Binding Networks Identify Targetable Protein Pockets for Mechanism-Based Drug Design
Abstract
1. Introduction
2. Results and Discussion
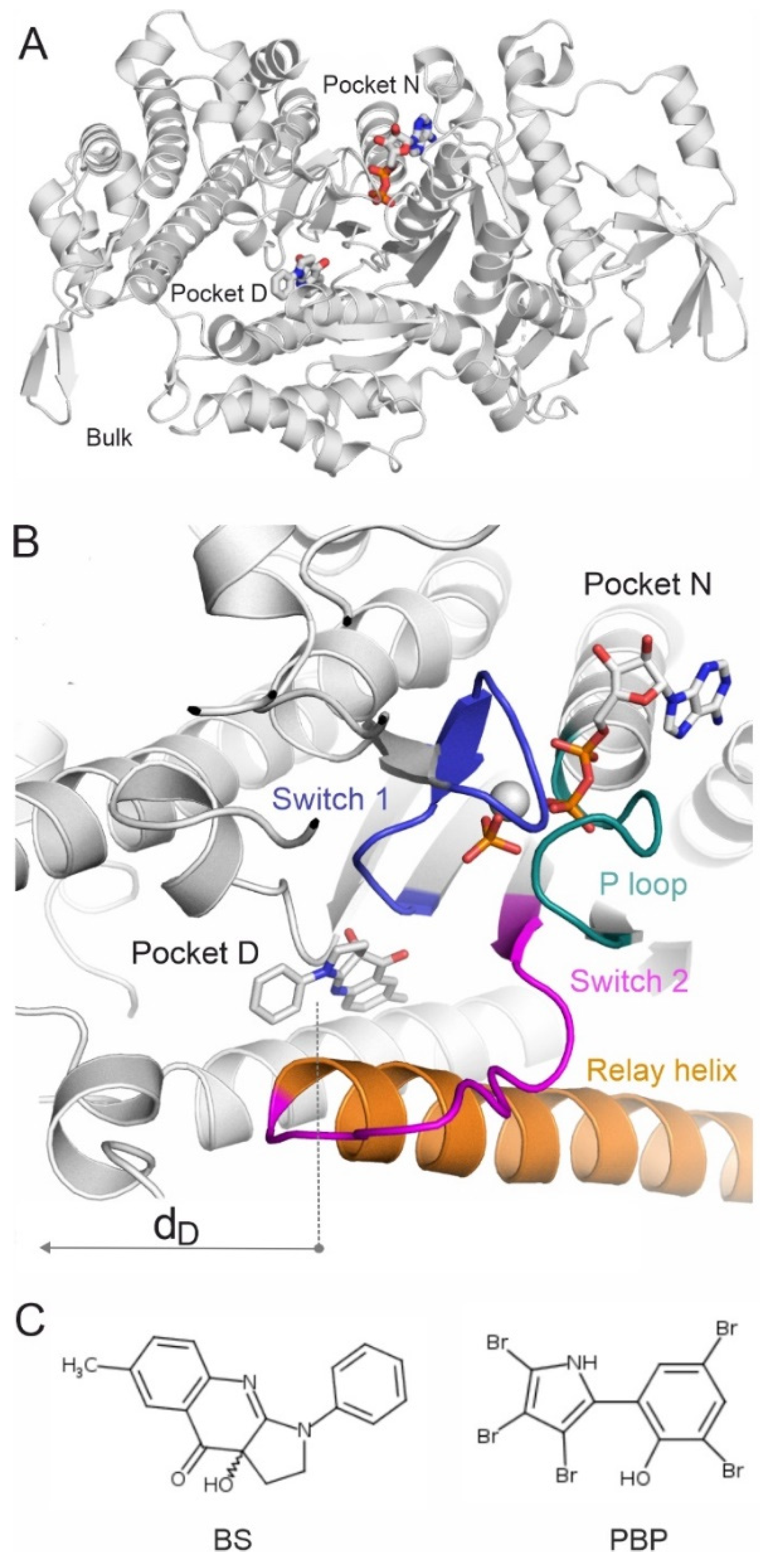
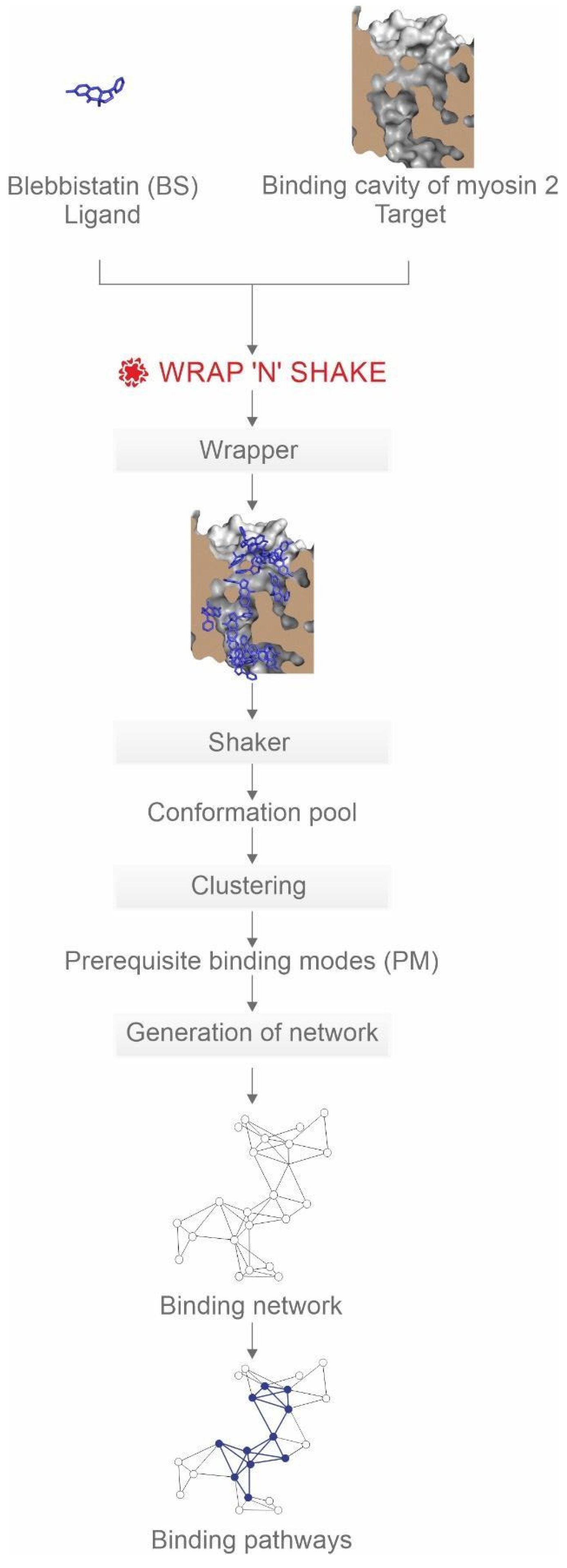
2.1. Systematic Mapping of Binding Modes
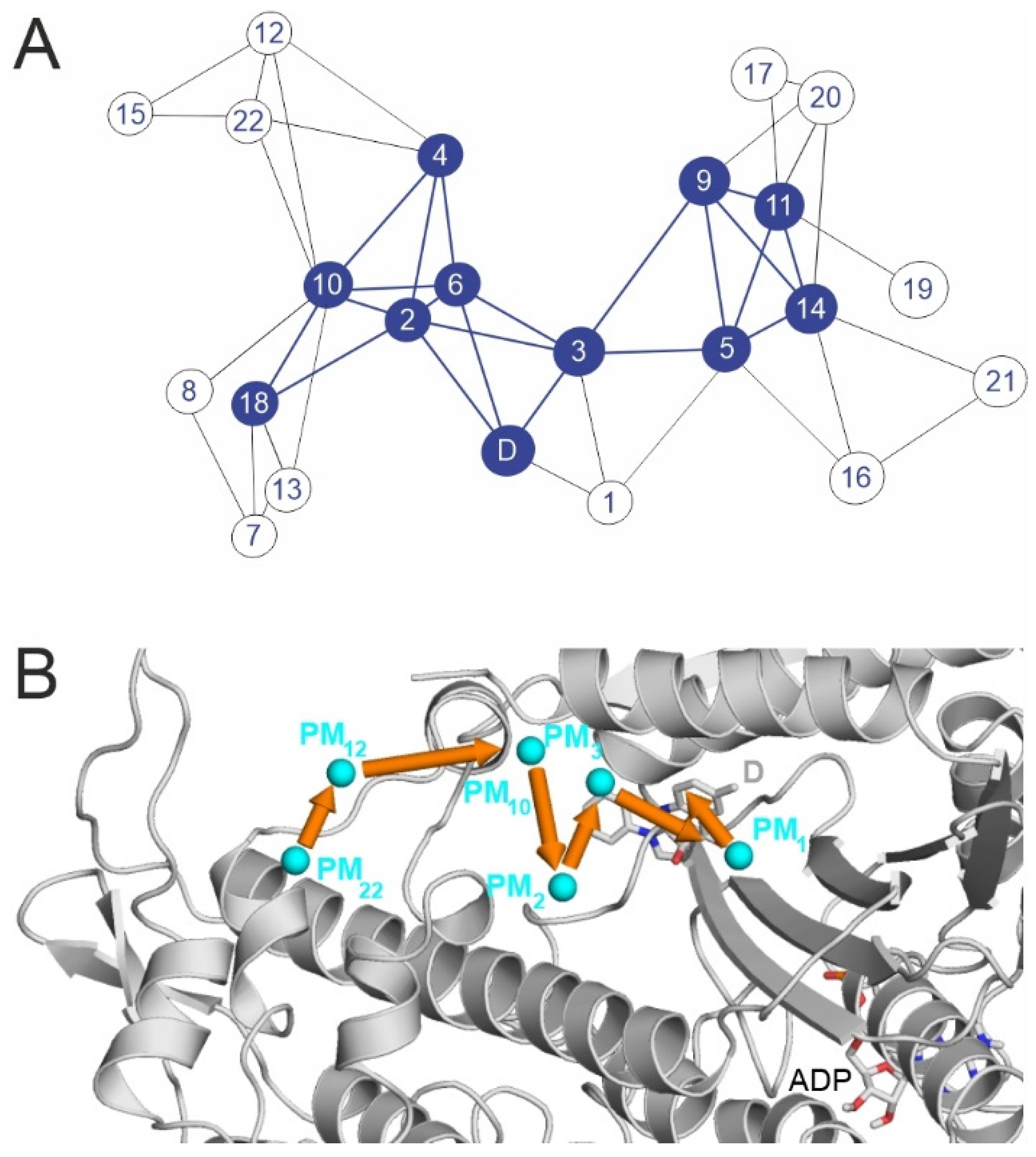
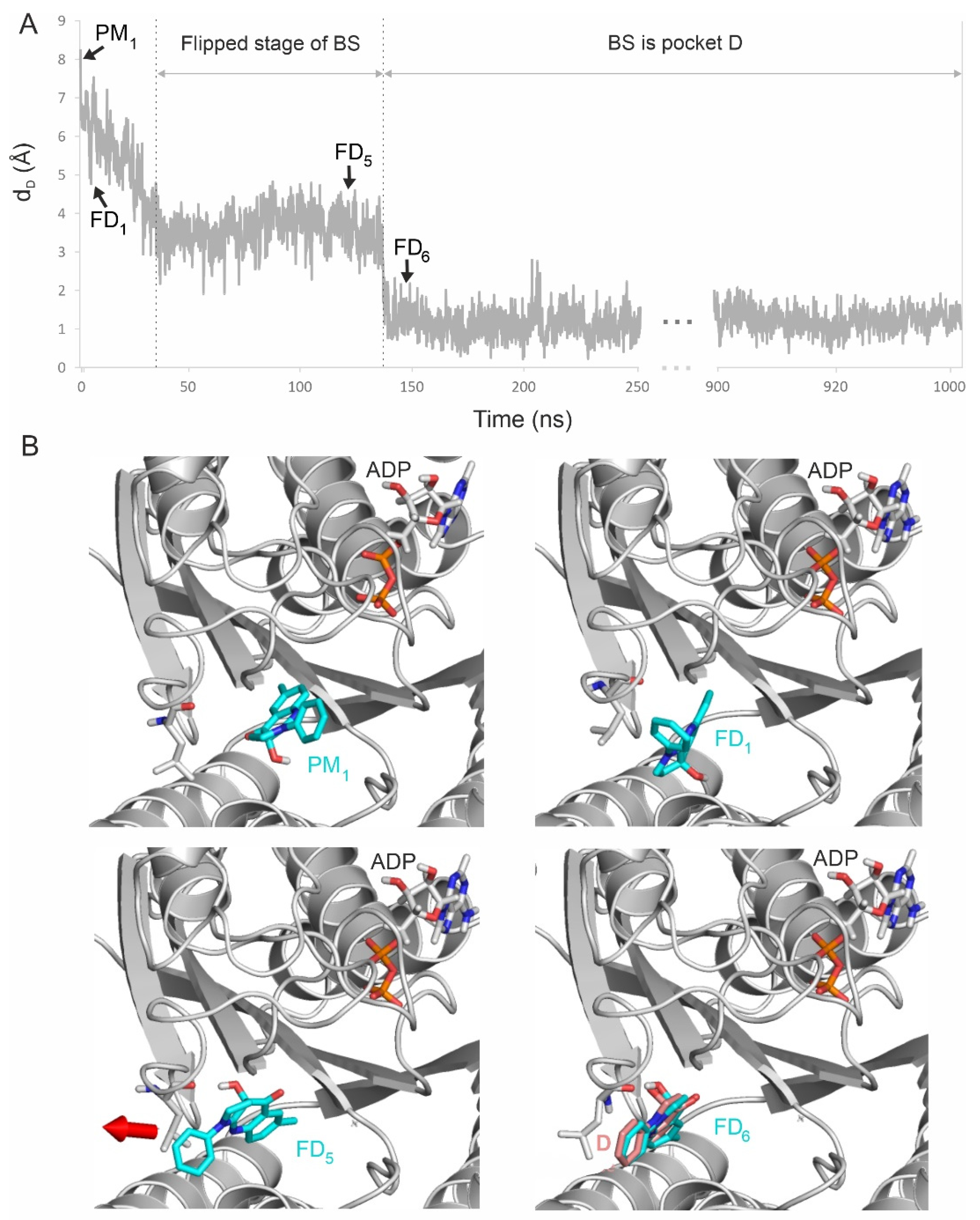
2.2. Binding Pathways from Binding Networks
2.3. Final Test: Docking to the Destination Pocket
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blundell, T.L.; Jhoti, H.; Abell, C. High-throughput crystallography for lead discovery in drug design. Nat. Rev. Drug Discov. 2002, 1, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Hui, R.; Edwards, A. High-throughput protein crystallization. J. Struct. Biol. 2003, 142, 154–161. [Google Scholar] [CrossRef]
- Cheng, Y.; Glaeser, R.M.; Nogales, E. How Cryo-EM Became so Hot. Cell 2017, 171, 1229–1231. [Google Scholar] [CrossRef]
- Houdusse, A.; Sweeney, H.L. How Myosin Generates Force on Actin Filaments. Trends Biochem. Sci. 2016, 41, 989–997. [Google Scholar] [CrossRef]
- Jiang, W.; Tang, L. Atomic cryo-EM structures of viruses. Curr. Opin. Struct. Biol. 2017, 46, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Zsidó, B.Z.; Börzsei, R.; Pintér, E.; Hetényi, C. Prerequisite binding modes determine the dynamics of action of covalent agonists of ion channel trpa1. Pharmaceuticals 2021, 14, 988. [Google Scholar] [CrossRef] [PubMed]
- Vogt, A.D.; Di Cera, E. Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry 2012, 51, 5894–5902. [Google Scholar] [CrossRef] [PubMed]
