Genome-Wide Analysis and Profile of UDP-Glycosyltransferases Family in Alfalfa (Medicago sativa L.) under Drought Stress
Abstract
:1. Introduction
2. Results
2.1. Identification of UGT Genes in Alfalfa
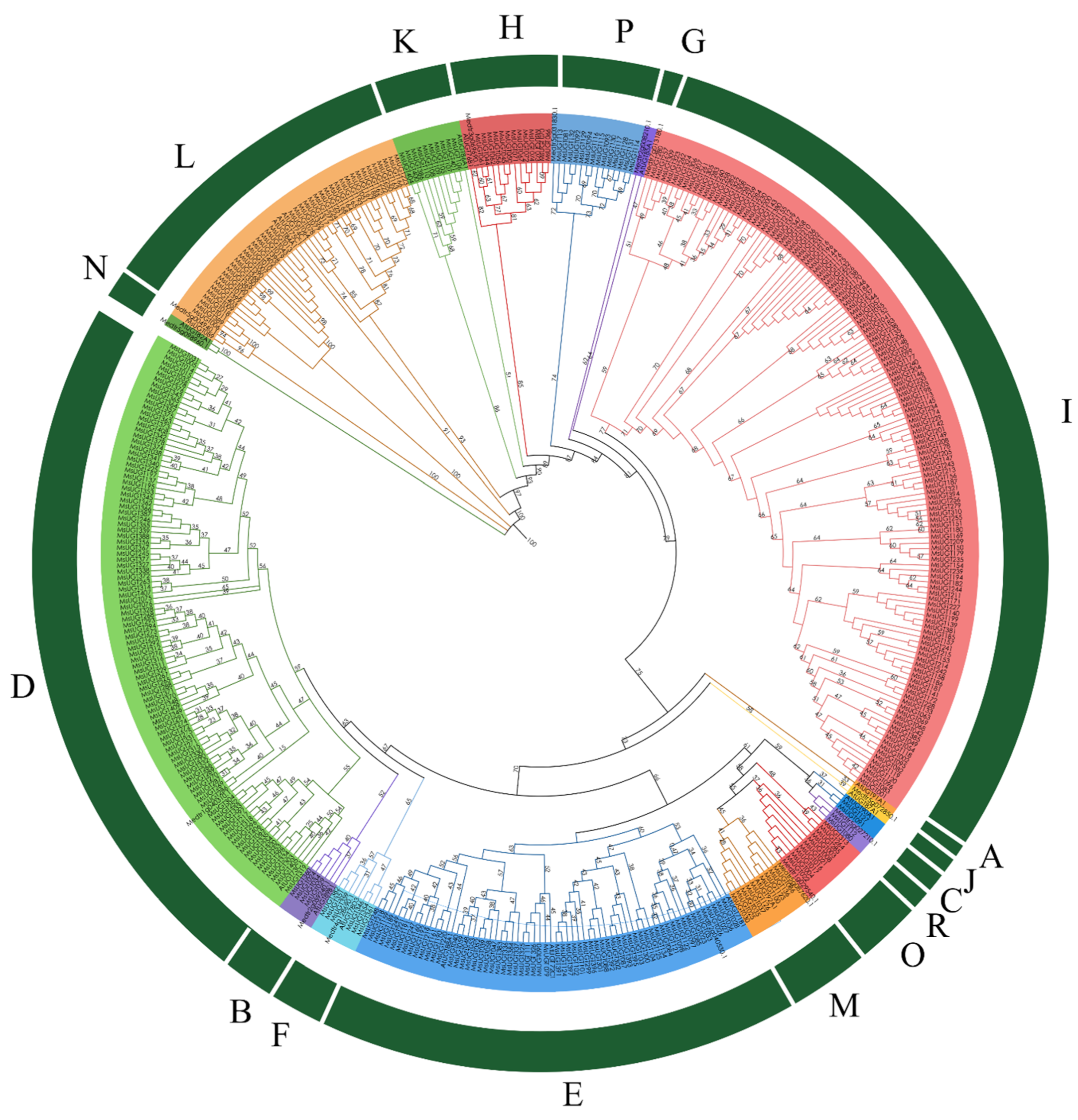
2.2. Phylogenetic Analysis of MsUGTs
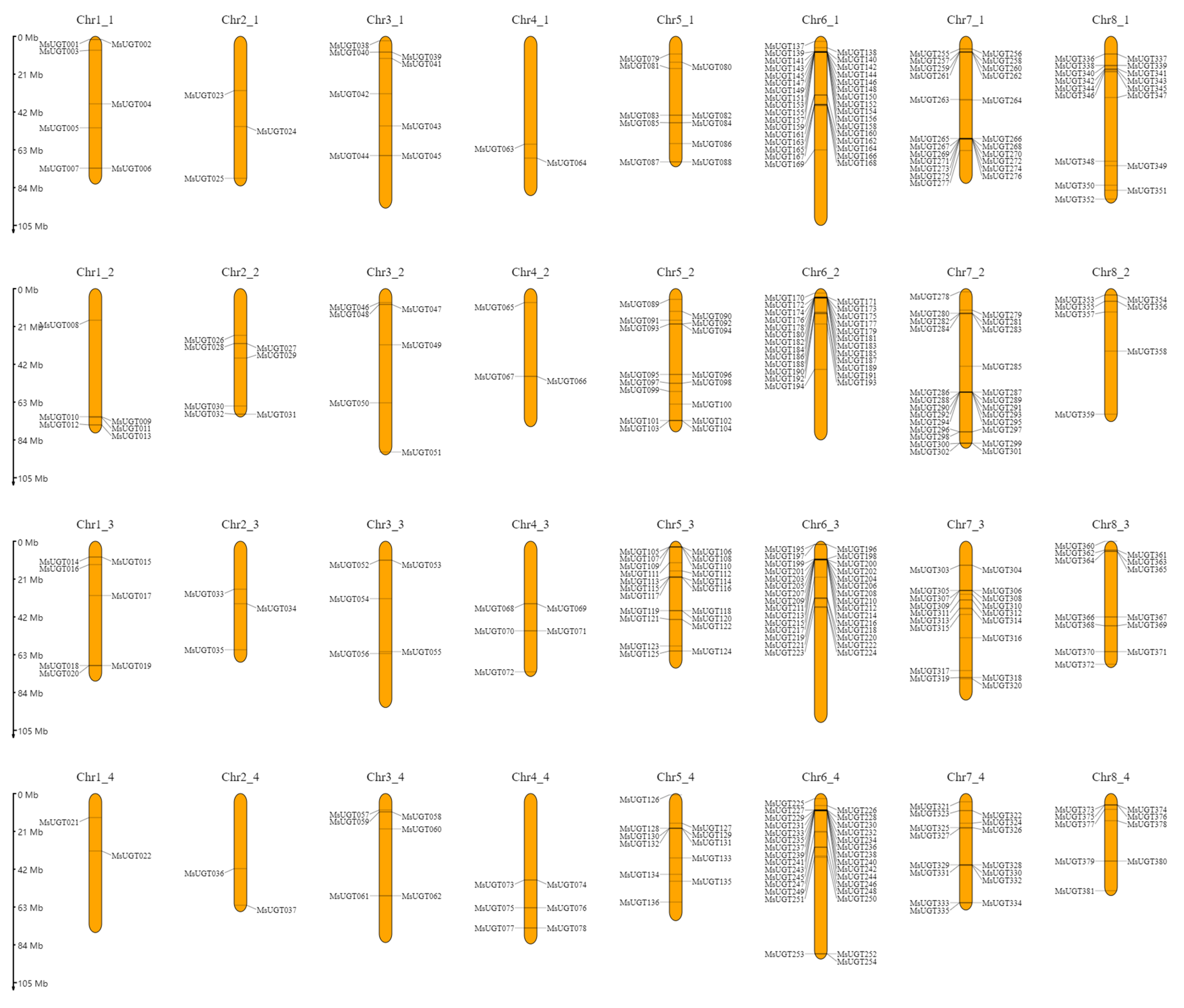
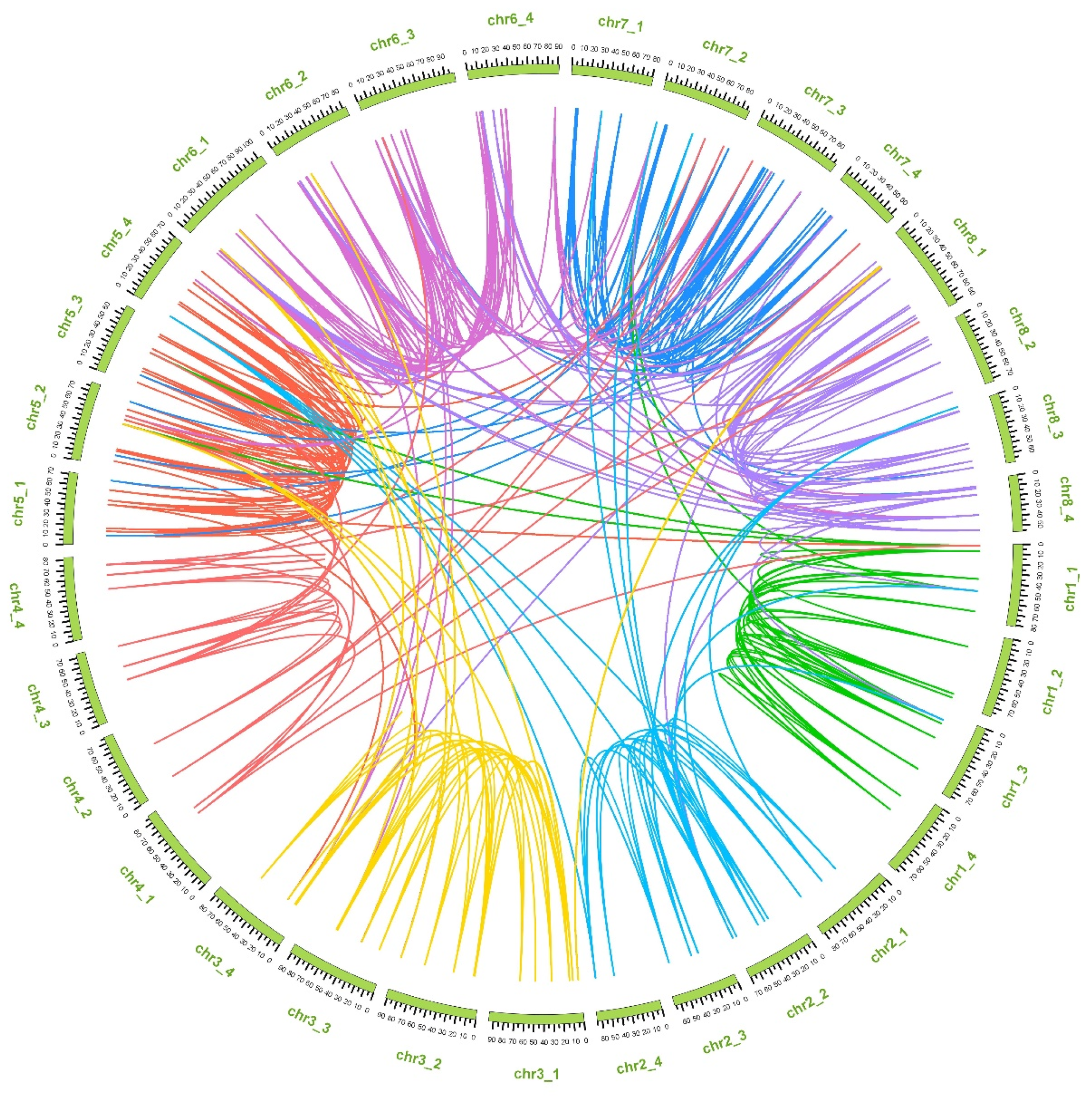
2.3. Genomic Localization and Synteny Analysis of MsUGT Genes
2.4. Conserved Motifs and Gene Structure of MsUGT Genes
2.5. Cis-Regulatory Elements Analysis in the MsUGT Gene Promoters
2.6. Expression Pattern of MsUGT Genes in Six Different Tissues
2.7. Expression Pattern Analysis of MsUGT Genes under Abiotic Stresses and ABA Treatments
2.8. Relative Water Content of Leaves under Drought Stress and ABA Treatments
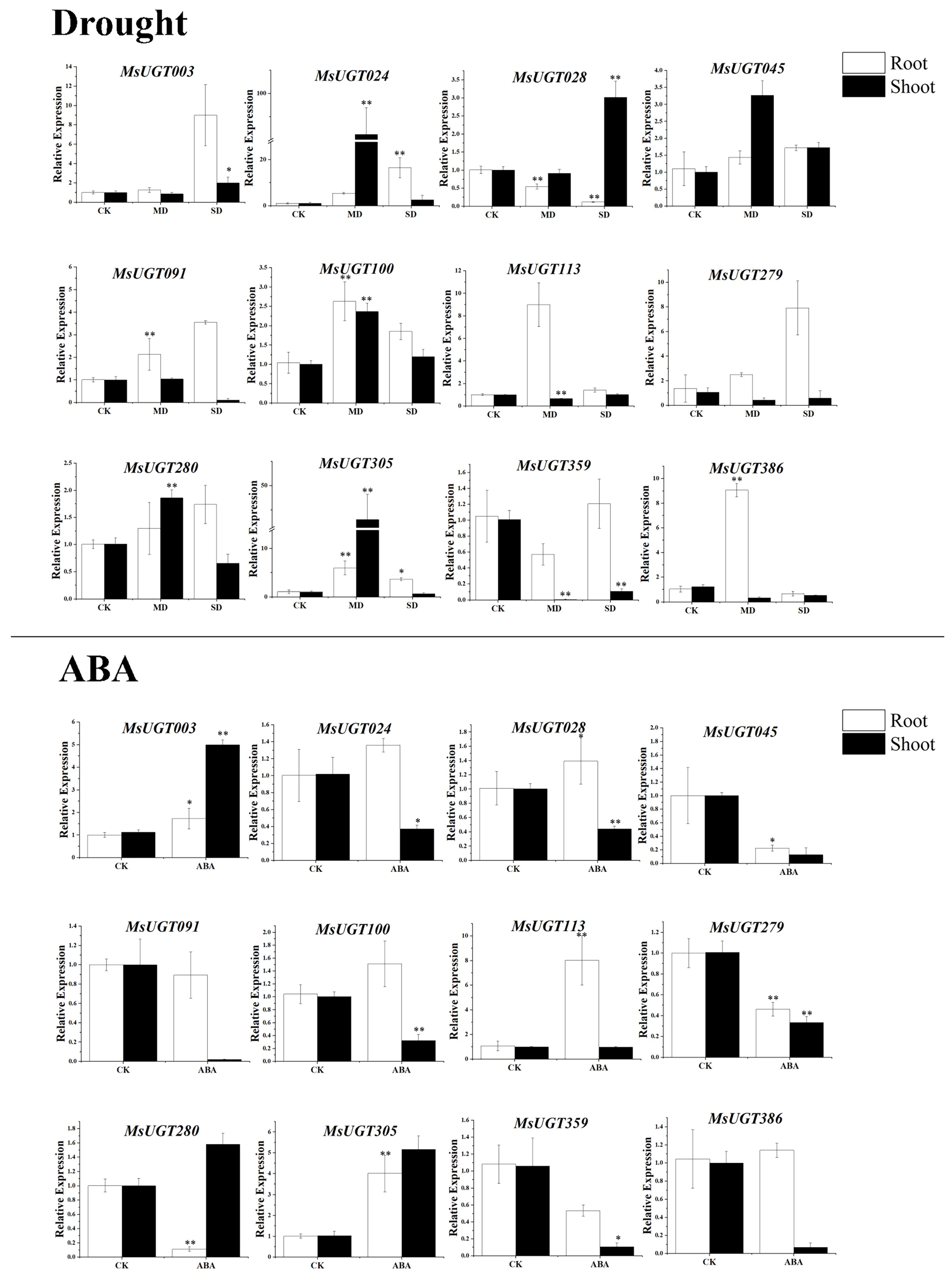
2.9. qRT-PCR Analysis of MsUGT Genes under Drought Stress and ABA Treatments
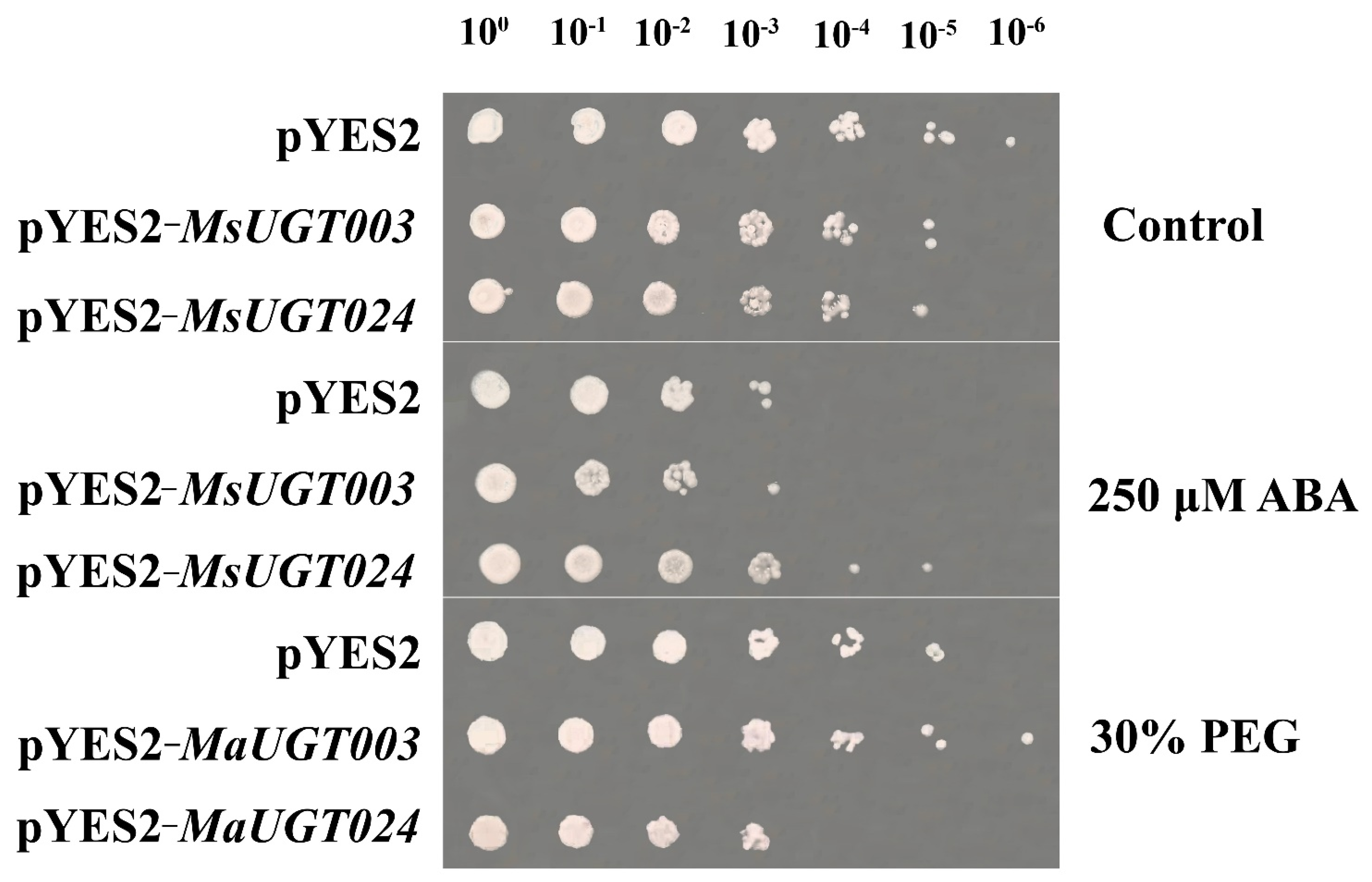
2.10. MsUGT003 and MsUGT024 in Response to Drought Tolerance and ABA Treatment in Yeast
3. Materials and Methods
3.1. Identification of UGT Genes in Alfalfa
3.2. Multiple Sequences Alignment and Phylogenetic Tree Construction
3.3. Chromosomal Locations of the MsUGT Genes and Gene Duplication Analysis
3.4. Gene Structure and Motifs’ Composition of the MsUGTs
3.5. Cis-Regulatory Element Analysis
3.6. Plant Materials, Relative Water Content, Drought and ABA Treatment
3.7. RNA seq and qRT-PCR Analysis
3.8. Heterologous Expression Validation in Yeast
