Outcome Prediction in Critically-Ill Patients with Venous Thromboembolism and/or Cancer Using Machine Learning Algorithms: External Validation and Comparison with Scoring Systems
Abstract
:1. Introduction
2. Related Work
3. Motivation of the Current Work
4. Materials and Methods
4.1. Ethics Statement
4.2. Dataset Description and Cohort Selection
4.2.1. MIMIC-III Database
4.2.2. eICU Database
4.3. Feature Pre-Analysis Selection
4.3.1. MIMIC-III
4.3.2. eICU
4.3.3. Traditional Clinical Scores
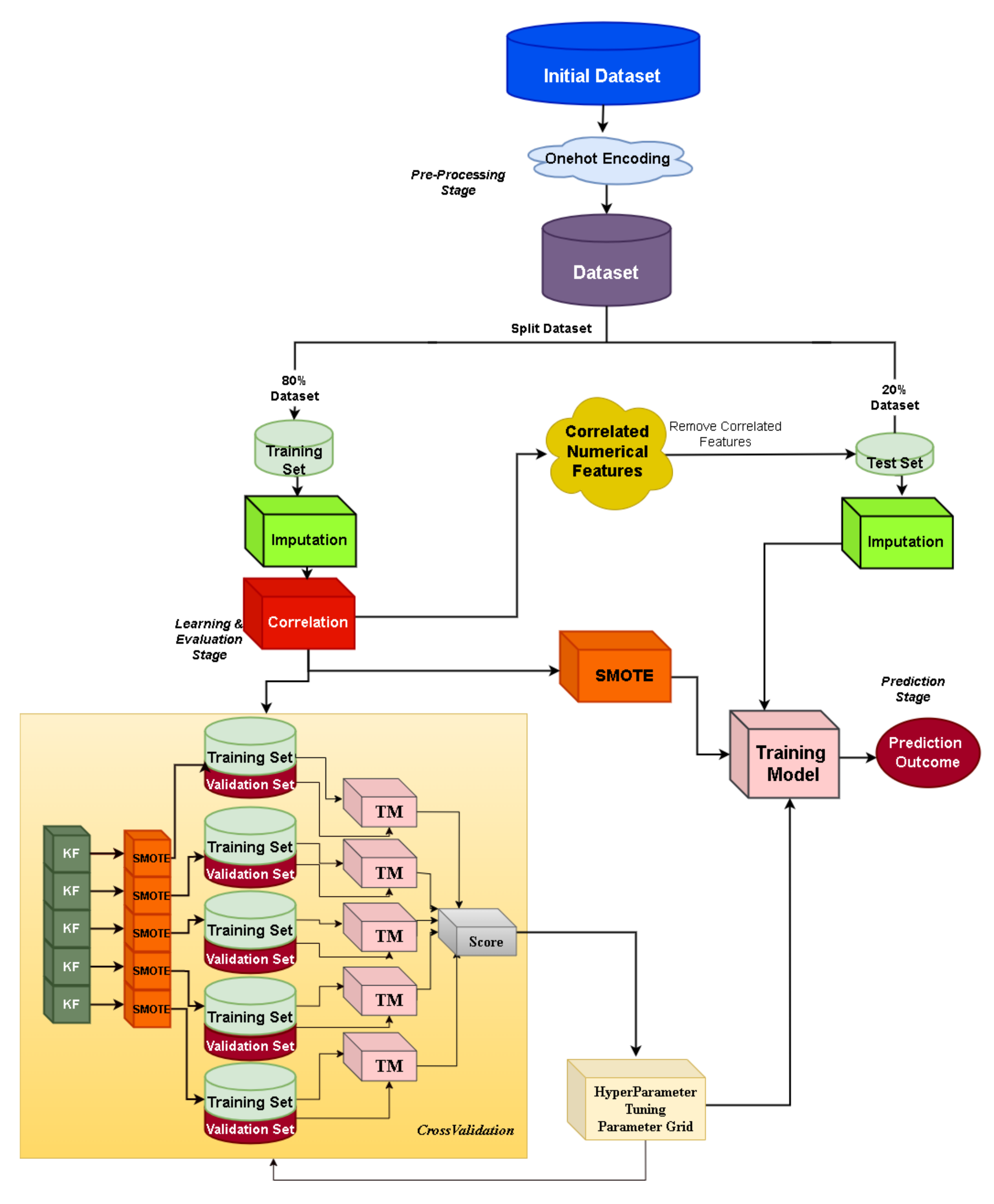
4.4. Data Manipulation and Transformation
4.5. Automated Machine Learning Framework: JADBio
4.6. Custom Machine Learning Framework: XGBoost
4.7. Machine Learning Models’ Performance Evaluation
5. Results
5.1. Automated Machine Learning
5.1.1. Prediction of Early Mortality in ICU Patients with Thrombosis
MIMIC-III Dataset
eICU Dataset
5.1.2. Prediction of Early Mortality in ICU Patients with Cancer
5.1.3. Prediction of Late Mortality in ICU Patients with Thrombosis
5.1.4. Prediction of Late Mortality in ICU Patients with Cancer
5.1.5. Model Validation
5.2. Custom Machine Learning Framework: XGBoost
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AC | Acute |
| ALP | Alkaline phosphatase |
| APACHE | Acute Physiology Age Chronic Health Evaluation |
| APS | Acute Physiology Score |
| AST | Aspartate transaminase |
| AUC | Area under the curve |
| AVG | Average |
| BUN | Blood urea nitrogen |
| COPD | Chronic obstructive pulmonary disease |
| CPK | Creatine phosphokinase |
| CV | CareVue |
| DRG | Disease-related group |
| DOACs | Direct oral anticoagulants |
| DVT | Deep vein thrombosis |
| FFP | Fresh frozen plasma |
| fiO2 | Fraction of inspired oxygen |
| FPR | False Positive Rate |
| HIPAAA | Health Insurance Portability and Accountability Act |
| GCS | Glasgow Coma Scale |
| ICD | International Classification Code |
| ICU | Intensive care |
| LMWH | Low-molecular-weight heparin |
| LOS | Length of stay |
| LODS | Logistic Organ Dysfunction Score |
| m | Month |
| mean1stRR | Mean value of respiratory rate on the 1st ICU day |
| MD | Missing data |
| ML | Machine learning |
| MODS | Multiple Organ Dysfunction Score |
| MV | Metavision |
| NOF | Number of features |
| NOS | Not otherwise specified |
| OASIS | Outcome and Assessment Information Set |
| PaO2 | Partial pressure of arterial oxygen |
| PaCO2 | Partial pressure of arterial carbon dioxide |
| PE | Pulmonary embolism |
| PESI | Pulmonary Embolism Severity Index |
| sPESI | Simplified Pulmonary Embolism Severity Index |
| PLT | Platelet |
| PT | Prothrombin time |
| RBC | Red blood cell |
| RDW | Red cell distribution width |
| RF | Random Forest |
| ROC | Receiver operating characteristics |
| SBP | Systolic blood pressure |
| SD | Standard deviation |
| SaO2 | Oxygen arterial saturation |
| SABER | Sequence Annotator for Biomedical Entities and Relations |
| SAPS | Simplified Acute Physiology Score |
| SES | Statistically Equivalent Signature |
| SIRS | Systemic Inflammatory Response Syndrome |
| SMOTE | Synthetic Minority Oversampling Technique |
| SOFA | Sequential Organ Failure |
| SVM | Support Vector Machine |
| TPR | True Positive Rate |
| VTE | Venous thromboembolism |
| WBC | White blood cells |
Appendix A