- Balint, M.; Jeszenoi, N.; Horvath, I.; Abraham, I.M.; Hetenyi, C. Dynamic changes in binding interaction networks of sex steroids establish their non-classical effects. Sci. Rep. 2017, 7, 14847. [Google Scholar] [CrossRef]
- Decherchi, S.; Berteotti, A.; Bottegoni, G.; Rocchia, W.; Cavalli, A. The ligand binding mechanism to purine nucleoside phosphorylase elucidated via molecular dynamics and machine learning. Nat. Commun. 2015, 6, 6155. [Google Scholar] [CrossRef]
- Buch, I.; Giorgino, T.; De Fabritiis, G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2011, 108, 10184–10189. [Google Scholar] [CrossRef]
- Dror, R.O.; Pan, A.C.; Arlow, D.H.; Borhani, D.W.; Maragakis, P.; Shan, Y.; Xu, H.; Shaw, D.E. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc. Natl. Acad. Sci. USA 2011, 108, 13118–13123. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Kim, E.T.; Eastwood, M.P.; Dror, R.O.; Seeliger, M.A.; Shaw, D.E. How does a drug molecule find its target binding site? J. Am. Chem. Soc. 2011, 133, 9181–9183. [Google Scholar] [CrossRef] [PubMed]
- Zsidó, B.Z.; Hetényi, C. The role of water in ligand binding. Curr. Opin. Struct. Biol. 2021, 67, 1–8. [Google Scholar] [CrossRef]
- Zsidó, B.Z.; Börzsei, R.; Szél, V.; Hetényi, C. Determination of Ligand Binding Modes in Hydrated Viral Ion Channels to Foster Drug Design and Repositioning. J. Chem. Inf. Model. 2021, 61, 4011–4022. [Google Scholar] [CrossRef] [PubMed]
