Effects of Ethanol on Expression of Coding and Noncoding RNAs in Murine Neuroblastoma Neuro2a Cells
Abstract
1. Introduction
2. Results
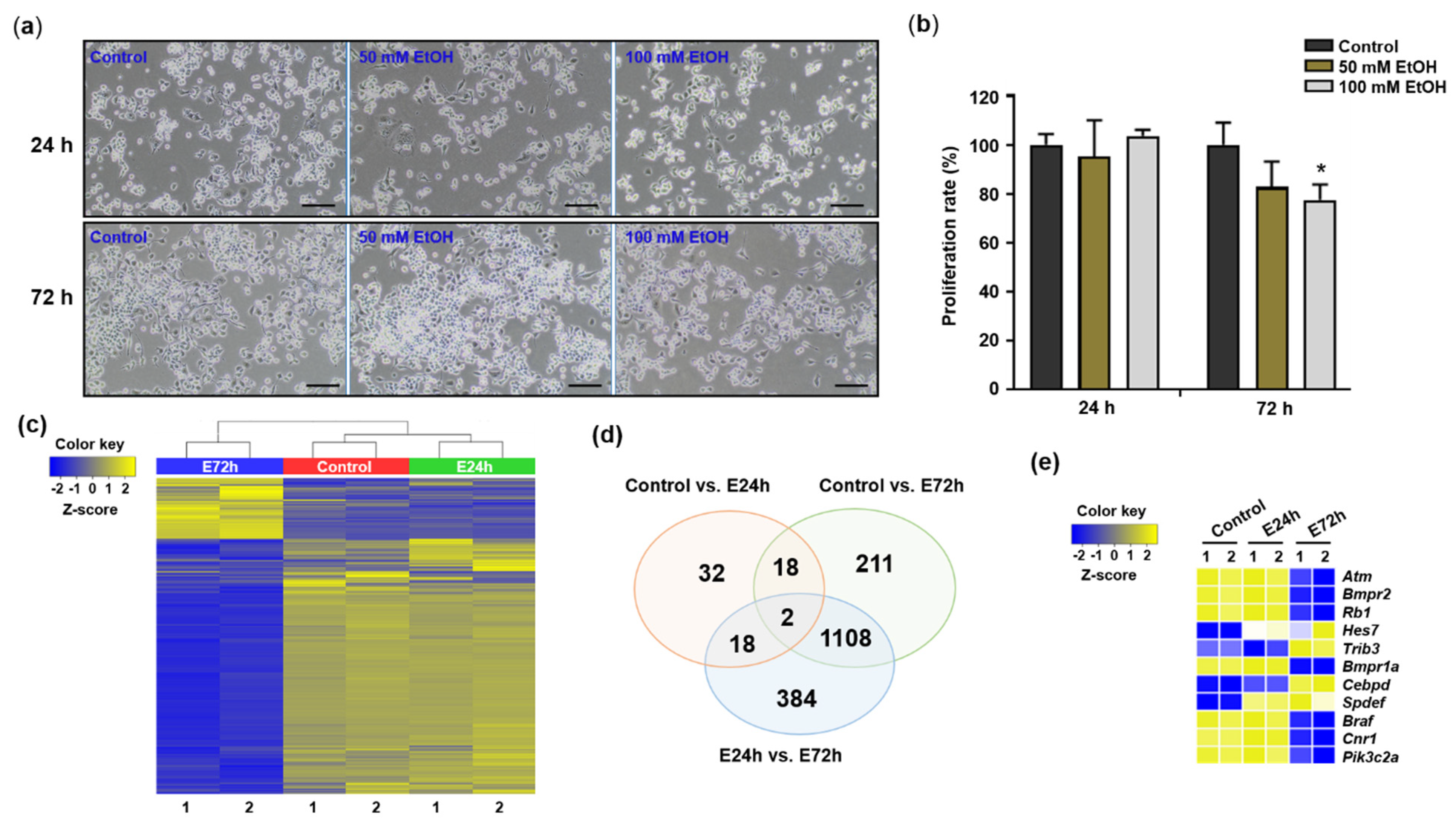
2.1. Identification of Genes Differentially Expressed in Response to Ethanol
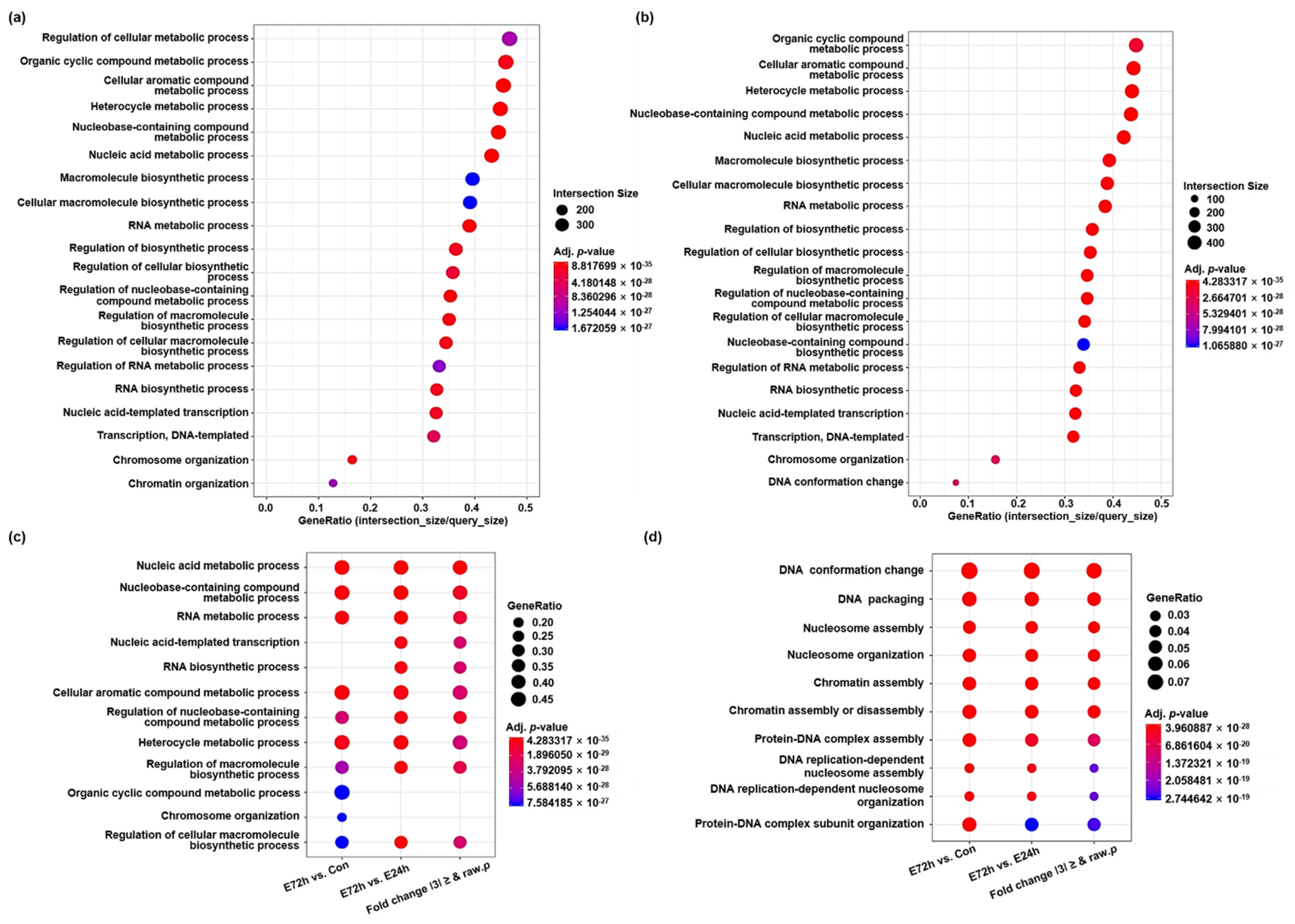
2.2. Functional Annotation and Pathway Network of DEGs
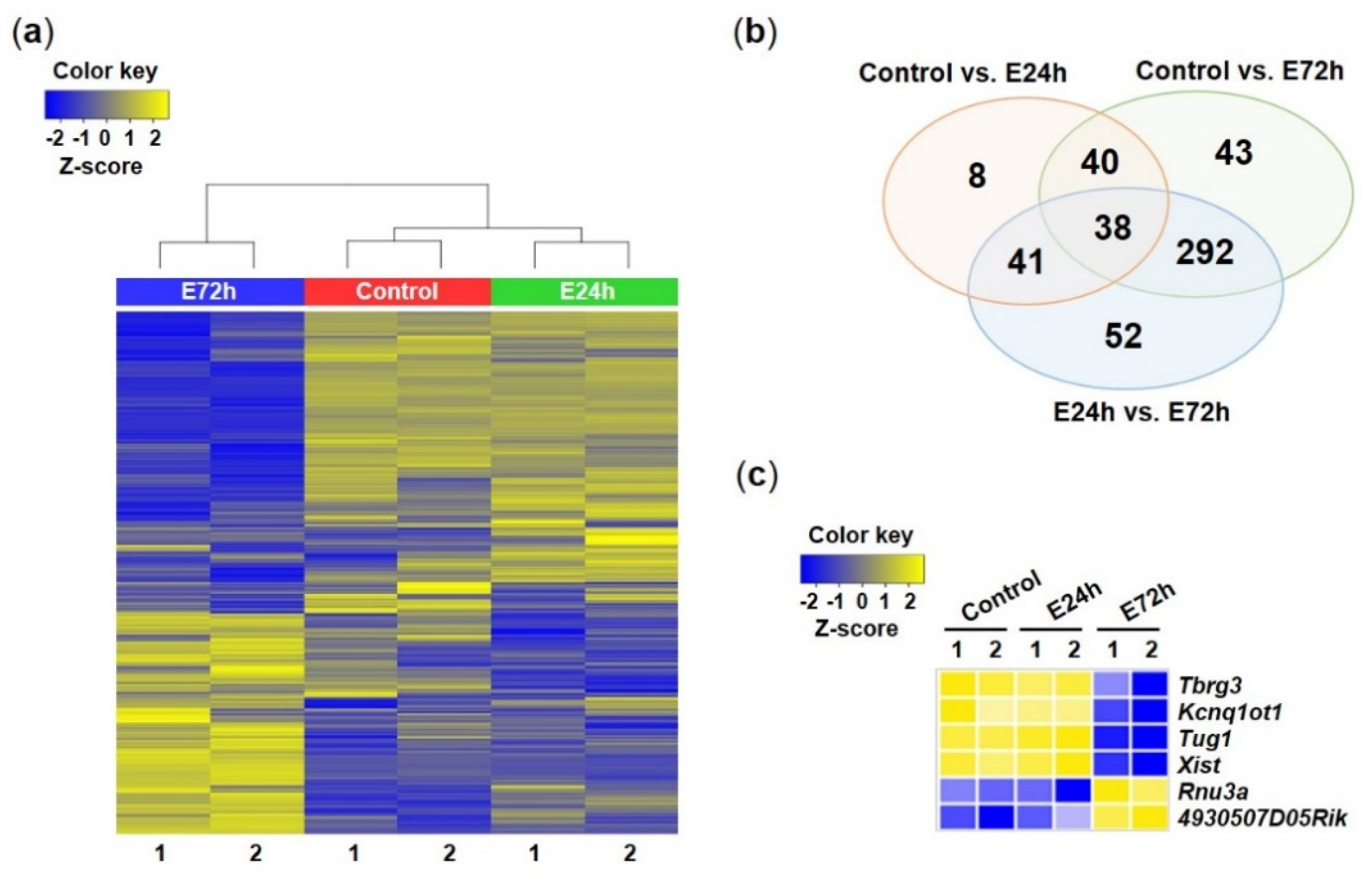
2.3. Identification of lncRNAs Differentially Expressed in Response to Ethanol
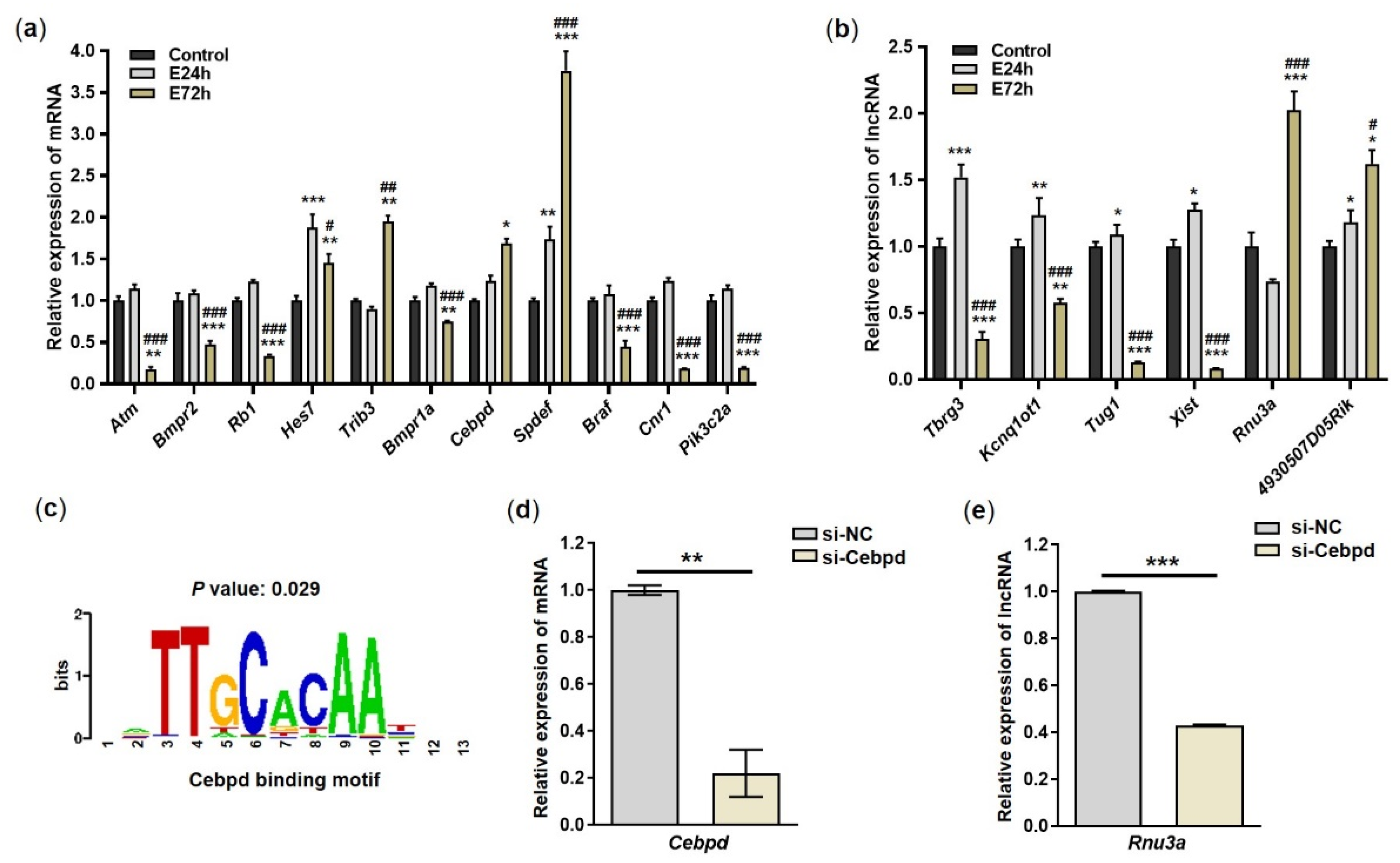
2.4. Validation of mRNAs and lncRNAs Based on RT-qPCR
2.5. Knock-Down of Cebpd
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Alcohol Treatment and Cell Proliferation Assay
4.3. Total RNA-Seq Library Preparation and Sequencing
4.4. Differential Gene Expression Analysis and Enrichment
4.5. RT-Quantitative PCR (RT-qPCR)
4.6. Knock-Down of Cebpd
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ADH | alcohol dehydrogenase |
| ALDH | aldehyde dehydrogenase |
| BP | biological process |
| CB1 | cannabinoid receptor 1 |
| CCK-8 | Cell Counting Kit-8 |
| CP | cellular component |
| DEGs | differentially expressed genes |
| DMEM | Dulbecco’s modified Eagle’s medium |
| ER | endoplasmic reticulum |
| ESE | epithelium-specific ETS |
| FBS | fetal bovine serum |
| FDR | false discovery rate |
| FPKM | fragments per kilobase million |
| GO | gene ontology |
| lncRNAs | long noncoding RNAs |
| MF | molecular function |
| miRNAs | microRNAs |
| ncRNAs | non-coding RNAs |
| NMDARs | N-methyl-D-aspartate receptors |
| NP | nucleus pulposus |
| OGD | oxygen and glucose deprivation |
| OGD/R | OGD/reoxygen |
| RIN | RNA integrity number |
| ROS | reactive oxygen species |
| RT-qPCR | RT-quantitative PCR |
| SEM | Standard error of the mean |
| si-NC | siRNA sequences for the negative control |
| siRNAs | Small interference RNAs |
References
- Fadda, F.; Rossetti, Z.L. Chronic ethanol consumption: From neuroadaptation to neurodegeneration. Prog. Neurobiol. 1998, 56, 385–431. [Google Scholar] [CrossRef]