| Group Name (as in eICU) | Table in eICU | Description | Number of Features | Missing Values (Merged with Patient) | Most Common Features |
|---|---|---|---|---|---|
| Patient (“patient”) | Patient | Basic demographic information, LOS, discharge status | 11 | 0 | Gender, age, ethnicity, length of stay, admission height, admission weight, discharge status |
| Diagnosis “admission _dx” | Diagnosis | Diagnoses documented during ICU stay | 61 | 395 | PE, DVT, hypertension acute respiratory failure, altered mental status, diabetes, pneumonia, acute renal failure, congestive heart, anemia failure, chronic kidney, cancer, disease, sepsis |
| APSIII score | Apache- ApsVar | Acute Physiology Score (APSIII) | 24 | 529 | Variables used in APS III score, e.g., Glasgow respiratory rate, heart rate, temperature, coma scale, paO2, etc. |
| APACHE IV score | Apache- PredVar | APACHE IV and IVa versions | 34 | 529 | Variables used in APACHE predictions, e.g., presence of cirrhosis, or metastatic carcinoma |
| Labs | Lab | Laboratory tests | 80 | 442 | Hematocrit hemoglobin, white blood cells, platelets, potassium, sodium, creatinine, nitrogen, chloride, glucose, calcium, bicarbonate, PT, INR, aPTT, blood urea MCH, MCHC, RDW |
| Vital signs | Physical Exam | Vital signs | 8 | 597 | Blood pressure diastolic and systolic current, highest, lowest, heart rate, GCS, fiO2, SatO2 in 1 h intervals |
| Vital Periodic | Vital signs | 10 | 433 | Heart rate, respiration, SatO2, blood pressure, temperature | |
| Medications prior to admission | Admission Drug | Medications taken prior to ICU admission | 36 | 3263 | Aspirin, furosemide bronchodilator metoprolol, warfarin, lisinopril, atorvastatin, inhaled corticosteroids insulin, amlodipin, levothyroxin, metformin, carbedilol, LMWH, clopidogrel, DOACs |
| Drugs | Infusion | Medications Transfusions Parenteral | 30 | 2503 | Heparin, epinephrine, norepinephrine, insulin, amiodarone, phenylephrine vasopressin, t-PA, diltiazem, dopamine |
| Treatment | Medications | 52 | 1040 | Medications in groups, e.g., anticoagulants antibiotics, antiarrhythmics vasopressor, ulcer prophylaxis and several procedures, e.g., mechanical ventilation, IVC filter, thrombolysis, dialysis, embolectomy | |
| Medical history | Pasthistory | Past history of chronic diseases | 111 | 422 | Hypertension, insulin-dependent diabetes, disease, cancer chronic heart failure, asthma, COPD, atrial fibrillation, stroke, DVT or PE within 6 months, dementia, peripheral vascular disease |
| 1st Admission | Endotrachial intubation | Min 1st RR | Metastatic Cancer | Albumin | Mean 1st RR | SBP | RDW |
|---|---|---|---|---|---|---|---|
| m1 | Metastatic cancer | OASIS Elective surgery | LOS | Renal insuf- ficiency | 1st day chloride max | SAPSII score | PaO2 |
| m3 | Metastatic cancer | Open heart surgery | ALP | OASIS Pre-ICU LOS | GCS | 1st day chloride max | Age |
| m6 | Metastatic cancer | AST | Albumin | RDW | Aorto- coronary bypass | OASIS Elective surgery | |
| m12 | Metastatic cancer | Age | SAPSII score | Etoposide | PaO2 | FFP | Lung biopsy |
| >m12 | Trans- fusions |
| F1 Score | Accuracy | Specificity | Sensitivity | ||
|---|---|---|---|---|---|
| Early/Late | Early/Late | Early/Late | Early/Late | Early/Late | |
| All features | 0.94 (0.91, 0.96) | 0.87 | 0.83 | 0.93 | 0.79 |
| All scores | 0.85 (0.81, 0.88) | 0.6 | 0.81 | 0.85 | 0.7 |