- Straight, A.F.; Cheung, A.; Limouze, J.; Chen, I.; Westwood, N.J.; Sellers, J.R.; Mitchison, T.J. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science 2003, 299, 1743–1747. [Google Scholar] [CrossRef]
- Ma, X.; Kovacs, M.; Conti, M.A.; Wang, A.; Zhang, Y.; Sellers, J.R.; Adelstein, R.S. Nonmuscle myosin II exerts tension but does not translocate actin in vertebrate cytokinesis. Proc. Natl. Acad. Sci. USA 2012, 109, 4509–4514. [Google Scholar] [CrossRef]
- Mogilner, A.; Keren, K. The Shape of Motile Cells. Curr. Biol. 2009, 19, R762–R771. [Google Scholar] [CrossRef]
- Takacs, B.; Billington, N.; Gyimesi, M.; Kintses, B.; Malnasi-Csizmadia, A.; Knight, P.J.; Kovacs, M. Myosin complexed with ADP and blebbistatin reversibly adopts a conformation resembling the start point of the working stroke. Proc. Natl. Acad. Sci. USA 2010, 107, 6799–6804. [Google Scholar] [CrossRef]
- Wakatsuki, T. Mechanics of cell spreading: Role of myosin II. J. Cell Sci. 2003, 116, 1617–1625. [Google Scholar] [CrossRef]
- Korobova, F.; Gauvin, T.J.; Higgs, H.N. Report A Role for Myosin II in Mammalian Mitochondrial Fission. Curr. Biol. 2014, 24, 409–414. [Google Scholar] [CrossRef]
- Wylie, S.R.; Chantler, P.D. Myosin IIA Drives Neurite Retraction. Mol. Biol. Cell 2003, 14, 4654–4666. [Google Scholar] [CrossRef] [PubMed]
- Beadle, C.; Assanah, M.C.; Monzo, P.; Vallee, R.; Rosenfeld, S.S.; Canoll, P. The Role of Myosin II in Glioma Invasion of the Brain. Mol. Biol. Cell 2008, 19, 3357–3368. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Lopez, C.; Allingham, J.S.; Lebl, T.; Lawson, C.P.A.T.; Brenk, R.; Sellers, J.R.; Rayment, I.; Westwood, N.J. The small molecule tool (S)-(−)-blebbistatin: Novel insights of relevance to myosin inhibitor design. Org. Biomol. Chem. 2008, 6, 2076–2084. [Google Scholar] [CrossRef] [PubMed]
- Allingham, J.S.; Smith, R.; Rayment, I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nat. Struct. Mol. Biol. 2005, 12, 378–379. [Google Scholar] [CrossRef]
- Fedorov, R.; Böhl, M.; Tsiavaliaris, G.; Hartmann, F.K.; Taft, M.H.; Baruch, P.; Brenner, B.; Martin, R.; Knölker, H.J.; Gutzeit, H.O.; et al. The mechanism of pentabromopseudilin inhibition of myosin motor activity. Nat. Struct. Mol. Biol. 2009, 16, 80–88. [Google Scholar] [CrossRef]
- Paszek, M.J.; Dufort, C.C.; Rossier, O.; Bainer, R.; Cassereau, L.; Rubashkin, M.G.; Magbanua, M.J.; Kurt, S. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 2015, 511, 319–325. [Google Scholar] [CrossRef]
- Derycke, L.; Stove, C.; Wever, O.D.E.; Dollé, L.; Colpaert, N.; Depypere, H.; Michalski, J.; Bracke, M. The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int. J. Dev. Biol. 2011, 55, 835–840. [Google Scholar] [CrossRef]
- Duxbury, M.S.; Ashley, S.W.; Whang, E.E. Inhibition of pancreatic adenocarcinoma cellular invasiveness by blebbistatin: A novel myosin II inhibitor. Biochem. Biophys. Res. Commun. 2004, 313, 992–997. [Google Scholar] [CrossRef]
- Kovacs, M.; Toth, J.; Hetenyi, C.; Malnasi-Csizmadia, A.; Sellers, J.R. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 2004, 279, 35557–35563. [Google Scholar] [CrossRef]
