Triiodothyronine Acts as a Smart Influencer on Hsp90 via a Triiodothyronine Binding Site
Abstract
:1. Introduction
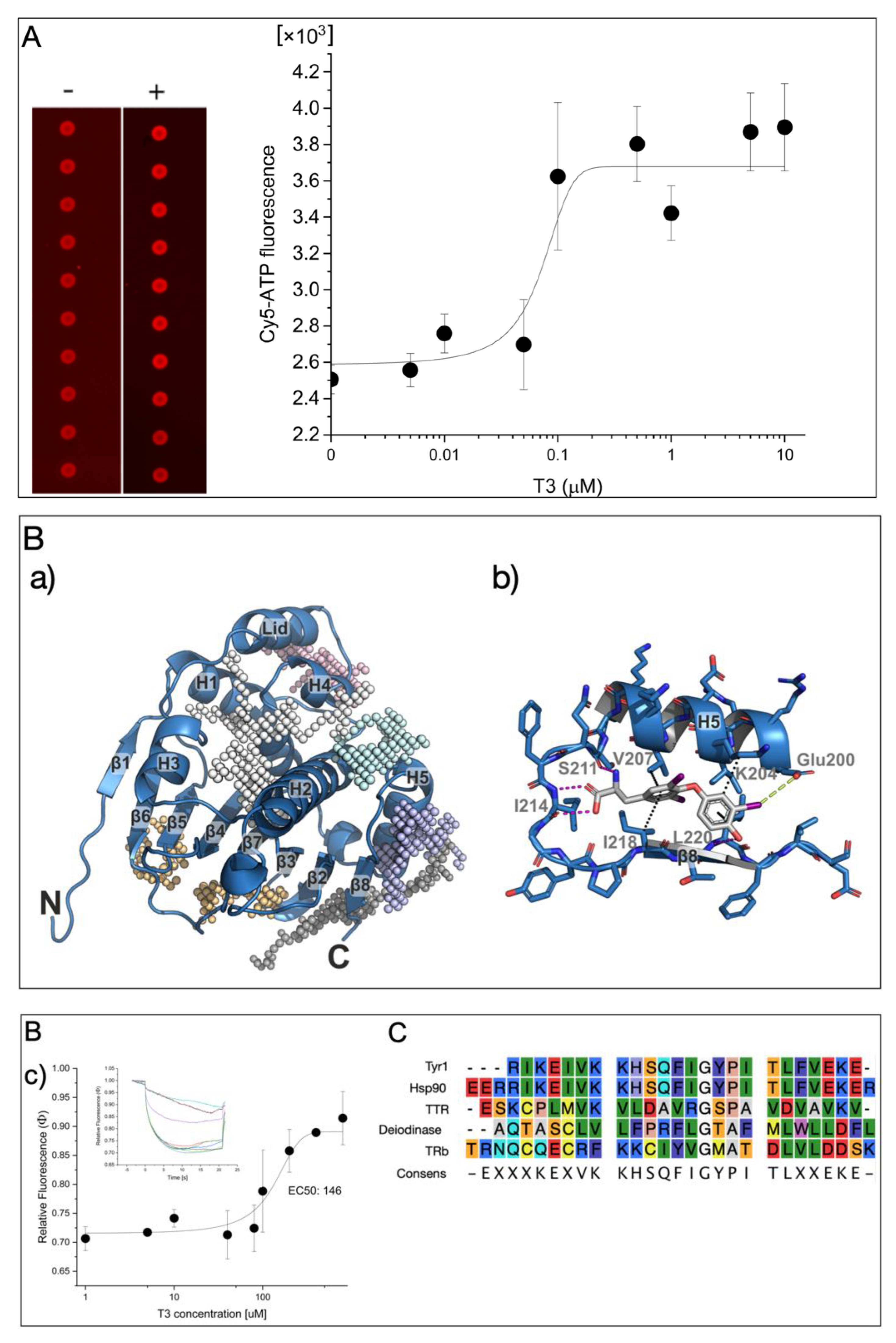
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Detection of Microarray-Based Hsp90 Activity
4.3. Molecular Docking
4.4. Microscale Thermophoresis Analysis (MST)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ortiga-Carvalho, T.M.; Chiamolera, M.I.; Pazos-Moura, C.C.; Wondisford, F.E. Hypothalamus-Pituitary-Thyroid Axis. Compr. Physiol. 2016, 6, 1387–1428. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.C.; da Conceição, R.R. The Deiodinase Trio and Thyroid Hormone Signaling. Methods Mol. Biol. 2018, 1801, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Ortiga-Carvalho, T.M.; Sidhaye, A.R.; Wondisford, F.E. Thyroid hormone receptors and resistance to thyroid hormone disorders. Nature reviews. Endocrinology 2014, 10, 582–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, C.P.; Münscher, A.; Machado, D.S.; Wondisford, F.E.; Ortiga-Carvalho, T.M. Complete activation of thyroid hormone receptor β by T3 is essential for normal cochlear function and morphology in mice. Cell. Physiol. Biochem. 2011, 28, 997–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortoglou, A.; Candiloros, H. The serum triiodothyronine to thyroxine (T3/T4) ratio in various thyroid disorders and after Levothyroxine replacement therapy. Hormones 2004, 3, 120–126. [Google Scholar] [CrossRef]
- Chiha, M.; Samarasinghe, S.; Kabaker, A.S. Thyroid storm: An updated review. J. Intensive Care Med. 2015, 30, 131–140. [Google Scholar] [CrossRef]
- Stepien, B.K.; Huttner, W.B. Transport, Metabolism, and Function of Thyroid Hormones in the Developing Mammalian Brain. Front. Endocrinol. 2019, 10, 209. [Google Scholar] [CrossRef]
- Eng, L.; Lam, L. Thyroid Function During the Fetal and Neonatal Periods. NeoReviews 2020, 21, e30–e36. [Google Scholar] [CrossRef]
- Leemans, M.; Couderq, S.; Demeneix, B.; Fini, J.B. Pesticides With Potential Thyroid Hormone-Disrupting Effects: A Review of Recent Data. Front. Endocrinol. 2019, 10, 743. [Google Scholar] [CrossRef] [Green Version]
- Kahaly, G.J. Management of Graves Thyroidal and Extrathyroidal Disease: An Update. J. Clin. Endocrinol. Metab. 2020, 105, 3704–3720. [Google Scholar] [CrossRef]
- Papamichael, K.; Delitheos, B.; Mourouzis, I.; Pantos, C.; Tiligada, E. L-Thyroxine induces thermotolerance in yeast. Cell Stress Chaperones 2019, 24, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Echeverría, P.C.; Bernthaler, A.; Dupuis, P.; Mayer, B.; Picard, D. An interaction network predicted from public data as a discovery tool: Application to the Hsp90 molecular chaperone machine. PLoS ONE 2011, 6, e26044. [Google Scholar] [CrossRef] [PubMed]