- Tabakoff, B.; Hoffman, P.L. The neurobiology of alcohol consumption and alcoholism: An integrative history. Pharmacol. Biochem. Behav. 2013, 113, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Heilig, M.; Goldman, D.; Berrettini, W.; O’Brien, C.P. Pharmacogenetic approaches to the treatment of alcohol addiction. Nat. Rev. Neurosci. 2011, 12, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Kauer, J.A.; Malenka, R.C. Synaptic plasticity and addiction. Nat. Rev. Neurosci. 2007, 8, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Koob, G.F.; Volkow, N.D. Neurocircuitry of addiction. Neuropsychopharmacology 2010, 35, 217–238. [Google Scholar] [CrossRef]
- Choi, M.R.; Han, J.S.; Chai, Y.G.; Jin, Y.B.; Lee, S.R.; Kim, D.J. Gene expression profiling in the hippocampus of adolescent rats after chronic alcohol administration. Basic Clin. Pharmacol. Toxicol. 2020, 126, 389–398. [Google Scholar] [CrossRef]
- Choi, M.R.; Jung, K.H.; Park, J.H.; Das, N.D.; Chung, M.K.; Choi, I.G.; Lee, B.C.; Park, K.S.; Chai, Y.G. Ethanol-induced small heat shock protein genes in the differentiation of mouse embryonic neural stem cells. Arch. Toxicol. 2011, 85, 293–304. [Google Scholar] [CrossRef]
- Mandal, C.; Park, J.H.; Choi, M.R.; Kim, S.H.; Badejo, A.C.; Chai, J.C.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. Transcriptomic study of mouse embryonic neural stem cell differentiation under ethanol treatment. Mol. Biol. Rep. 2015, 42, 1233–1239. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Kreek, M.J. mTORC1 pathway is involved in the kappa opioid receptor activation-induced increase in excessive alcohol drinking in mice. Pharmacol. Biochem. Behav. 2020, 195, 172954. [Google Scholar] [CrossRef]
- Bogenpohl, J.W.; Smith, M.L.; Farris, S.P.; Dumur, C.I.; Lopez, M.F.; Becker, H.C.; Grant, K.A.; Miles, M.F. Cross-Species Co-analysis of Prefrontal Cortex Chronic Ethanol Transcriptome Responses in Mice and Monkeys. Front. Mol. Neurosci. 2019, 12, 197. [Google Scholar] [CrossRef]