| APS II | 0.81 (0.76, 0.85) | 0.55 | 0.8 | 0.85 | 0.6 |
| Comorbidity | 0.78 (0.74, 0.82) | 0.5 | 0.736 | 0.76 | 0.65 |
| LODS/MLODS | 0.77 (0.72, 0.81) | 0.5 | 0.77 | 0.8 | 0.6 |
| OASIS | 0.8 (0.76, 0.84) | 0.55 | 0.76 | 0.77 | 0.7 |
| SAPS | 0.8 (0.76, 0.84) | 0.53 | 0.78 | 0.82 | 0.6 |
| SAPSII | 0.85 (0.81, 0.89) | 0.61 | 0.81 | 0.83 | 0.73 |
| SIRS | 0.64 (0.59, 0.68) | 0.39 | 0.53 | 0.5 | 0.71 |
| SOFA | 0.76 (0.72, 0.81) | 0.5 | 0.7 | 0.7 | 0.71 |
| F1 Score | Accuracy | Specificity | Sensitivity | ||
|---|---|---|---|---|---|
| Early/Late | Early/Late | Early/Late | Early/Late | Early/Late | |
| All features | 0.83 (0.79, 0.87) | 0.72 | 0.74 | 0.75 | 0.78 |
| All scores | 0.76 (0.71, 0.81) | 0.64 | 0.69 | 0.7 | 0.7 |
| APS II | 0.65 (0.59, 0.7) | 0.56 | 0.62 | 0.53 | 0.69 |
| Comorbidity | 0.74 (0.69, 0.79) | 0.62 | 0.69 | 0.69 | 0.68 |
| LODS/MLODS | 0.57 (0.51, 0.62) | 0.46 | 0.62 | 0.56 | 0.53 |
| OASIS | 0.68 (0.63, 0.69) | 0.59 | 0.63 | 0.51 | 0.77 |
| SAPS | 0.68 (0.63, 0.72) | 0.59 | 0.62 | 0.55 | 0.73 |
| SAPSII | 0.76 (0.72, 0.81) | 0.63 | 0.7 | 0.78 | 0.63 |
| SIRS | 0.49 (0.48, 0.5) | 0.54 | 0.62 | 0.34 | 0.65 |
| SOFA | 0.58 (0.52, 0.63) | 0.52 | 0.62 | 0.35 | 0.73 |
References
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Crimmins, E.M. Lifespan and healthspan: Past, present, and promise. Gerontologist 2015, 55, 901–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajat, C.; Stein, E. The global burden of multiple chronic conditions: A narrative review. Prev. Med. Rep. 2018, 12, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Boonyawat, K.; Chantrathammachart, P.; Numthavaj, P.; Nanthatanti, N.; Phusanti, S.; Phuphuakrat, A.; Niparuck, P.; Angchaisuksiri, P. Incidence of thromboembolism in patients with COVID-19: A systematic review and meta-analysis. Thromb. J. 2020, 18, 34. [Google Scholar] [CrossRef]
- Martin, K.A.; Molsberry, R.; Cuttica, M.J.; Desai, K.R.; Schimmel, D.R.; Khan, S.S. Time trends in pulmonary embolism mortality rates in the United States, 1999 to 2018. J. Am. Heart Assoc. 2020, 9, e016784. [Google Scholar] [CrossRef]
- Zimmerman, J.E.; Kramer, A.A. A history of outcome prediction in the ICU. Curr. Opin. Crit. Care 2014, 20, 550–556. [Google Scholar] [CrossRef]
- Teixeira, C.; Kern, M.; Rosa, R.G. What outcomes should be evaluated in critically ill patients? Rev. Bras. Ter. Intensiv. 2021, 33, 312–319. [Google Scholar] [CrossRef]
- Sadeghi, R.; Banerjee, T.; Romine, W. Early hospital mortality prediction using vital signals. Smart Health 2018, 9, 265–274. [Google Scholar] [CrossRef]
- El-Rashidy, N.; El-Sappagh, S.; Abuhmed, T.; Abdelrazek, S.; El-Bakry, H.M. Intensive care unit mortality prediction: An improved patient-specific stacking ensemble model. IEEE Access 2020, 8, 133541–133564. [Google Scholar] [CrossRef]
- Choi, M.H.; Kim, D.; Choi, E.J.; Jung, Y.J.; Choi, Y.J.; Cho, J.H.; Jeong, S.H. Mortality prediction of patients in intensive care units using machine learning algorithms based on electronic health records. Sci. Rep. 2022, 12, 7180. [Google Scholar] [CrossRef]
- Patel, S.; Singh, G.; Zarbiv, S.; Ghiassi, K.; Rachoin, J.S. Mortality Prediction Using SaO2/FiO2 Ratio Based on eICU Database Analysis. Crit. Care Res. Pract. 2021, 2021, 6672603. [Google Scholar] [CrossRef]