- Picard, D.; Khursheed, B.; Garabedian, M.J.; Fortin, M.G.; Lindquist, S.; Yamamoto, K.R. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature 1990, 348, 166–168. [Google Scholar] [CrossRef]
- Yuno, A.; Lee, M.J.; Lee, S.; Tomita, Y.; Rekhtman, D.; Moore, B.; Trepel, J.B. Clinical Evaluation and Biomarker Profiling of Hsp90 Inhibitors. Methods Mol. Biol. 2018, 1709, 423–441. [Google Scholar] [CrossRef] [PubMed]
- Schax, E.; Walter, J.G.; Märzhäuser, H.; Stahl, F.; Scheper, T.; Agard, D.A.; Eichner, S.; Kirschning, A.; Zeilinger, C. Microarray-based screening of heat shock protein inhibitors. J. Biotechnol. 2014, 180, 1–9. [Google Scholar] [CrossRef]
- Harris, R.; Olson, A.J.; Goodsell, D.S. Automated prediction of ligand-binding sites in proteins. Proteins 2008, 70, 1506–1517. [Google Scholar] [CrossRef]
- Li, J.; Sun, L.; Xu, C.; Yu, F.; Zhou, H.; Zhao, Y.; Zhang, J.; Cai, J.; Mao, C.; Tang, L.; et al. Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90. Acta Biochim. Biophys. Sin. 2012, 44, 300–306. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Yue, Q.; Stahl, F.; Plettenburg, O.; Kirschning, A.; Warnecke, A.; Zeilinger, C. The Noncompetitive Effect of Gambogic Acid Displaces Fluorescence-Labeled ATP but Requires ATP for Binding to Hsp90/HtpG. Biochemistry 2018, 57, 2601–2605. [Google Scholar] [CrossRef]
- Wallin, G.; Brönnegård, M.; Grimelius, L.; McGuire, J.; Tørring, O. Expression of the thyroid hormone receptor, the oncogenes c-myc and H-ras, and the 90 kD heat shock protein in normal, hyperplastic, and neoplastic human thyroid tissue. Thyroid 1992, 2, 307–313. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nature reviews. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, L.; Warnecke, A.; Weder, J.; Preller, M.; Zeilinger, C. Triiodothyronine Acts as a Smart Influencer on Hsp90 via a Triiodothyronine Binding Site. Int. J. Mol. Sci. 2022, 23, 7150. https://doi.org/10.3390/ijms23137150
Fan L, Warnecke A, Weder J, Preller M, Zeilinger C. Triiodothyronine Acts as a Smart Influencer on Hsp90 via a Triiodothyronine Binding Site. International Journal of Molecular Sciences. 2022; 23(13):7150. https://doi.org/10.3390/ijms23137150
Chicago/Turabian StyleFan, Lu, Athanasia Warnecke, Julia Weder, Matthias Preller, and Carsten Zeilinger. 2022. "Triiodothyronine Acts as a Smart Influencer on Hsp90 via a Triiodothyronine Binding Site" International Journal of Molecular Sciences 23, no. 13: 7150. https://doi.org/10.3390/ijms23137150
APA StyleFan, L., Warnecke, A., Weder, J., Preller, M., & Zeilinger, C. (2022). Triiodothyronine Acts as a Smart Influencer on Hsp90 via a Triiodothyronine Binding Site. International Journal of Molecular Sciences, 23(13), 7150. https://doi.org/10.3390/ijms23137150