- Smith, M.L.; Lopez, M.F.; Wolen, A.R.; Becker, H.C.; Miles, M.F. Brain regional gene expression network analysis identifies unique interactions between chronic ethanol exposure and consumption. PLoS ONE 2020, 15, e0233319. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Louro, R.; Smirnova, A.S.; Verjovski-Almeida, S. Long intronic noncoding RNA transcription: Expression noise or expression choice? Genomics 2009, 93, 291–298. [Google Scholar] [CrossRef]
- Heo, J.B.; Lee, Y.S.; Sung, S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 2013, 21, 685–693. [Google Scholar] [CrossRef]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Sushma; Divakar, A.; Kanchan, S.; Jha, G.; Mishra, S.; Sharma, D.; Rath, S.K. Alcohol induced impairment/abnormalities in brain: Role of MicroRNAs. Neurotoxicology 2021, 87, 11–23. [Google Scholar] [CrossRef]
- Choi, M.R.; Han, J.S.; Jin, Y.B.; Lee, S.R.; Choi, I.Y.; Lee, H.; Cho, H.; Kim, D.J. Differential expression of microRNAs in the hippocampi of male and female rodents after chronic alcohol administration. Biol. Sex. Differ. 2020, 11, 65. [Google Scholar] [CrossRef]
- Kong, H.; Yin, F.; He, F.; Omran, A.; Li, L.; Wu, T.; Wang, Y.; Peng, J. The Effect of miR-132, miR-146a, and miR-155 on MRP8/TLR4-Induced Astrocyte-Related Inflammation. J. Mol. Neurosci. 2015, 57, 28–37. [Google Scholar] [CrossRef]
- Miranda, R.C. MicroRNAs and Fetal Brain Development: Implications for Ethanol Teratology during the Second Trimester Period of Neurogenesis. Front. Genet. 2012, 3, 77. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.C.; Bake, S.; Balaraman, S.; Rawlings, J.; Holgate, R.R.; Dubois, D.; Miranda, R.C. MiR-153 targets the nuclear factor-1 family and protects against teratogenic effects of ethanol exposure in fetal neural stem cells. Biol. Open 2014, 3, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.; McMichael, G.O.; Vornholt, E.S.; Cresswell, K.; Williamson, V.; Chatzinakos, C.; Mamdani, M.; Hariharan, S.; Kendler, K.S.; Kalsi, G.; et al. Assessing the Role of Long Noncoding RNA in Nucleus Accumbens in Subjects With Alcohol Dependence. Alcohol. Clin. Exp. Res. 2020, 44, 2468–2480. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, M.; Shen, Z.; Bu, F.; Yu, H.; Pan, X.; Yang, Y.; Meng, X.; Huang, C.; Li, J. The Long Non-coding RNA MEG3/miR-let-7c-5p Axis Regulates Ethanol-Induced Hepatic Steatosis and Apoptosis by Targeting NLRC5. Front. Pharmacol. 2018, 9, 302. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ross, R.A.; Zhao, S.; Tu, W.; Liangpunsakul, S.; Wang, L. LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis. Hepatol. Commun. 2017, 1, 513–523. [Google Scholar] [CrossRef]
- Zorumski, C.F.; Mennerick, S.; Izumi, Y. Acute and chronic effects of ethanol on learning-related synaptic plasticity. Alcohol 2014, 48, 1–17. [Google Scholar] [CrossRef]
- Dasgupta, B.; Milbrandt, J. Resveratrol stimulates AMP kinase activity in neurons. Proc. Natl. Acad. Sci. USA 2007, 104, 7217–7222. [Google Scholar] [CrossRef]
- De Girolamo, L.A.; Billett, E.E.; Hargreaves, A.J. Effects of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine on differentiating mouse N2a neuroblastoma cells. J. Neurochem. 2000, 75, 133–140. [Google Scholar] [CrossRef]
- Katiyar, P.; Banerjee, S.; Nathani, S.; Roy, P. Triclosan-induced neuroinflammation develops caspase-independent and TNF-α signaling pathway associated necroptosis in Neuro-2a cells. Curr. Res. Toxicol. 2022, 3, 100072. [Google Scholar] [CrossRef]
- Chen, G.; Bower, K.A.; Xu, M.; Ding, M.; Shi, X.; Ke, Z.J.; Luo, J. Cyanidin-3-glucoside reverses ethanol-induced inhibition of neurite outgrowth: Role of glycogen synthase kinase 3 Beta. Neurotox. Res. 2009, 15, 321–331. [Google Scholar] [CrossRef]
- Saito, M.; Saito, M.; Cooper, T.B.; Vadasz, C. Ethanol-induced changes in the content of triglycerides, ceramides, and glucosylceramides in cultured neurons. Alcohol. Clin. Exp. Res. 2005, 29, 1374–1383. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, S.N. Neuroglial and neuroblastoma cell lines are capable of metabolizing ethanol via an alcohol-dehydrogenase-independent pathway. Alcohol. Clin. Exp. Res. 1987, 11, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Zhang, S.Y.; Xu, X.M.; Zhao, S.; Zhu, K.H.; Zhang, W.W.; Zhao, L.L. Alcohol inhibits the proliferation of Neuro2a cells via promoting the asymmetric cell division through