Bioenergetics and Reactive Nitrogen Species in Bacteria
Abstract
1. Introduction
2. Bacterial Aerobic Respiratory Chains
3. •NO and Bacterial Terminal Oxidases
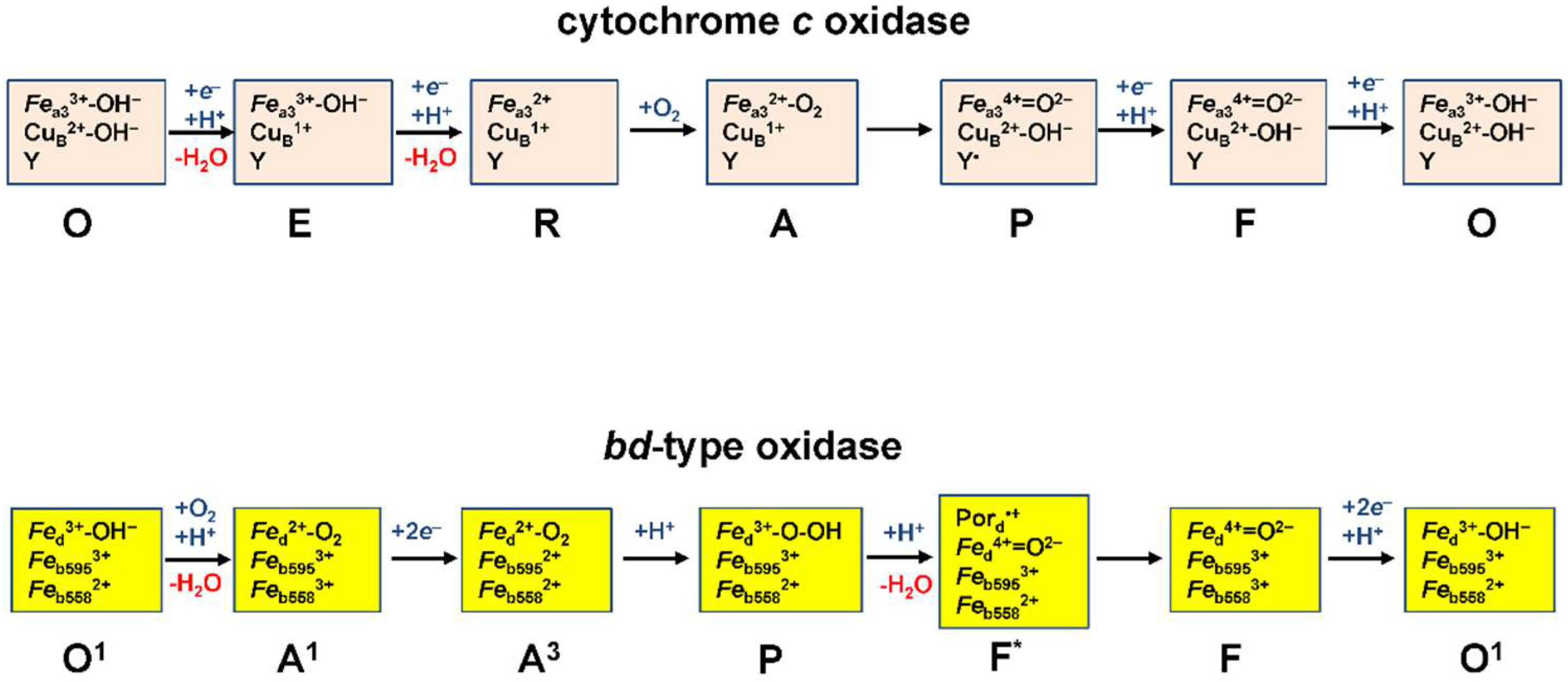
3.1. •NO and Bacterial