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- O’Connell, E. Towards adaptation of water resource systems to climatic and socio-economic Chang. Water Resour. Manag. 2017, 31, 2965–2984. [Google Scholar] [CrossRef]
- Brodersen, C.; Roddy, A.; Wason, J.; McElrone, A. Functional status of xylem through time. Annu. Rev. Plant Biol. 2019, 70, 407–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aditi, G.; Andrés, R.; Ana, I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Salemi, F.; Esfahani, M.; Tran, L.S. Mechanistic insights into enhanced tolerance of early growth of alfalfa (Medicago sativa L.) under low water potential by seed-priming with ascorbic acid or polyethylene glycol solution. Ind. Crops Prod. 2019, 137, 436–445. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Meech, R.; Hu, D.G.; McKinnon, R.A.; Mubarokah, S.N.; Haines, A.Z.; Nair, P.C.; Rowland, A.; Mackenzie, P.I. The UDP-Glycosyltransferase (UGT) superfamily: New members, new functions, and novel paradigms. Physiol. Rev. 2019, 99, 1153–1222. [Google Scholar] [CrossRef]
- Mackenzie, P.I.; Owens, I.S.; Burchell, B.; Bock, K.W.; Bairoch, A.; Belanger, A.; Gigleux, S.F.; Green, M.; Hum, D.W.; Iyanagi, T. The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenet. Genom. 1997, 7, 255. [Google Scholar] [CrossRef]
- Yonekura, S.K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Jones, P.; Vogt, T. Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 2001, 213, 164–174. [Google Scholar] [CrossRef]
- Bowles, D. A multigene family of glycosyltransferases in a model plant, Arabidopsis thaliana. Biochem. Soc. Trans. 2002, 30, 301–306. [Google Scholar] [CrossRef]