- Holmgren, G.; Andersson, P.; Jakobsson, A.; Frigyesi, A. Artificial neural networks improve and simplify intensive care mortality prognostication: A national cohort study of 217,289 first-time intensive care unit admissions. J. Intens. Care 2019, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.M.; Knuiman, M.; Finn, J.; Webb, S.A. Estimating long-term survival of critically ill patients: The PREDICT model. PLoS ONE 2008, 3, e3226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorsen-Meyer, H.C.; Nielsen, A.B.; Nielsen, A.P.; Kaas-Hansen, B.S.; Toft, P.; Schierbeck, J.; Strøm, T.; Chmura, P.J.; Heimann, M.; Dybdahl, L.; et al. Dynamic and explainable machine learning prediction of mortality in patients in the intensive care unit: A retrospective study of high-frequency data in electronic patient records. Lancet Digit. Health 2020, 2, e179–e191. [Google Scholar] [CrossRef]
- Simpson, A.; Puxty, K.; McLoone, P.; Quasim, T.; Sloan, B.; Morrison, D.S. Comorbidity and survival after admission to the intensive care unit: A population-based study of 41,230 patients. J. Intens. Care Soc. 2021, 22, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Le Gall, J.R.; Lemeshow, S.; Saulnier, F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA 1993, 270, 2957–2963. [Google Scholar] [CrossRef]
- Knaus, W.A.; Wagner, D.P.; Draper, E.A.; Zimmerman, J.E.; Bergner, M.; Bastos, P.G.; Sirio, C.A.; Murphy, D.J.; Lotring, T.; Damiano, A.; et al. The APACHE III prognostic system: Risk prediction of hospital mortality for critically III hospitalized adults. Chest 1991, 100, 1619–1636. [Google Scholar] [CrossRef] [Green Version]
- Lopes Ferreira, F.; Peres Bota, D.; Bross, A.; Mélot, C.; Vincent, J.L. Serial evaluation of the SOFA score to predict outcome in critically ill patients. JAMA 2001, 286, 1754–1758. [Google Scholar] [CrossRef] [Green Version]
- Vincent, J.L.; Moreno, R. Clinical review: Scoring systems in the critically ill. Crit. Care 2010, 14, 207. [Google Scholar] [CrossRef] [Green Version]
- Rapsang, A.G.; Shyam, D.C. Scoring systems in the intensive care unit: A compendium. Indian J. Crit. Care Med.-Peer-Rev. Off. Publ. Indian Soc. Crit. Care Med. 2014, 18, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.M.; Martin, C.M.; Morrison, T.L.; Sibbald, W.J. Interobserver variability in data collection of the APACHE II score in teaching and community hospitals. Crit. Care Med. 1999, 27, 1999–2004. [Google Scholar] [CrossRef] [PubMed]
- Cosgriff, C.V.; Celi, L.A.; Ko, S.; Sundaresan, T.; Armengol de la Hoz, M.Á.; Kaufman, A.R.; Stone, D.J.; Badawi, O.; Deliberato, R.O. Developing well-calibrated illness severity scores for decision support in the critically ill. NPJ Digit. Med. 2019, 2, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafiq, A.; Lodhi, H.; Ahmed, Z.; Bajwa, A. Is the pulmonary embolism severity index being routinely used in clinical practice? Thrombosis 2015, 2015, 175357. [Google Scholar] [CrossRef]
- Aujesky, D.; Roy, P.M.; Le Manach, C.P.; Verschuren, F.; Meyer, G.; Obrosky, D.S.; Stone, R.A.; Cornuz, J.; Fine, M.J. Validation of a model to predict adverse outcomes in patients with pulmonary embolism. Eur. Heart J. 2006, 27, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Aujesky, D.; Perrier, A.; Roy, P.M.; Stone, R.; Cornuz, J.; Meyer, G.; Obrosky, D.; Fine, M. Validation of a clinical prognostic model to identify low-risk patients with pulmonary embolism. J. Intern. Med. 2007, 261, 597–604. [Google Scholar] [CrossRef]