- Kepiro, M.; Varkuti, B.H.; Vegner, L.; Voros, G.; Hegyi, G.; Varga, M.; Malnasi-Csizmadia, A. Para-nitroblebbistatin, the non-cytotoxic and photostable myosin II inhibitor. Angew. Chem. Int. Ed. 2014, 53, 8211–8215. [Google Scholar] [CrossRef]
- Rauscher, A.; Gyimesi, M.; Kovacs, M.; Malnasi-Csizmadia, A. Targeting Myosin by Blebbistatin Derivatives: Optimization and Pharmacological Potential. Trends Biochem. Sci. 2018, 43, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Ekpenyong, A.E.; Golfier, S.; Li, W.; Chalut, K.J.; Otto, O.; Elgeti, J.; Guck, J.; Lautenschläger, F. Myosin II activity softens cells in suspension. Biophys. J. 2015, 108, 1856–1869. [Google Scholar] [CrossRef] [PubMed]
- Sayyad, W.A.; Amin, L.; Fabris, P.; Ercolini, E.; Torre, V. The role of myosin-II in force generation of DRG filopodia and lamellipodia. Sci. Rep. 2015, 5, 7842. [Google Scholar] [CrossRef]
- Eddinger, T.J.; Meer, D.P.; Miner, A.S.; Joel, M.; Rovner, A.S.; Ratz, P.H. Potent Inhibition of Arterial Smooth Muscle Tonic Contractions by the Selective Myosin II Inhibitor, Blebbistatin. Med. Econ. 2013, 90, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Balint, M.; Jeszenoi, N.; Horvath, I.; van der Spoel, D.; Hetenyi, C. Systematic exploration of multiple drug binding sites. J. Cheminform. 2017, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Aldeghi, M.; Heifetz, A.; Bodkin, M.J.; Knapp, S.; Biggin, P.C. Predictions of ligand selectivity from absolute binding free energy calculations. J. Am. Chem. Soc. 2017, 139, 946–957. [Google Scholar] [CrossRef]