Heme–Copper Terminal Oxidases
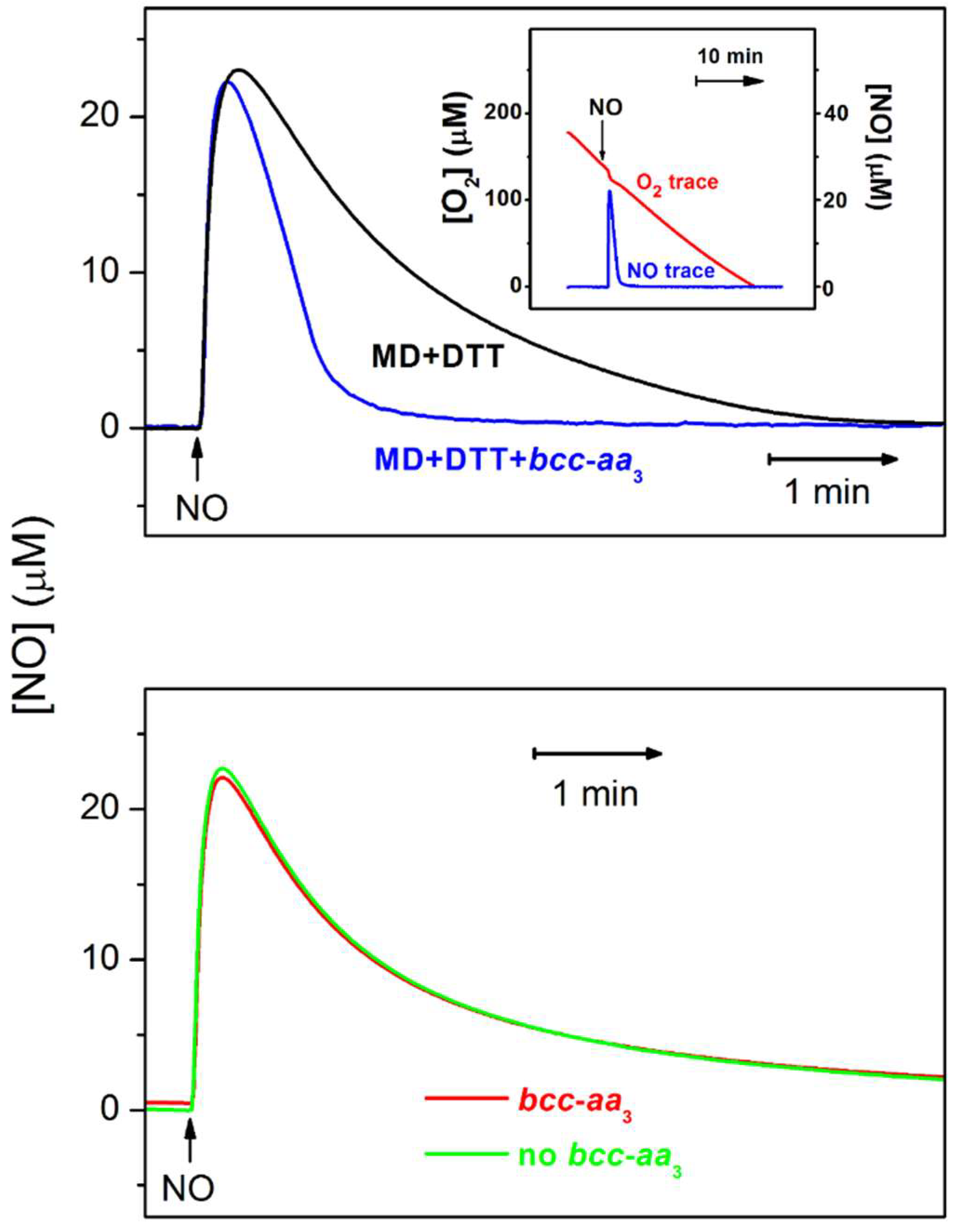
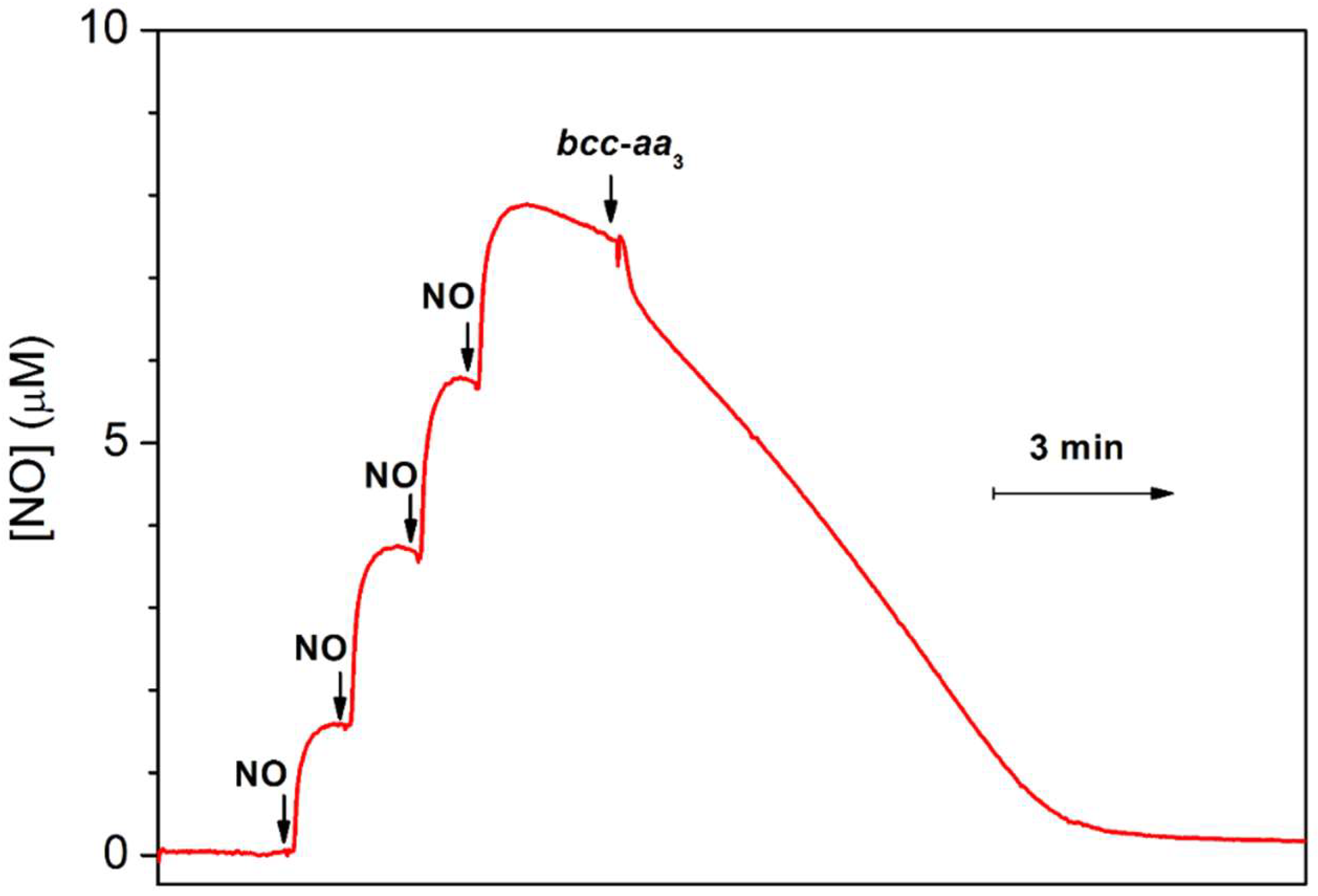