- Jiménez, D.; Yusen, R.D.; Otero, R.; Uresandi, F.; Nauffal, D.; Laserna, E.; Conget, F.; Oribe, M.; Cabezudo, M.A.; Diaz, G. Prognostic models for selecting patients with acute pulmonary embolism for initial outpatient therapy. Chest 2007, 132, 24–30. [Google Scholar] [CrossRef]
- Donzé, J.; Le Gal, G.; Fine, M.J.; Roy, P.M.; Sanchez, O.; Verschuren, F.; Cornuz, J.; Meyer, G.; Perrier, A.; Righini, M.; et al. Prospective validation of the pulmonary embolism severity index. Thromb. Haemost. 2008, 100, 943–948. [Google Scholar]
- Kohn, C.G.; Mearns, E.S.; Parker, M.W.; Hernandez, A.V.; Coleman, C.I. Prognostic accuracy of clinical prediction rules for early post-pulmonary embolism all-cause mortality: A bivariate meta-analysis. Chest 2015, 147, 1043–1062. [Google Scholar] [CrossRef]
- Churpek, M.; Yuen, T.; Winslow, C.; Meltzer, D.; Kattan, M.; Edelson, D. Multicenter Comparison of Machine Learning Methods and Conventional Regression for Predicting Clinical Deterioration on the Wards. Crit. Care Med. 2016, 44, 368–374. [Google Scholar] [CrossRef] [Green Version]
- Pirracchio, R.; Petersen, M.; Carone, M.; Rigon, M.; Chevret, S.; Laan, M. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): A population-based study. Lancet Respir. Med. 2014, 3, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Runnan, S.; Gao, M.; Tao, Y.; Chen, Q.; Wu, G.; Guo, X.; Xia, Z.; You, G.; Hong, Z.; Huang, K. Prognostic nomogram for 30-day mortality of deep vein thrombosis patients in intensive care unit. BMC Cardiovasc. Disord. 2021, 21, 11. [Google Scholar] [CrossRef]
- Purushotham, S.; Meng, C.; Che, Z.; Liu, Y. Benchmarking deep learning models on large healthcare datasets. J. Biomed. Inform. 2018, 83, 112–134. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Xu, C.; Nguyen, C.; Geng, Y.; Pfob, A.; Sidey-Gibbons, C. Machine Learning-based Short-term Mortality Prediction Models for Cancer Patients Using Electronic Health Record Data: A Systematic Review and Critical Appraisal (Preprint). JMIR Med. Inform. 2021, 10, e33182. [Google Scholar] [CrossRef]
- Staudinger, T.; Stoiser, B.; Müllner, M.; Locker, G.; Laczika, K.; Knapp, S.; Burgmann, H.; Wilfing, A.; Kofler, J.; Thalhammer, F.; et al. Outcome and prognostic factors in critically ill cancer patients admitted to the intensive care unit. Crit. Care Med. 2000, 28, 1322–1328. [Google Scholar] [CrossRef]
- Boer, S.; de Keizer, N.; Jonge, E. Performance of prognostic models in critically ill cancer patients—A review. Crit. Care 2005, 9, R458–R463. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.E.; Pollard, T.J.; Lu, S.; Lehman, L.-w.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.E.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [Green Version]
- Pollard, T.; Johnson, A.; Raffa, J.; Celi, L.A.; Mark, R.G.; Badawi, O. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci. Data 2018, 5, 180178. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [Green Version]
- Bader Lab. Saber (Sequence Annotator for Biomedical Entities and Relations). 2022. Available online: https://baderlab.github.io/saber/ (accessed on 19 April 2022).