- Wang, F.; Su, Y.; Chen, N.; Shen, S. Genome-wide analysis of the UGT gene family and identification of flavonoids in Broussonetia papyrifera. Molecules 2021, 26, 3449. [Google Scholar] [CrossRef] [PubMed]
- Humphries, A.W.; Ovalle, C.; Hughes, S.; Del, P.A.; Inostroza, L.; Barahona, V.; Yu, L.; Yerzhanova, S.; Rowe, T.; Hill, J.; et al. Characterization and pre-breeding of diverse alfalfa wild relatives originating from drought-stressed environments. Crop Sci. 2020, 61, 69–88. [Google Scholar] [CrossRef]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef]
- Yu, J.; Hu, F.; Dossa, K.; Wang, Z.; Ke, T. Genome-wide analysis of UDP-glycosyltransferase super family in Brassica rapa and Brassica oleracea reveals its evolutionary history and functional characterization. BMC Genomics 2017, 18, 474. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurthy, P.; Tsukamoto, C.; Ishimoto, M. Reconstruction of the evolutionary histories of UGT gene superfamily in Legumes Clarifies the functional divergence of duplicates in specialized metabolism. Int. J. Mol. Sci. 2020, 21, 1855. [Google Scholar] [CrossRef] [Green Version]
- Duan, Z.; Yan, Q.; Wu, F.; Wang, Y.; Wang, S.; Zong, X.; Zhou, P.; Zhang, J. Genome-wide analysis of the UDP-Glycosyltransferase family reveals its roles in coumarin biosynthesis and abiotic stress in Melilotus albus. Int. J. Mol. Sci. 2021, 22, 10826. [Google Scholar] [CrossRef]
