Investigation of Structural Features of Two Related Lipases and the Impact on Fatty Acid Specificity in Vegetable Fats
Abstract
:1. Introduction
2. Results and Discussion
2.1. Lipases Activity Profile
2.2. Lipases Sequence Validation
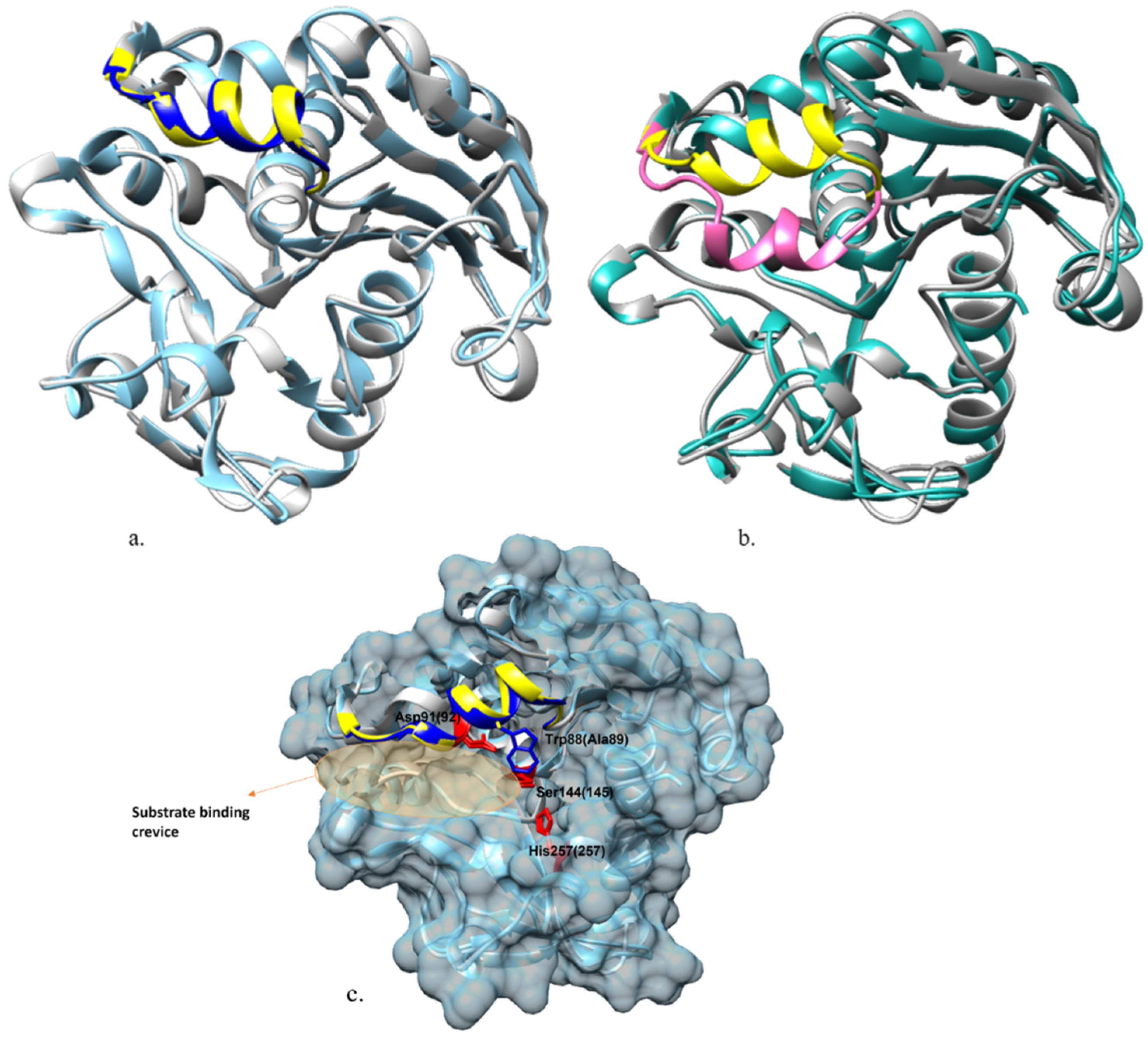
2.3. Homology Modeling of Open-Lid Structure of ROL
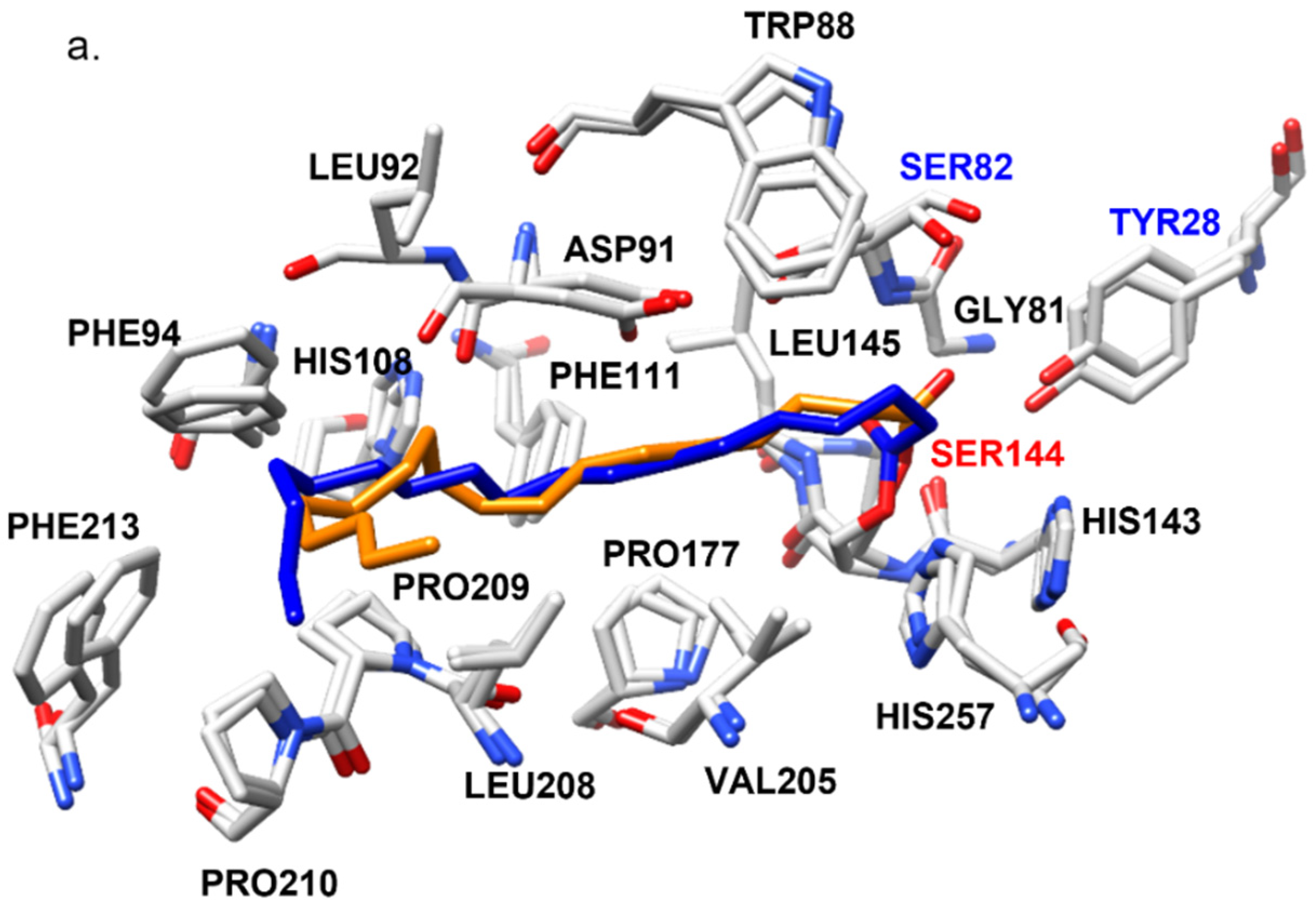
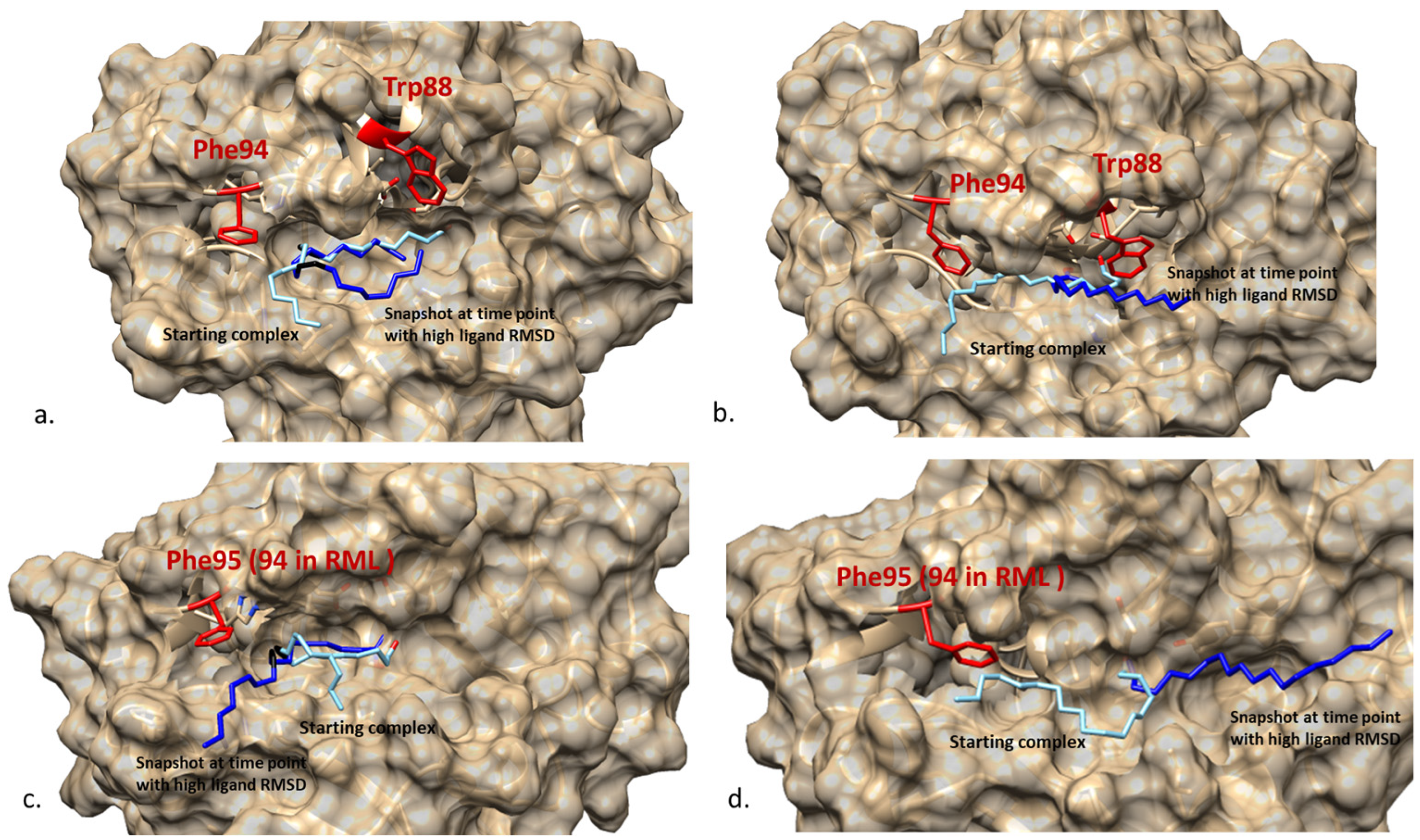
2.4. Structural Features of ROL and RML Related to Substrate Interactions
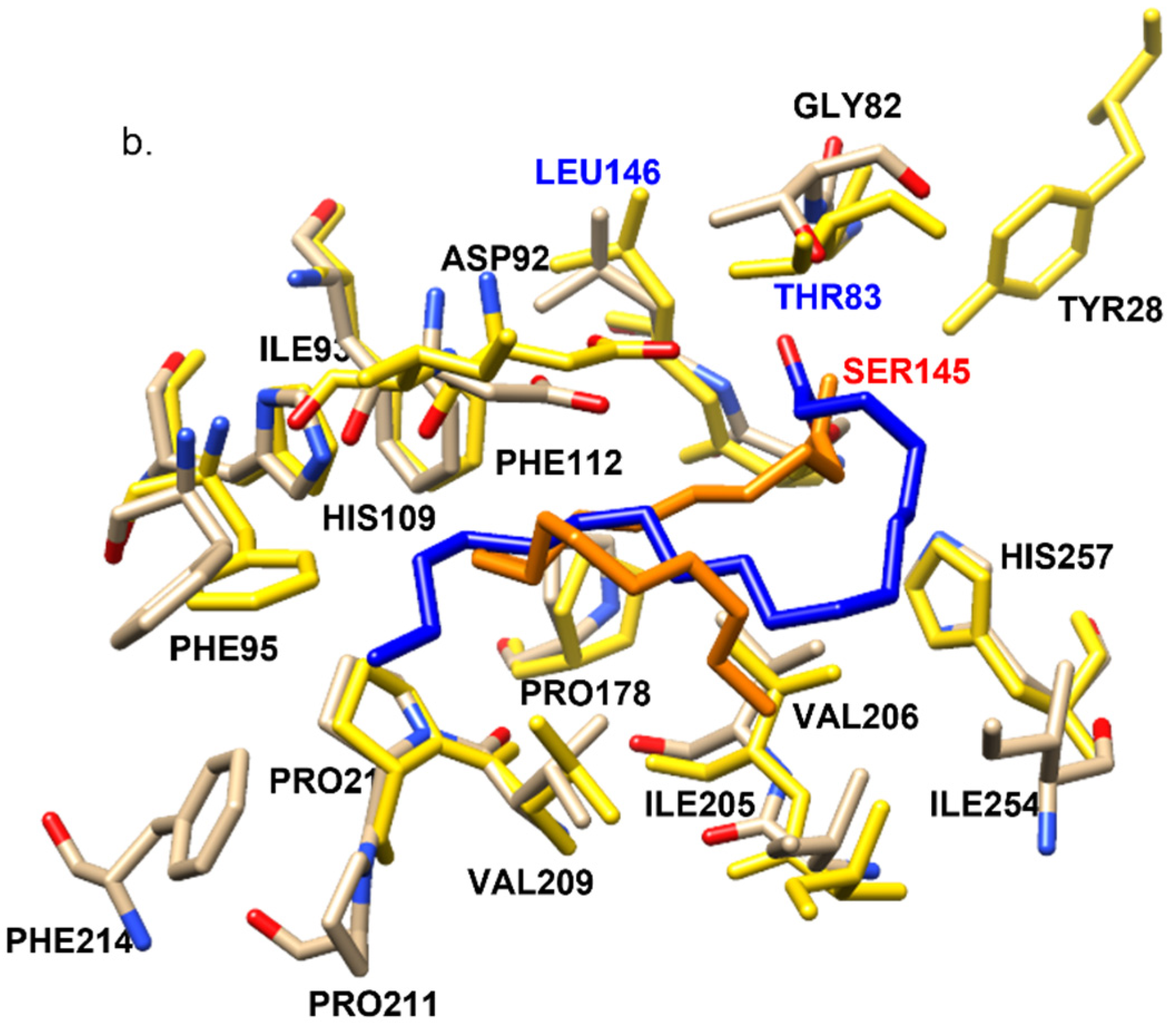
2.5. Differences in Interactions That May Explain the Higher Relative Activity of RML VES/VEO over ROL
2.6. MD Simulation
3. Materials and Methods
3.1. Material
3.2. Lipase Immobilization
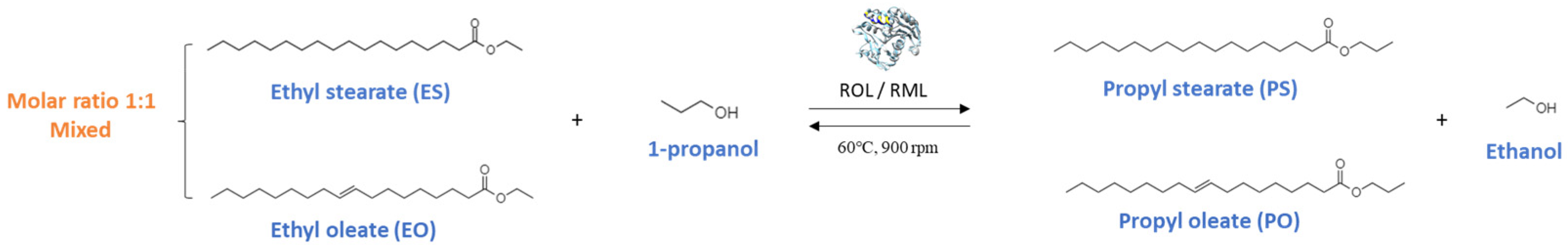
3.3. Transesterification Reaction
3.4. GC Analysis