- Danilatou, V.; Antonakaki, D.; Tzagkarakis, C.; Kanterakis, A.; Katos, V.; Kostoulas, T. Automated Mortality Prediction in Critically-ill Patients with Thrombosis using Machine Learning. In Proceedings of the 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, OH, USA, 26–28 October 2020; pp. 247–254. [Google Scholar]
- Tsamardinos, I.; Charonyktakis, P.; Borboudakis, G.; Lakiotaki, K.; Zenklusen, J.C. Just Add Data: Automated Predictive Modeling for Knowledge Discovery and Feature Selection. Nat. Precis. Oncol. 2022, 6, 38. [Google Scholar] [CrossRef]
- Tsamardinos, I.; Charonyktakis, P.; Lakiotaki, K.; Borboudakis, G.; Zenklusen, J.C.; Juhl, H.; Chatzaki, E.; Lagani, V. Just add data: Automated predictive modeling and biosignature discovery. BioRxiv 2020. [Google Scholar] [CrossRef]
- Tsamardinos, I.; Greasidou, E.; Borboudakis, G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. Mach. Learn. 2018, 107, 1895–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagani, V.; Athineou, G.; Farcomeni, A.; Tsagris, M.; Tsamardinos, I. Feature selection with the R package MXM: Discovering statistically-equivalent feature subsets. arXiv 2016, arXiv:1611.03227. [Google Scholar] [CrossRef] [Green Version]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian Optimization of Machine Learning Algorithms. In Proceedings of the 25th International Conference on Neural Information Processing Systems, 2012, NIPS’12, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 2951–2959. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, L.; Anstey, M.; Feng, M.; Toledano, R.; Kogan, S.; Howell, M.; Clardy, P.; Celli, L.; Talmor, D.; Novack, V. Quantifying the Mortality Impact of Do-Not-Resuscitate Orders in the ICU. Crit. Care Med. 2017, 45, 1019–1027. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD’16, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.S.; Soares, J.P.; Abreu, P.H.; Araujo, H.; Santos, J. Cross-validation for imbalanced datasets: Avoiding overoptimistic and overfitting approaches [research frontier]. IEEE Comput. Intell. Mag. 2018, 13, 59–76. [Google Scholar] [CrossRef]
- Ghosheh, G.; Li, J.; Zhu, T. A review of Generative Adversarial Networks for Electronic Health Records: Applications, evaluation measures and data sources. arXiv 2022, arXiv:2203.07018. [Google Scholar]
- Shillan, D.; Sterne, J.A.; Champneys, A.; Gibbison, B. Use of machine learning to analyse routinely collected intensive care unit data: A systematic review. Crit. Care 2019, 23, 284. [Google Scholar] [CrossRef] [Green Version]
- Science, T. The Multiple faces of ‘Feature Importance’ in XGBoost. 2022. Available online: shorturl.at/oGU12 (accessed on 21 April 2022).