down-regulation of the expression of centrosome protein-J. Toxicol. Lett. 2018, 294, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Gery, S.; Tanosaki, S.; Hofmann, W.K.; Koppel, A.; Koeffler, H.P. C/EBPdelta expression in a BCR-ABL-positive cell line induces growth arrest and myeloid differentiation. Oncogene 2005, 24, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Noritake, K.; Aki, T.; Uemura, K. Possible roles of AMPK and macropinocytosis in the defense responses against Δ(9)-THC toxicity on HL-1 cardiomyocytes. Toxicol. Rep. 2021, 8, 980–987. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, X.; Zhou, Y.; Jiang, Y.; Wu, R.; Lu, C. Pterostilbene attenuates RIPK3-dependent hepatocyte necroptosis in alcoholic liver disease via SIRT2-mediated NFATc4 deacetylation. Toxicology 2021, 461, 152923. [Google Scholar] [CrossRef]
- Edenberg, H.J. The genetics of alcohol metabolism: Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health 2007, 30, 5–13. [Google Scholar]
- Abrahao, K.P.; Salinas, A.G.; Lovinger, D.M. Alcohol and the Brain: Neuronal Molecular Targets, Synapses, and Circuits. Neuron 2017, 96, 1223–1238. [Google Scholar] [CrossRef]
- Cui, C.; Koob, G.F. Titrating Tipsy Targets: The Neurobiology of Low-Dose Alcohol. Trends Pharmacol. Sci. 2017, 38, 556–568. [Google Scholar] [CrossRef]
- Yang, C.; Hao, J.; Kong, D.; Cui, X.; Zhang, W.; Wang, H.; Guo, X.; Ma, S.; Liu, X.; Pu, P.; et al. ATM-mediated Mad1 Serine 214 phosphorylation regulates Mad1 dimerization and the spindle assembly checkpoint. Carcinogenesis 2014, 35, 2007–2013. [Google Scholar] [CrossRef][Green Version]
- Cirotti, C.; Filomeni, G. ATM plays antioxidant, boosting mitophagy via denitrosylation. Autophagy 2021, 17, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.; Nakamura, T. Tribbles in disease: Signaling pathways important for cellular function and neoplastic transformation. Cancer Sci. 2011, 102, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Han, Y.; Zhao, Y.; Li, Q.; Jin, H.; Qin, J. Inhibition of TRIB3 Protects Against Neurotoxic Injury Induced by Kainic Acid in Rats. Front. Pharmacol. 