3.5. Lipase Sequence Validation
3.6. Homology Modeling
3.7. Substrate Docking
3.8. Molecular Dynamic Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodrigues, R.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as a biocatalyst in fats and oils modification. J. Mol. Catal. B Enzym. 2010, 66, 15–32. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Trono, D. Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef] [Green Version]
- Bornscheuer, U.T. Alteration of lipase properties by protein engineering methods. OCL—Ol. Corps Gras Lipides 2008, 15, 184–188. [Google Scholar] [CrossRef] [Green Version]
- Adlercreutz, P. Immobilisation and application of lipases in organic media. Chem. Soc. Rev. 2013, 42, 6406–6436. [Google Scholar] [CrossRef] [Green Version]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Harel, M.; Remington, J.S.; Silman, I.; Schrag, J.; et al. The α/β hydrolase fold. Protein Eng. Des. Sel. 1992, 5, 197–211. [Google Scholar] [CrossRef] [Green Version]
- Widmann, M.; Juhl, P.B.; Pleiss, J. Structural classification by the Lipase Engineering Database: A case study of Candida antarctica lipase A. BMC Genom. 2010, 11, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, C.; Shimada, Y. Lipases. Encycl. Microbiol. 2009, 385–392. [Google Scholar] [CrossRef]
- Šinkūnienė, D.; Adlercreutz, P. Effects of regioselectivity and lipid class specificity of lipases on transesterification, exemplified by biodiesel production. JAOCS J. Am. Oil Chem. Soc. 2014, 91, 1283–1290. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Xu, X. Production of specific-structured triacylglycerols by lipase-catalyzed reactions: A review. Eur. J. Lipid Sci. Technol. 