- Veronicavon, S.P.; Zhang, W.; Kanawati, B.; Geist, B.; Faus, K.T.; Schmitt, K.P.; Schaffner, A.R. The arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 2011, 23, 4124–4145. [Google Scholar] [CrossRef] [Green Version]
- Gatti, M.; Cambon, F.; Tassy, C.; Macadre, C.; Guerard, F.; Langin, T.; Dufresne, M. The brachypodium distachyon UGT Bradi5gUGT03300 confers type II fusarium head blight resistance in wheat. Plant Pathol. 2019, 68, 334–343. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, B.; Jin, S.; Qu, X.; Li, Y.; Hou, B. Ectopic expression of Arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PLoS ONE 2013, 8, e59924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priest, D.M.; Ambrose, S.J.; Vaistij, F.E.; Elias, L.; Higgins, G.S.; Ross, A.R.; Abrams, S.R.; Bowles, D.J. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006, 46, 492–502. [Google Scholar] [CrossRef]
- Ma, Y.; Cao, J.; Chen, Q.; He, J.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. Abscisic acid receptors maintain abscisic acid homeostasis by modulating UGT71C5 glycosylation activity. J. Integr. Plant Biol. 2021, 63, 543–552. [Google Scholar] [CrossRef]
- Sun, Y.; Ji, K.; Liang, B.; Du, Y.; Jiang, L.; Wang, J.; Kai, W.; Zhang, Y.; Zhai, X.; Chen, P.; et al. Suppressing ABA uridine diphosphate glucosyltransferase (SlUGT75C1) alters fruit ripening and the stress response in tomato. Plant J. 2017, 91, 574–589. [Google Scholar] [CrossRef] [Green Version]
- Russell, A.E.; Laird, D.A.; Mallarino, A.P. Nitrogen fertilization and cropping system impacts on soil quality in midwestern mollisols. Soil Sci. Soc. Am. J. 2006, 70, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Radovic, J.; Sokolovic, D.; Markovic, J. Alfalfa-most important perennial forage legume in animal husbandry. Biotech. Anim. Husb. 2009, 25, 465–475. [Google Scholar] [CrossRef]