2019, 10, 585. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sarkar, T.R.; Zhou, M.; Sharan, S.; Ritt, D.A.; Veenstra, T.D.; Morrison, D.K.; Huang, A.M.; Sterneck, E. CCAAT/enhancer binding protein delta (C/EBPdelta, CEBPD)-mediated nuclear import of FANCD2 by IPO4 augments cellular response to DNA damage. Proc. Natl. Acad. Sci. USA 2010, 107, 16131–16136. [Google Scholar] [CrossRef]
- Wang, S.M.; Lim, S.W.; Wang, Y.H.; Lin, H.Y.; Lai, M.D.; Ko, C.Y.; Wang, J.M. Astrocytic CCAAT/Enhancer-binding protein delta contributes to reactive oxygen species formation in neuroinflammation. Redox Biol. 2018, 16, 104–112. [Google Scholar] [CrossRef]
- Chen, Y.; Qin, C.; Huang, J.; Tang, X.; Liu, C.; Huang, K.; Xu, J.; Guo, G.; Tong, A.; Zhou, L. The role of astrocytes in oxidative stress of central nervous system: A mixed blessing. Cell Prolif. 2020, 53, e12781. [Google Scholar] [CrossRef]
- Ko, C.Y.; Wang, W.L.; Wang, S.M.; Chu, Y.Y.; Chang, W.C.; Wang, J.M. Glycogen synthase kinase-3β-mediated CCAAT/enhancer-binding protein delta phosphorylation in astrocytes promotes migration and activation of microglia/macrophages. Neurobiol. Aging 2014, 35, 24–34. [Google Scholar] [CrossRef]
- Luk, I.Y.; Reehorst, C.M.; Mariadason, J.M. ELF3, ELF5, EHF and SPDEF Transcription Factors in Tissue Homeostasis and Cancer. Molecules 2018, 23, 2191. [Google Scholar] [CrossRef]
- Cheng, X.H.; Black, M.; Ustiyan, V.; Le, T.; Fulford, L.; Sridharan, A.; Medvedovic, M.; Kalinichenko, V.V.; Whitsett, J.A.; Kalin, T.V. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet. 2014, 10, e1004656. [Google Scholar] [CrossRef]
- Feldman, R.J.; Sementchenko, V.I.; Gayed, M.; Fraig, M.M.; Watson, D.K. Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res. 2003, 63, 4626–4631. [Google Scholar]
- Ceccarini, J.; Hompes, T.; Verhaeghen, A.; Casteels, C.; Peuskens, H.; Bormans, G.; Claes, S.; Van Laere, K. Changes in cerebral CB1 receptor availability after acute and chronic alcohol abuse and monitored abstinence. J. Neurosci. 2014, 34, 2822–2831. [Google Scholar] [CrossRef] [PubMed]