2000, 102, 287–303. [Google Scholar] [CrossRef]
- Zorn, K.; Oroz-Guinea, I.; Brundiek, H.; Dörr, M.; Bornscheuer, U.T. Alteration of Chain Length Selectivity of Candida antarctica Lipase A by Semi-Rational Design for the Enrichment of Erucic and Gondoic Fatty Acids. Adv. Synth. Catal. 2018, 360, 4115–4131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albayati, S.H.; Masomian, M.; Ishak, S.N.H.; Ali, M.S.B.M.; Thean, A.L.; Shariff, F.B.M.; Noor, N.D.B.M.; Rahman, R.N.Z.R.A. Main structural targets for engineering lipase substrate specificity. Catalysts 2020, 10, 747. [Google Scholar] [CrossRef]
- López-Fernández, J.; Benaiges, M.; Valero, F. Rhizopus oryzae lipase, a promising industrial enzyme: Biochemical characteristics, production and biocatalytic applications. Catalysts 2020, 10, 1277. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzym. Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Lipp, M.; Simoneau, C.; Ulberth, F.; Anklam, E.; Crews, C.; Brereton, P.; De Greyt, W.; Schwack, W.; Wiedmaier, C. ORIGINAL ARTICLE: Composition of Genuine Cocoa Butter and Cocoa Butter Equivalents. J. Food Compos. Anal. 2001, 14, 399–408. [Google Scholar] [CrossRef]
- Naik, B.; Kumar, V. Cocoa Butter and Its Alternatives: A Review Biofortification of Finger Millet with Iron and Zinc View Project Studies on Different Type of Fungi in Patientswith Special Focus on Mucormycosis View Project Cocoa Butter and Its Alternatives: A Reveiw. 2014. Available online: www.jakraya.com/journal/jbet (accessed on 1 May 2021).