3.1.1. •NO-Metabolizing Activity of the Mycobacterial bcc-aa3 Supercomplex in Turnover
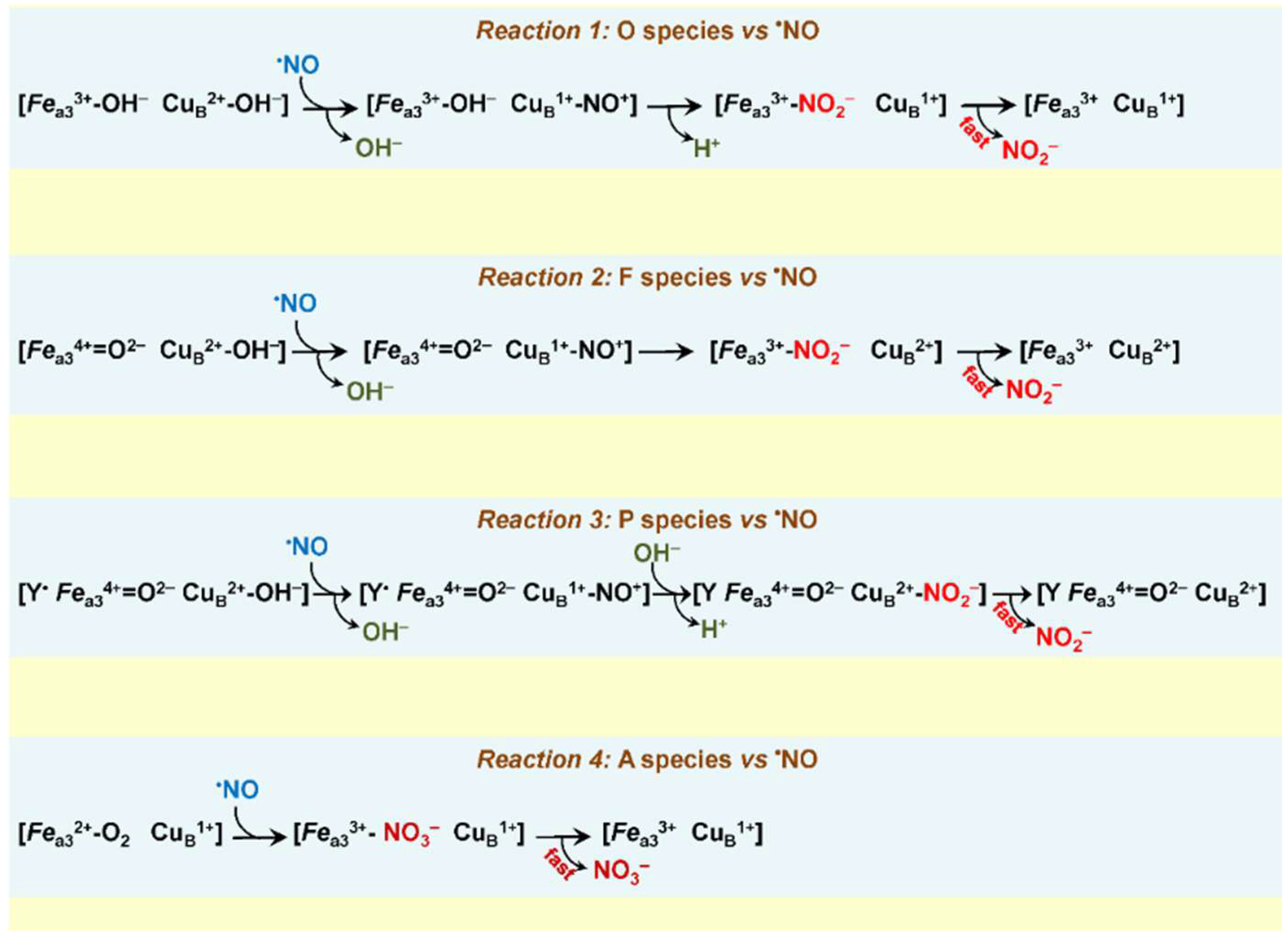