- Chu, W.T.; Wang, J. Energy landscape topography reveals the underlying link between binding specificity and activity of enzymes. Sci. Rep. 2016, 6, 27808. [Google Scholar] [CrossRef] [PubMed]
- Moraca, F.; Amato, J.; Ortuso, F.; Artese, A.; Pagano, B.; Novellino, E.; Alcaro, S.; Parrinello, M.; Limongelli, V. Ligand binding to telomeric G-quadruplex DNA investigated by funnel-metadynamics simulations. Proc. Natl. Acad. Sci. USA 2017, 114, E2136–E2145. [Google Scholar] [CrossRef]
- Plattner, N.; Doerr, S.; De Fabritiis, G.; Noé, F. Complete protein-protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat. Chem. 2017, 9, 1005–1011. [Google Scholar] [CrossRef]
- Csermely, P.; Korcsmáros, T.; Kiss, H.J.M.; London, G.; Nussinov, R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery: A comprehensive review. Pharmacol. Ther. 2013, 138, 333–408. [Google Scholar] [CrossRef]
- Hulovatyy, Y.; Chen, H.; Milenković, T. Exploring the structure and function of temporal networks with dynamic graphlets. Bioinformatics 2015, 31, i171–i180. [Google Scholar] [CrossRef] [PubMed]
- Jeszenoi, N.; Balint, M.; Horvath, I.; Van Der Spoel, D.; Hetenyi, C. Exploration of Interfacial Hydration Networks of Target-Ligand Complexes. J. Chem. Inf. Model. 2016, 56, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Brysbaert, G.; Blossey, R.; Lensink, M.F. The inclusion of water molecules in residue interaction networks identifies additional central residues. Front. Mol. Biosci. 2018, 5, 88. [Google Scholar] [CrossRef] [PubMed]