- Fernando, S.M.; Mok, G.; Castellucci, L.A.; Dowlatshahi, D.; Rochwerg, B.; McIsaac, D.I.; Carrier, M.; Wells, P.S.; Bagshaw, S.M.; Fergusson, D.A.; et al. Impact of anticoagulation on mortality and resource utilization among critically ill patients with major bleeding. Crit. Care Med. 2020, 48, 515–524. [Google Scholar] [CrossRef]
- Fernandez, R.; Cano, S.; Catalan, I.; Rubio, O.; Subira, C.; Masclans, J.; Rognoni, G.; Ventura, L.; Macharete, C.; Winfield, L.; et al. High red blood cell distribution width as a marker of hospital mortality after ICU discharge: A cohort study. J. Intensive Care 2018, 6, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.C.; Chow, W.W.; Lau, J.K.; Chow, V.; Ng, A.C.; Kritharides, L. Red blood cell transfusion and outcomes in acute pulmonary embolism. Respirology 2018, 23, 935–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arihan, O.; Wernly, B.; Lichtenauer, M.; Franz, M.; Kabisch, B.; Muessig, J.; Masyuk, M.; Lauten, A.; Schulze, P.C.; Hoppe, U.C.; et al. Blood Urea Nitrogen (BUN) is independently associated with mortality in critically ill patients admitted to ICU. PloS ONE 2018, 13, e0191697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvagno, G.L.; Sanchis-Gomar, F.; Picanza, A.; Lippi, G. Red blood cell distribution width: A simple parameter with multiple clinical applications. Crit. Rev. Clin. Lab. Sci. 2015, 52, 86–105. [Google Scholar] [CrossRef]
- Yang, J.; Yang, J. Association between blood eosinophils and mortality in critically ill patients with acute exacerbation of chronic obstructive pulmonary disease: A retrospective cohort study. Int. J. Chronic Obstr. Pulm. Dis. 2021, 16, 281. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Ghidini, M.; Ghidini, A.; Sgroi, G.; Vavassori, I.; Petrò, D.; Cabiddu, M.; Aiolfi, A.; Bonitta, G.; Zaniboni, A.; et al. Red blood cell transfusions and the survival in patients with cancer undergoing curative surgery: A systematic review and meta-analysis. Surg. Today 2021, 51, 1535–1557. [Google Scholar] [CrossRef]
- AHRQ. Clinical Classifications Software (CCS) for ICD-9-CM. 2022. Available online: https://cutt.ly/7H0o4f8 (accessed on 19 April 2022).
- Busse, R.; Geissler, A.; Aaviksoo, A.; Cots, F.; Häkkinen, U.; Kobel, C.; Mateus, C.; Or, Z.; O’Reilly, J.; Serdén, L.; et al. Diagnosis related groups in Europe: Moving towards transparency, efficiency, and quality in hospitals? BMJ 2013, 346, f3197. [Google Scholar] [CrossRef] [Green Version]
- Ali, L.; Bukhari, S.A.C. An Approach Based on Mutually Informed Neural Networks to Optimize the Generalization Capabilities of Decision Support Systems Developed for Heart Failure Prediction. IRBM 2020, 42, 345–352. [Google Scholar] [CrossRef]
- Lee, K.; Kha, H.; Nguyen, V.; Chen, Y.C.; Cheng, S.J.; Chen, C.Y. Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 9254. [Google Scholar] [CrossRef]









| VTE | VTE | Cancer | ||||
|---|---|---|---|---|---|---|
| Characteristic | eICU | p-Value | MIMIC-III | p-Value | MIMIC-III | p-Value |
| Thrombosis - PE - DVT | 3724 2739 (62.4%) 2220 (50.6%) | 0.71 0.95 | 2468 960 (38.9%) 1543 (62.5%) | 0.92 - | 5128 - - | |
| Sex - Female - Male - Unknown | 1788 (48 %) 1934 (52 %) 1 | 0.24 0.24 | 1024 (41.5%) 1444 (58.5%) - | 0.53 0.53 | 2192 (41.2%) 3126 (58.8%) - | 0.07 0.07 |
| Ethnicity - White - Black - Other | 2845 (76.4%) 522 (14%) 357 (9.6%) | 0.36 0.16 | 1801 (73%) 246 (10%) 421 (17%) | 0.27 0.47 | 4049 (76%) 378 (7.1 %) 891 (16.8%) | 0.57 0.83 |
| Age, years Average (SD) Min Max | 60.28 (16.26) 15 90 | <0.001 | 62.64 (16.7) 17.4 98.7 | <0.001 | 66.2 (14.2) 18.9 98.9 | <0.001 |
| Cancer diagnosis | 409 (9.3 %) | <0.001 | 605 (24.5%) | <0.001 | ||