- Mitrirattanakul, S.; López-Valdés, H.E.; Liang, J.; Matsuka, Y.; Mackie, K.; Faull, K.F.; Spigelman, I. Bidirectional alterations of hippocampal cannabinoid 1 receptors and their endogenous ligands in a rat model of alcohol withdrawal and dependence. Alcohol. Clin. Exp. Res. 2007, 31, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, S.; Oliva, J.M.; Pérez-Rial, S.; Palomo, T.; Manzanares, J. Chronic ethanol consumption regulates cannabinoid CB1 receptor gene expression in selected regions of rat brain. Alcohol Alcohol. 2004, 39, 88–92. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clark, M.B.; Mattick, J.S. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. 2011, 22, 366–376. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.T. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 2018, 9, 613–626. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Liu, Z.; Lin, B.; Deng, X.; Xiao, Q.; Chen, Z.; Ye, H.; Chen, D.; Su, Y.; et al. Downregulation of Kcnq1ot1 attenuates β-cell proliferation and insulin secretion via the miR-15b-5p/Ccnd1 and Ccnd2 axis. Acta Diabetol. 2022, 59, 885–899. [Google Scholar] [CrossRef]
- Ding, R.; Wei, S.; Huang, M. Long non-coding RNA KCNQ1OT1 overexpression promotes osteogenic differentiation of staphylococcus aureus-infected human bone mesenchymal stem cells by sponging microRNA miR-29b-3p. Bioengineered 2022, 13, 5855–5867. [Google Scholar] [CrossRef]
- Wang, J.; Niu, Y.; Tao, H.; Xue, M.; Wan, C. Knockdown of lncRNA TUG1 inhibits hippocampal neuronal apoptosis and participates in aerobic exercise-alleviated vascular cognitive impairment. Biol. Res. 2020, 53, 53. [Google Scholar] [CrossRef]
- Du, J.; Li, W.; Wang, B. Long non-coding RNA TUG1 aggravates cerebral ischemia and reperfusion injury by sponging miR-493-3p/miR-410-3p. Open Med. (Wars) 2021, 16, 919–930. [Google Scholar] [CrossRef]
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 645647. [Google Scholar] [CrossRef]
