A.; Wang, Z.-F.; Fischer, P.D.; Coote, P.W.; Das, K.M.P.; Malets, Y.S.; Radchenko, D.S.; Moroz, Y.S.; Scott, D.A.; et al. An open-source drug discovery platform enables ultra-large virtual screens. Nature 2020, 580, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.; Thornton, B.P.; Tabernero, L. A New Paradigm for KIM-PTP Drug Discovery: Identification of Allosteric Sites with Potential for Selective Inhibition Using Virtual Screening and LEI Analysis. Int. J. Mol. Sci. 2021, 22, 12206. [Google Scholar] [CrossRef]
- Mullard, A. DNA tags help the hunt for drugs. Nature 2016, 530, 367–369. [Google Scholar] [CrossRef]
- Burslem, G.; Crews, C.M. Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell 2020, 181, 102–114. [Google Scholar] [CrossRef]
- Yang, N.J.; Hinner, M.J. Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins. In Site-Specific Protein Labeling: Methods and Protocols; Gautier, A., Hinner, M.J., Eds.; Springer: New York, NY, USA, 2015; Volume 1266, pp. 29–53. [Google Scholar]
- Klein, V.G.; Townsend, C.E.; Testa, A.; Zengerle, M.; Maniaci, C.; Hughes, S.J.; Chan, K.-H.; Ciulli, A.; Lokey, R.S. Understanding and Improving the Membrane Permeability of VH032-Based PROTACs. ACS Med. Chem. Lett. 2020, 11, 1732–1738. [Google Scholar] [CrossRef]
- Su, S.; Wu, J.; Gao, Y.; Luo, Y.; Yang, D.; Wang, P. The pharmacological properties of chrysophanol, the recent advances. Biomed. Pharmacother. 2020, 125, 110002. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, T.; Haile, T.; Nigusse, T.; Dhanaraju, M.D. Nanotechnology: An effective tool for enhancing bioavailability and bioactivity of phytomedicine. Asian Pac. J. Trop. Biomed. 2014, 4, S1–S7. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Dong, Y.; Shang, B.; Zhang, B.; Crans, D.C.; Yang, X. Convergent Protein Phosphatase Inhibitor Design for PTP1B and TCPTP: Exchangeable Vanadium Coordination Complexes on Graphene Quantum Dots. Adv. Funct. Mater. 2021, 32, 2108645. [Google Scholar] [CrossRef]

| Compound | Company | IC50 or Ki (µM) | Clinical Phase | Continued or Not | Indications | Trials’ Registration Number | |
|---|---|---|---|---|---|---|---|
| PTP1B | TCPTP | ||||||
| Ertiprotafib | Pfizer | IC50 > 20 | Unknown | Phase II | × | T2DM | Unknown |
| JTT-551 | Tobacco | Ki = 0.22 | Ki = 9.30 | Pre-clinical | × | T2DM, obesity | Unknown |
| KQ-791 | Kaneq Bioscience | Unknown | Unknown | Phase I | Unknown | T2DM | ClinicalTrials.gov: NCT02445911 |
| TTP-814 | TransTech Pharma | Unknown | Unknown | Phase II | × | T2DM | Unknown |
| Trodusquemine | DepYmed | IC50 = 1.0 | IC50 = 224 | Phase I | × | T2DM, obesity Metastatic breast cancer | ClinicalTrials.gov: NCT00606112 ClinicalTrials.gov: NCT02524951 |
| DPM-1001 | DepYmed | IC50 = 0.10 | Unknown | Pre-clinical | √ | T2DM, obesity | Unknown |
| IONIS 113715 | IONIS Pharmaceuticals | IC50 < 0.01 | Unknown | Phase II | × | T2DM | ClinicalTrials.gov: NCT00330330 |
| IONIS PTP1BRx | IONIS Pharmaceuticals | Unknown | Unknown | Phase II | √ | T2DM | ClinicalTrials.gov: NCT01918865 |
| AC type | 2-(oxalyl-amino)-benzoic acid (2-OBA) derivatives |
| 2-carboxymethyl-benzoic acid-derived inhibitors | |
| difluoromethylphosphonic acid (DFMP)-based inhibitors | |
| isothiazolidinone (IZD)-contained inhibitors | |
| monocyclic thiophene-based inhibitors | |
| AB type | monocyclic, bicyclic and tricyclic thiophene inhibitors |
| sulfamic acid moiety-contained inhibitors | |
| other novel AB type inhibitors | |
| ABC type | a series of OBA derivatives |
| DFMP group-contained PTP1B inhibitors | |
| sulfamic acid moiety-contained PTP1B inhibitors | |
| ADC type | N-(2,5-diethoxy-phenyl)-methanesul-fonamide derivatives |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Mathieu, C.; Berthelet, J.; Zhang, W.; Dupret, J.-M.; Rodrigues Lima, F. Human Protein Tyrosine Phosphatase 1B (PTP1B): From Structure to Clinical Inhibitor Perspectives. Int. J. Mol. Sci. 2022, 23, 7027. https://doi.org/10.3390/ijms23137027
Liu R, Mathieu C, Berthelet J, Zhang W, Dupret J-M, Rodrigues Lima F. Human Protein Tyrosine Phosphatase 1B (PTP1B): From Structure to Clinical Inhibitor Perspectives. International Journal of Molecular Sciences. 2022; 23(13):7027. https://doi.org/10.3390/ijms23137027
Chicago/Turabian StyleLiu, Rongxing, Cécile Mathieu, Jérémy Berthelet, Wenchao Zhang, Jean-Marie Dupret, and Fernando Rodrigues Lima. 2022. "Human Protein Tyrosine Phosphatase 1B (PTP1B): From Structure to Clinical Inhibitor Perspectives" International Journal of Molecular Sciences 23, no. 13: 7027. https://doi.org/10.3390/ijms23137027
APA StyleLiu, R., Mathieu, C., Berthelet, J., Zhang, W., Dupret, J.-M., & Rodrigues Lima, F. (2022). Human Protein Tyrosine Phosphatase 1B (PTP1B): From Structure to Clinical Inhibitor Perspectives. International Journal of Molecular Sciences, 23(13), 7027. https://doi.org/10.3390/ijms23137027