- Ernest, S. Adaptations to herbivory in alfalfa (Medicago sativa). Can. J. Bot. 1996, 74, 807–822. [Google Scholar] [CrossRef]
- Long, R.; Zhang, F.; Zhang, Z.; Li, M.; Chen, L.; Wang, X.; Liu, W.; Zhang, T.; Yu, L.; He, F.; et al. Genome assembly of alfalfa cultivar zhongmu-4 and identification of SNPs associated with agronomic traits. Genom. Proteom. Bioinf. 2022. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yang, X.; Lu, M.; Chen, J.; Shi, T. Gene expression and evolution of Family-1 UDP-glycosyltransferases—insights from an aquatic flowering plant (Sacred lotus). Aquat. Bot. 2020, 166, 103270. [Google Scholar] [CrossRef]
- Ross, J.; Yi, L.; Lim, E.K.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, reviews3004.1. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Chen, Y.; Lu, C.; Hwang, J. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef]
- Jiang, C.; Ying, K.; Qian, W.; Yu, S.; Guan, L. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas 2015, 37, 91–97. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkitdeveloped for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Guo, A.; Zhu, Q.; Chen, X.; Luo, J. GSDS: A gene structure display serve. Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van, P.Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Y.; Nan, Z. Relative and absolute quantification expression analysis of CsSAMDC gene as a case. China Biotech. 2009, 29, 86–91. [Google Scholar]