- Brysbaert, G.; Lorgouilloux, K.; Vranken, W.F.; Lensink, M.F. RINspector: A Cytoscape app for centrality analyses and DynaMine flexibility prediction. Bioinformatics 2018, 34, 294–296. [Google Scholar] [CrossRef]
- Kunstmann, S.; Gohlke, U.; Broeker, N.K.; Roske, Y.; Heinemann, U.; Santer, M.; Barbirz, S. Solvent Networks Tune Thermodynamics of Oligosaccharide Complex Formation in an Extended Protein Binding Site. J. Am. Chem. Soc. 2018, 140, 10447–10455. [Google Scholar] [CrossRef]
- Fisher, A.J.; Smith, C.A.; Thoden, J.B.; Smith, R.; Sutoh, K.; Holden, H.M.; Rayment, I. X-ray Structures of the Myosin Motor Domain of Dictyostelium discoideum Complexed with MgADP.BeFx and MgADP.AlF4-. Biochemistry 1995, 34, 8960–8972. [Google Scholar] [CrossRef]
- Fischer, S.; Windshügel, B.; Horak, D.; Holmes, K.C.; Smith, J.C. Structural mechanism of the recovery stroke in the myosin molecular motor. Proc. Natl. Acad. Sci. USA 2005, 102, 6873–6878. [Google Scholar] [CrossRef]
- Koppole, S.; Smith, J.C.; Fischer, S. The Structural Coupling between ATPase Activation and Recovery Stroke in the Myosin II Motor. Structure 2007, 15, 825–837. [Google Scholar] [CrossRef][Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, 162–173. [Google Scholar] [CrossRef]
- LLC. Schrödinger Release 2019-3: Maestro, Schrödinger; LLC: New York, NY, USA, 2019; Available online: https://www.schrodinger.com/maestro. (accessed on 27 June 2022).