| LOS, days Mean (SD) Median: | 11.22 (12.24) 7.01 | 0.002 | 7.06 (10.06), 153.9 days | <0.001 | 4.4 (6.5) 2.2 | <0.001 |
| Mortality (%): Early: Late: “Survivor”: | 267 (7.16 %) N/A 3457 (92.84%) | 348 (14.1%) 817 (33.1%) 1303 (52.8%) | 902 (17%) 2659 (49.9%) 1757 (33.1%) | |||
| Death time (days) Average (SD): Median: | N/A | 390 (647) 83 | 328.5 (536.55) 365 |
| F1 Score | Accuracy | Specificity | Sensitivity | ||
|---|---|---|---|---|---|
| All features | 0.87 (0.84, 0.9) | 0.42 | 0.92 | 0.95 | 0.42 |
| Labs | 0.87 (0.83, 0.9) | 0.43 | 0.92 | 0.95 | 0.50 |
| Vital Signs | 0.83 (0.78, 0.87) | 0.37 | 0.92 | 0.95 | 0.38 |
| APSIII | 0.79 (0.75, 0.84) | 0.32 | 0.92 | 0.96 | 0.29 |
| APACHE IVa | 0.78 (0.73, 0.82) | 0.30 | 0.89 | 0.92 | 0.38 |
| Drugs | 0.82 (0.77, 0.86) | 0.53 | 0.94 | 0.99 | 0.23 |
| Diagnosis | 0.73 (0.68, 0.77) | 0.26 | 0.82 | 0.92 | 0.30 |
| Medical History | 0.60 (0.56, 0.64) | 0.27 | 0.93 | 0.98 | 0.06 |
| Medication prior to admission | 0.55 (0.5, 0.59) | 0.41 | 0.92 | 0.99 | 0.01 |
| Mortality Prediction | Accuracy | F1 Score | Specificity | Sensitivity | |
|---|---|---|---|---|---|
| m0 | 0.94 (0.92, 0.96) | 0.88 | 0.82 | 0.91 | 0.82 |
| m1 | 0.88 (0.85, 0.91) | 0.85 | 0.89 | 0.84 | 0.78 |
| m3 | 0.84 (0.8, 0.87) | 0.79 | 0.85 | 0.74 | 0.79 |
| m6 | 0.78 (0.74, 0.82) | 0.72 | 0.49 | 0.72 | 0.71 |
| m12 | 0.76 (0.71, 0.8) | 0.72 | 0.5 | 0.72 | 0.7 |
| m > 12 | 0.74 (0.69, 0.74) | 0.68 | 0.76 | 0.64 | 0.74 |
| Early Mortality | Late Mortality | ||
|---|---|---|---|
| MIMIC-III | eICU | MIMIC-III | |
| 0.93 | 0.87 | 0.82 | |
| Accuracy | 0.89 | 0.92 | 0.76 |
| F1 score | 0.72 | 0.97 | 0.60 |
| Sensitivity | 0.67 | 0.99 | 0.49 |
| Specificity | 0.95 | 0.1 | 0.90 |
| F1 Score | Accuracy | Specificity | Sensitivity | ||
|---|---|---|---|---|---|
| MIMIC-III | 0.93 [0.91, 0.96] | 0.74 | 0.77 | 0.93 | 0.74 |
| eICU | 0.89 [0.87, 0.91] | 0.56 | 0.86 | 0.89 | 0.7 |
| XGBoost | All Features |
|---|---|
| 0.84 (0.83–0.85) | |
| Accuracy | 0.87 |
| F1 score | 0.63 |
| Sensitivity | 0.29 |
| Specificity | 0.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danilatou, V.; Nikolakakis, S.; Antonakaki, D.; Tzagkarakis, C.; Mavroidis, D.; Kostoulas, T.; Ioannidis, S. Outcome Prediction in Critically-Ill Patients with Venous Thromboembolism and/or Cancer Using Machine Learning Algorithms: External Validation and Comparison with Scoring Systems. Int. J. Mol. Sci. 2022, 23, 7132. https://doi.org/10.3390/ijms23137132
Danilatou V, Nikolakakis S, Antonakaki D, Tzagkarakis C, Mavroidis D, Kostoulas T, Ioannidis S. Outcome Prediction in Critically-Ill Patients with Venous Thromboembolism and/or Cancer Using Machine Learning Algorithms: External Validation and Comparison with Scoring Systems. International Journal of Molecular Sciences. 2022; 23(13):7132. https://doi.org/10.3390/ijms23137132
Chicago/Turabian StyleDanilatou, Vasiliki, Stylianos Nikolakakis, Despoina Antonakaki, Christos Tzagkarakis, Dimitrios Mavroidis, Theodoros Kostoulas, and Sotirios Ioannidis. 2022. "Outcome Prediction in Critically-Ill Patients with Venous Thromboembolism and/or Cancer Using Machine Learning Algorithms: External Validation and Comparison with Scoring Systems" International Journal of Molecular Sciences 23, no. 13: 7132. https://doi.org/10.3390/ijms23137132
APA StyleDanilatou, V., Nikolakakis, S., Antonakaki, D., Tzagkarakis, C., Mavroidis, D., Kostoulas, T., & Ioannidis, S. (2022). Outcome Prediction in Critically-Ill Patients with Venous Thromboembolism and/or Cancer Using Machine Learning Algorithms: External Validation and Comparison with Scoring Systems. International Journal of Molecular Sciences, 23(13), 7132. https://doi.org/10.3390/ijms23137132