- Wang, J.; Hou, B. Glycosyltransferases: Key players involved in the modification of plant secondary metabolites. Front. Biol. China 2009, 4, 39–46. [Google Scholar] [CrossRef]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genomics 2012, 13, 175. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Lu, Q.; Liu, R.; Gong, J.; Gong, W.; Liu, A.; Ge, Q.; Li, J.; Shang, H.; Li, P.; et al. Genome-wide characterization of the UDP-glycosyltransferase gene family in upland cotton. Biotech 2019, 9, 453. [Google Scholar] [CrossRef]
- Mamoon, R.H.; Amjad, N.M.; Bao, L.; Hussain, S.Z.; Lee, J.M.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-wide analysis of Family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Niu, L.; Yang, Q.; Dong, B.; Wang, L.; Dong, M.; Fan, X.; Jian, Y.; Meng, D.; Fu, Y. Genome-wide identification and characterization of UGT family in pigeonpea (Cajanus cajan) and expression analysis in abiotic stress. Trees 2019, 33, 987–1002. [Google Scholar] [CrossRef]
- Cui, L.; Yao, S.; Dai, X.; Yin, Q.; Liu, Y.; Jiang, X.; Wu, Y.; Qian, Y.; Pang, Y.; Gao, L.; et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 2016, 67, 2285–2297. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.; Sun, Y.; Guo, T.; Shi, C.; Zhang, Y.; Kan, Y.; Xiang, Y.; Zhang, H.; Yang, Y.; Li, Y.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Dare, A.P.; Yauk, Y.K.; Tomes, S.; McGhie, T.K.; Rebstock, R.S.; Cooney, J.M.; Atkinson, R.G. Silencing a phloretin-specific glycosyltransferase perturbs both general phenylpropanoid biosynthesis and plant development. Plant J. 2017, 91, 237–250. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Lim, E.K.; Bowles, D.J. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2014, 23, 2915–2922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Medina, C.A.; Boge, B.; Hu, J.; Fransen, S.; Norberg, S.; Yu, L.X. Identification of genetic loci associated with forage quality in response to water deficit in autotetraploid alfalfa (Medicago sativa L.). BMC Plant Biol. 2020, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yu, H.; Meng, X.; Lin, J.; Li, Y.; Hou, B. Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biol. 2018, 20, 10–19. [Google Scholar] [CrossRef]
- Sauter, A.; Dietz, K.J.; Hartung, W. A possible stress physiological role of abscisic acid conjugates in root-to-shoot signalling. Plant Cell Environ. 2002, 25, 223–228. [Google Scholar] [CrossRef] [Green Version]


| Primer Name | Sequences |
|---|---|
| MsUGT280-F | TTTGTCCACAAGAACCACTATGAG |
| MsUGT280-R | TCCTTGGAAGACCACTTCGAG |
| MsUGT305-F | TTGATTCTGAAGGAAATCCCAC |
| MsUGT305-R | AATTGGTAGGAACTTTGGTGAC |
| MsUGT386-F | ACTGACCAACCAACAAATGCT |
| MsUGT386-R | TGCTAGAACTACCATCCTCCT |
| MsUGT045-F | TCTTACCCTCACTTCACTTTCC |
| MsUGT045-R | CCTCCATTCGAGATTCAGTCCT |
| MsUGT113-F | CCATTAGAGGAGACGAGGAC |
| MsUGT113-R | TATTGAGGTGGTGATGAAGAGG |
| MsUGT279-F | TAATACCGCTCAAAGGATTGCAG |
| MsUGT279-R | ATGAAGGTAAAGGTCCAATGGT |
| MsUGT359-F | CTCCAAGGAAGTCCATGATGTC |
| MsUGT359-R | GTGGTGTTGACAAGTTGAATGG |