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Chemaxon. Marvin Sketch; v 6.3.0; Chemaxon: Budapest, Hungary, 2014. [Google Scholar]
- Stewart, J.J.P. MOPAC2009, 2009; Steward Computational Chemistry: Colorado Springs, CO, USA, 2008. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 28, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Hetényi, C.; Van Der Spoel, D. Blind docking of drug-sized compounds to proteins with up to a thousand residues. FEBS Lett. 2006, 580, 1447–1450. [Google Scholar] [CrossRef] [PubMed]
- Jeszenoi, N.; Horváth, I.; Bálint, M.; Van Der Spoel, D.; Hetényi, C. Mobility-based prediction of hydration structures of protein surfaces. Bioinformatics 2015, 31, 1959–1965. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinforma. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.Y. RED Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011, 39, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian; Version 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Krishnan, R.; Binkley, J.S.; Seeger, R.; Pople, J.A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980, 72, 650–654. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Boutet, S.; Lomb, L.; Williams, G.J.; Barends, T.R.; Aquila, A.; Doak, R.B.; Weierstall, U.; DePonte, D.P.; Steinbrener, J.; Shoeman, R.L.; et al. High-resolution protein structure determination by serial femtosecond crystallography. Science 2012, 337, 362–364. [Google Scholar] [CrossRef]
- Choinowski, T.; Hauser, H.; Piontek, K. Structure of sterol carrier protein 2 at 1.8 Å resolution reveals a hydrophobic tunnel suitable for lipid binding. Biochemistry 2000, 39, 1897–1902. [Google Scholar] [CrossRef]
- Logothetis, D.E.; Kurachi, Y.; Galper, J.; Neer, E.J.; Clapham, D.E. The βγ subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature 1987, 325, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Thuresson, E.D.; Lakkides, K.M.; Rieke, C.J.; Sun, Y.; Wingerd, B.A.; Micielli, R.; Mulichak, A.M.; Malkowski, M.G.; Garavito, R.M.; Smith, W.L. Prostaglandin endoperoxide H synthase-1: The functions of cyclooxygenase active site residues in the binding, positioning, and oxygenation of arachidonic acid. J. Biol. Chem. 2001, 276, 10347–10357. [Google Scholar] [CrossRef] [PubMed]
- Acharya, R.; Carnevale, V.; Fiorin, G.; Levine, B.G.; Polishchuk, A.L.; Balannik, V.; Samish, I.; Lamb, R.A.; Pinto, L.H.; DeGrado, W.F.; et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc. Natl. Acad. Sci. USA 2010, 107, 15075–15080. [Google Scholar] [CrossRef]
- Thomaston, J.L.; Polizzi, N.F.; Konstantinidi, A.; Wang, J.; Kolocouris, A.; Degrado, W.F. Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters. J. Am. Chem. Soc. 2018, 140, 15219–15226. [Google Scholar] [CrossRef] [PubMed]
- Mandala, V.; McKay, M.; Shcherbakov, A.; Dregni, A.; Kolocouris, A.; Hong, M. Structure and Drug Binding of the SARS-CoV-2 Envelope Protein in Phospholipid Bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef] [PubMed]


| Structural Change | Starting Time (ns) | Distance (Å) | |||
|---|---|---|---|---|---|
| 0 ns | 1000 ns | Experimental | |||
| Formation of H-bond (G240…BS) | 40 | 8.3 a | 3.2 a | 2.8 a | |
| Formation of salt bridge (E459…R238) | 71 | 8.3 b | 4.0 b | 4.2 b | |
| Movement of BS | 137 | 8.0 c | 0.9 c | 0 c | |
| Formation of H-bond (L262…BS) | 137 | 6.6 d | 2.5 d | 2.5 d | |
| Flipping of L262 | 137 | 3.4 c | 1.8 c | 0 c | |
| Flipping of Y261 | 143 | 1.9 a | 1.4 a | 0 a | |
| Flipping of S456 | 447 | 4.87 a | 1.9 a | 0 a | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bálint, M.; Zsidó, B.Z.; van der Spoel, D.; Hetényi, C. Binding Networks Identify Targetable Protein Pockets for Mechanism-Based Drug Design. Int. J. Mol. Sci. 2022, 23, 7313. https://doi.org/10.3390/ijms23137313
Bálint M, Zsidó BZ, van der Spoel D, Hetényi C. Binding Networks Identify Targetable Protein Pockets for Mechanism-Based Drug Design. International Journal of Molecular Sciences. 2022; 23(13):7313. https://doi.org/10.3390/ijms23137313
Chicago/Turabian StyleBálint, Mónika, Balázs Zoltán Zsidó, David van der Spoel, and Csaba Hetényi. 2022. "Binding Networks Identify Targetable Protein Pockets for Mechanism-Based Drug Design" International Journal of Molecular Sciences 23, no. 13: 7313. https://doi.org/10.3390/ijms23137313
APA StyleBálint, M., Zsidó, B. Z., van der Spoel, D., & Hetényi, C. (2022). Binding Networks Identify Targetable Protein Pockets for Mechanism-Based Drug Design. International Journal of Molecular Sciences, 23(13), 7313. https://doi.org/10.3390/ijms23137313