- Pleiss, J.; Fischer, M.; Schmid, R.D. Anatomy of lipase binding sites: The scissile fatty acid binding site. Chem. Phys. Lipids 1998, 93, 67–80. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Darden, T.; Nabuurs, S.B.; Finkelstein, A.; Vriend, G. Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins Struct. Funct. Genet. 2004, 57, 678–683. [Google Scholar] [CrossRef]
- Pleiss, J.; Fischer, M.; Peiker, M.; Thiele, C.; Schmid, R.D. Lipase Engineering Database Understanding and Exploiting Sequence-Structure-Function Relationships. 2000. Available online: www.elsevier.comrlocatermolcatb (accessed on 1 May 2021).
- Beer, H.; Wohlfahrt, G.; McCarthy, J.; Schomburg, D.; Schmid, R. Analysis of the catalytic mechanism of a fungal lipase using computer-aided design and structural mutants. Protein Eng. 1996, 9, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Gitlesen, T.; Bauer, M.; Adlercreutz, P. Adsorption of lipase on polypropylene powder. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1997, 1345, 188–196. [Google Scholar] [CrossRef]
- Cottrell, J.S. Protein identification by peptide mass fingerprinting. Pept. Res. 1994, 7, 115–124. [Google Scholar] [CrossRef] [PubMed]
- DPerkins, N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2007, 1, 2856–2860. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Derewenda, Z.S.; Dodson, E.J.; Dodson, G.G.; Turkenburg, J.P. Structure and molecular model refinement of Rhizomucor miehei triacyglyceride lipase: A case study of the use of simulated annealing in partial model refinement. Acta Crystallogr. Sect. B 1992, 48, 307–319. [Google Scholar] [CrossRef]
- Derewenda, U.; Brzozowski, A.M.; Lawson, D.M.; Derewenda, Z.S. Catalysis at the Interface: The Anatomy of a Conformational Change in a Triglyceride Lipase. 1992. Available online: https://pubs.acs.org/sharingguidelines (accessed on 1 May 2021).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. 2000. Available online: http://www.rcsb.org/pdb/status.html (accessed on 1 May 2021).
- Kohno, M.; Funatsu, J.; Mikami, B.; Kugimiya, W.; Matsuo, T.; Morita, Y. The Crystal Structure of Lipase II from Rhizopus Niveus at 2.2 A Resolution. 1996. Available online: https://academic.oup.com/jb/article/120/3/505/824776 (accessed on 1 May 2021).
- AWaterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Steinegger, M.; Meier, M.; Mirdita, M.; Vöhringer, H.; Haunsberger, S.J.; Söding, J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 2019, 20, 473. [Google Scholar] [CrossRef] [Green Version]
- Bianco, G.; Forli, S.; Goodsell, D.S.; Olson, A.J. Covalent docking using autodock: Two-point attractor and flexible side chain methods. Protein Sci. 2016, 25, 295–301. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Enzyme | Enzyme Dosage/mg | Activity towards EO: VEO/(mM/min) | Activity towards ES: VES/(mM/min) | Relative Activity: VES/VEO * |
|---|---|---|---|---|
| Immobilized ROL | 50 | 16.5 ± 1.5 | 10.8 ± 1.0 | 0.65 |
| 60 | 21.2 ± 3.7 | 13.2 ± 2.6 | 0.62 | |
| 70 | 24.3 ± 1.2 | 15.1 ± 0.6 | 0.62 | |
| Immobilized RML | 10 | 9.8 ± 1.9 | 7.2 ± 1.4 | 0.73 |
| 20 | 20.9 ± 0.8 | 15.9 ± 0.7 | 0.76 | |
| 30 | 28.4 ± 1.5 | 21.7 ± 0.9 | 0.76 |
| (a) | |
| Docking complex | Binding energy/kcal/mol |
| Oleic acid docked in RML (RMOL) | 8.43 |
| Stearic acid docked in RML (RMST) | 7.86 |
| Oleic acid docked in ROL (ROOL) | 6.52 |
| Stearic acid docked in ROL (ROST) | 5.63 |
| (b) | |
| Docking complex | Binding energy/kcal/mol |
| Oleic acid docked in RMLW88A | 6.07 |
| Stearic acid docked in RMLW88A | 5.70 |
| Oleic acid docked in ROLA89W | 7,81 |
| Stearic acid docked in ROLA89W | 6.10 |
| ROOL | ROST | RMOL | RMST | |
|---|---|---|---|---|
| Tyr28(28) | ||||
| Gly81(82) | ||||
| Ser82(Thr83) | ||||
| Trp88(Ala89) | ||||
| Asp91(92) | ||||
| Leu92(Ile93) | ||||
| Phe94(95) | ||||
| His108(109) | ||||
| Phe111(112) | ||||
| His143(144) | ||||
| Leu145(146) | ||||
| Pro177(178) | ||||
| Ile204(205) | ||||
| Val205(206) | ||||
| Leu208(Val209) | ||||
| Pro209(210) | ||||
| Pro210(211) | ||||
| Phe213(214) | ||||
| Val254(Ile254) | ||||
| His257(257) |
| Reaction No. | Lipase Name | Loading Dosage (mg) |
|---|---|---|
| 1 | Immobilized ROL | 50 |
| 2 | Immobilized ROL | 60 |
| 3 | Immobilized ROL | 70 |
| 4 | Immobilized RML | 10 |
| 5 | Immobilized RML | 20 |
| 6 | Immobilized RML | 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Z.; Olofsson, K.; Linares-Pastén, J.A.; Nordberg Karlsson, E. Investigation of Structural Features of Two Related Lipases and the Impact on Fatty Acid Specificity in Vegetable Fats. Int. J. Mol. Sci. 2022, 23, 7072. https://doi.org/10.3390/ijms23137072
Dong Z, Olofsson K, Linares-Pastén JA, Nordberg Karlsson E. Investigation of Structural Features of Two Related Lipases and the Impact on Fatty Acid Specificity in Vegetable Fats. International Journal of Molecular Sciences. 2022; 23(13):7072. https://doi.org/10.3390/ijms23137072
Chicago/Turabian StyleDong, Zehui, Kim Olofsson, Javier A. Linares-Pastén, and Eva Nordberg Karlsson. 2022. "Investigation of Structural Features of Two Related Lipases and the Impact on Fatty Acid Specificity in Vegetable Fats" International Journal of Molecular Sciences 23, no. 13: 7072. https://doi.org/10.3390/ijms23137072
APA StyleDong, Z., Olofsson, K., Linares-Pastén, J. A., & Nordberg Karlsson, E. (2022). Investigation of Structural Features of Two Related Lipases and the Impact on Fatty Acid Specificity in Vegetable Fats. International Journal of Molecular Sciences, 23(13), 7072. https://doi.org/10.3390/ijms23137072