| MsUGT003-F | AAGCTTCTTTACATCTTCGCGAG |
| MsUGT003-R | CATTTGTGCCATCATGAAATCGAC |
| MsUGT091-F | CACTTAACATCCACAAACTTTCAC |
| MsUGT091-R | CCTTTAGTAAGCTCGTCATGG |
| MsUGT024-F | TTCGGATGAGTTGGGAAGAG |
| MsUGT024-R | CAGAACTTCCACCTTCTCTAACAG |
| MsUGT100-F | GGAGCATTTCTAAGTCATTGTGG |
| MsUGT100-R | CACCCTTCTCTATTGTTGCCTC |
| MsUGT028-F | TTTCTTCCATTTCCATCTCCC |
| MsUGT028-R | GTATCCTAAATTGTTGTCACTGTC |
| MsMsUGT003-pYES2-F | cttggtaccgagctcggatccATGGTTCCGTCTTTTGAAG |
| MsMsUGT003-pYES2-R | tacatgatgcggccctctagaCTATCTAGTAATATGAGCGATGAAAG |
| MsMsUGT024-pYES2-F | cttggtaccgagctcggatccATGACGTTTCAACCAGGC |
| MsMsUGT024-pYES2-R | tacatgatgcggccctctagaCTAAAATTGTAACACAAGTTGTTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ao, B.; Han, Y.; Wang, S.; Wu, F.; Zhang, J. Genome-Wide Analysis and Profile of UDP-Glycosyltransferases Family in Alfalfa (Medicago sativa L.) under Drought Stress. Int. J. Mol. Sci. 2022, 23, 7243. https://doi.org/10.3390/ijms23137243
Ao B, Han Y, Wang S, Wu F, Zhang J. Genome-Wide Analysis and Profile of UDP-Glycosyltransferases Family in Alfalfa (Medicago sativa L.) under Drought Stress. International Journal of Molecular Sciences. 2022; 23(13):7243. https://doi.org/10.3390/ijms23137243
Chicago/Turabian StyleAo, Bao, Yangyang Han, Shengsheng Wang, Fan Wu, and Jiyu Zhang. 2022. "Genome-Wide Analysis and Profile of UDP-Glycosyltransferases Family in Alfalfa (Medicago sativa L.) under Drought Stress" International Journal of Molecular Sciences 23, no. 13: 7243. https://doi.org/10.3390/ijms23137243