- Shieh, T.M.; Liu, C.J.; Hsia, S.M.; Ningrum, V.; Liao, C.C.; Lan, W.C.; Shih, Y.H. Lack of Salivary Long Non-Coding RNA XIST Expression Is Associated with Increased Risk of Oral Squamous Cell Carcinoma: A Cross-Sectional Study. J. Clin. Med. 2021, 10, 4622. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Yang, L.; Huang, R.; Lin, L.; Shen, Y.; Cheng, L.; Jin, L.; Wang, S.; Zhu, R. Activation of CB2R with AM1241 ameliorates neurodegeneration via the Xist/miR-133b-3p/Pitx3 axis. J. Cell. Physiol. 2020, 235, 6032–6042. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Gribskov, M.; Nakai, K.; Schönbach, C. Encyclopedia of Bioinformatics and Computational Biology; Academic Press: Oxford, UK, 2019; pp. 230–240. [Google Scholar]
- Veras, M.A.; McCann, M.R.; Tenn, N.A.; Séguin, C.A. Transcriptional profiling of the murine intervertebral disc and age-associated changes in the nucleus pulposus. Connect. Tissue Res. 2020, 61, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, M.R.; Cho, S.; Kim, D.-J.; Choi, J.-S.; Jin, Y.-B.; Kim, M.; Chang, H.J.; Jeon, S.H.; Yang, Y.D.; Lee, S.-R. Effects of Ethanol on Expression of Coding and Noncoding RNAs in Murine Neuroblastoma Neuro2a Cells. Int. J. Mol. Sci. 2022, 23, 7294. https://doi.org/10.3390/ijms23137294
Choi MR, Cho S, Kim D-J, Choi J-S, Jin Y-B, Kim M, Chang HJ, Jeon SH, Yang YD, Lee S-R. Effects of Ethanol on Expression of Coding and Noncoding RNAs in Murine Neuroblastoma Neuro2a Cells. International Journal of Molecular Sciences. 2022; 23(13):7294. https://doi.org/10.3390/ijms23137294
Chicago/Turabian StyleChoi, Mi Ran, Sinyoung Cho, Dai-Jin Kim, Jung-Seok Choi, Yeung-Bae Jin, Miran Kim, Hye Jin Chang, Seong Ho Jeon, Young Duk Yang, and Sang-Rae Lee. 2022. "Effects of Ethanol on Expression of Coding and Noncoding RNAs in Murine Neuroblastoma Neuro2a Cells" International Journal of Molecular Sciences 23, no. 13: 7294. https://doi.org/10.3390/ijms23137294
APA StyleChoi, M. R., Cho, S., Kim, D.-J., Choi, J.-S., Jin, Y.-B., Kim, M., Chang, H. J., Jeon, S. H., Yang, Y. D., & Lee, S.-R. (2022). Effects of Ethanol on Expression of Coding and Noncoding RNAs in Murine Neuroblastoma Neuro2a Cells. International Journal of Molecular Sciences, 23(13), 7294. https://doi.org/10.3390/ijms23137294