3.1.2. •NO Reductase Activity of Heme–Copper Oxidases
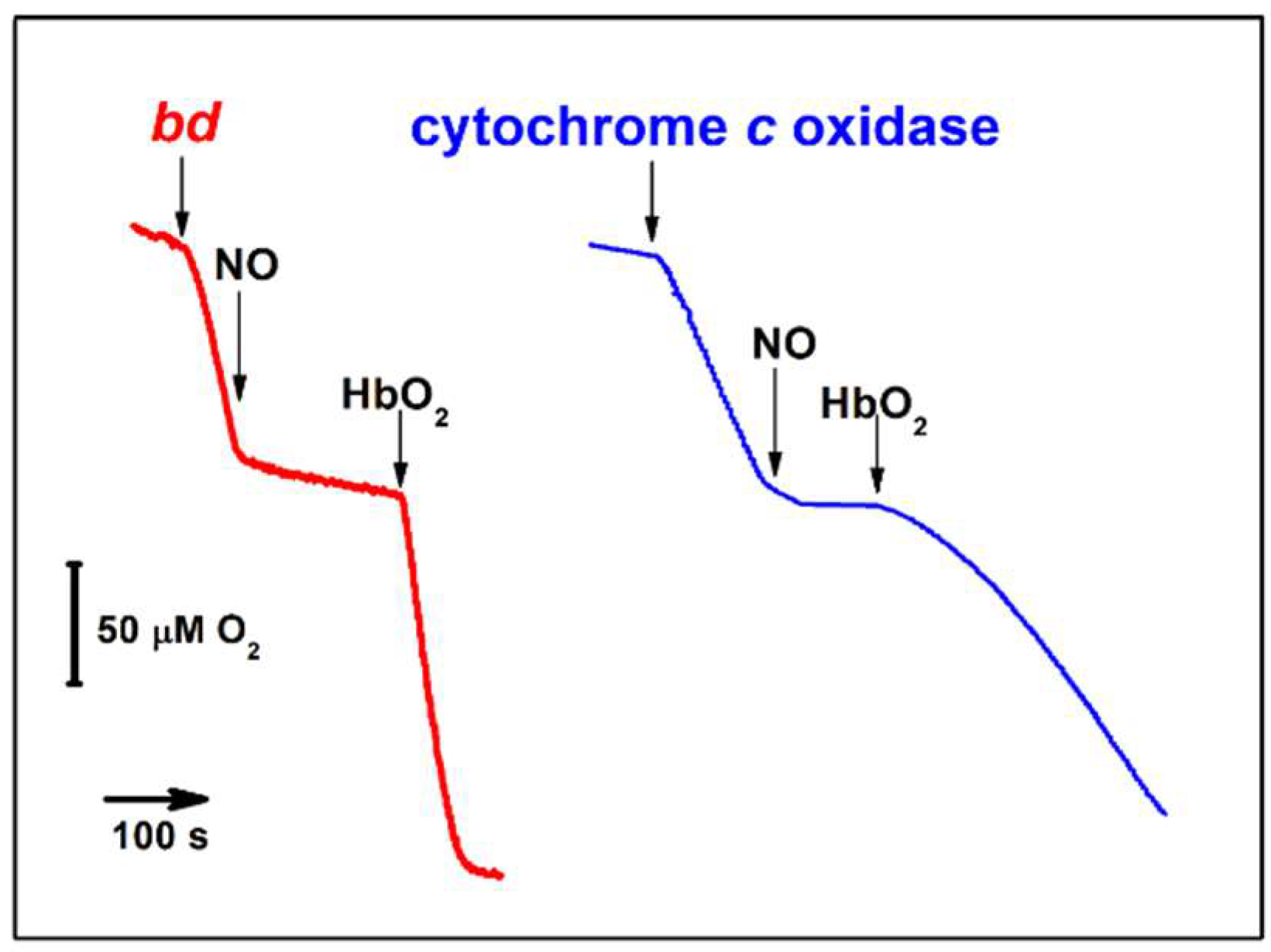
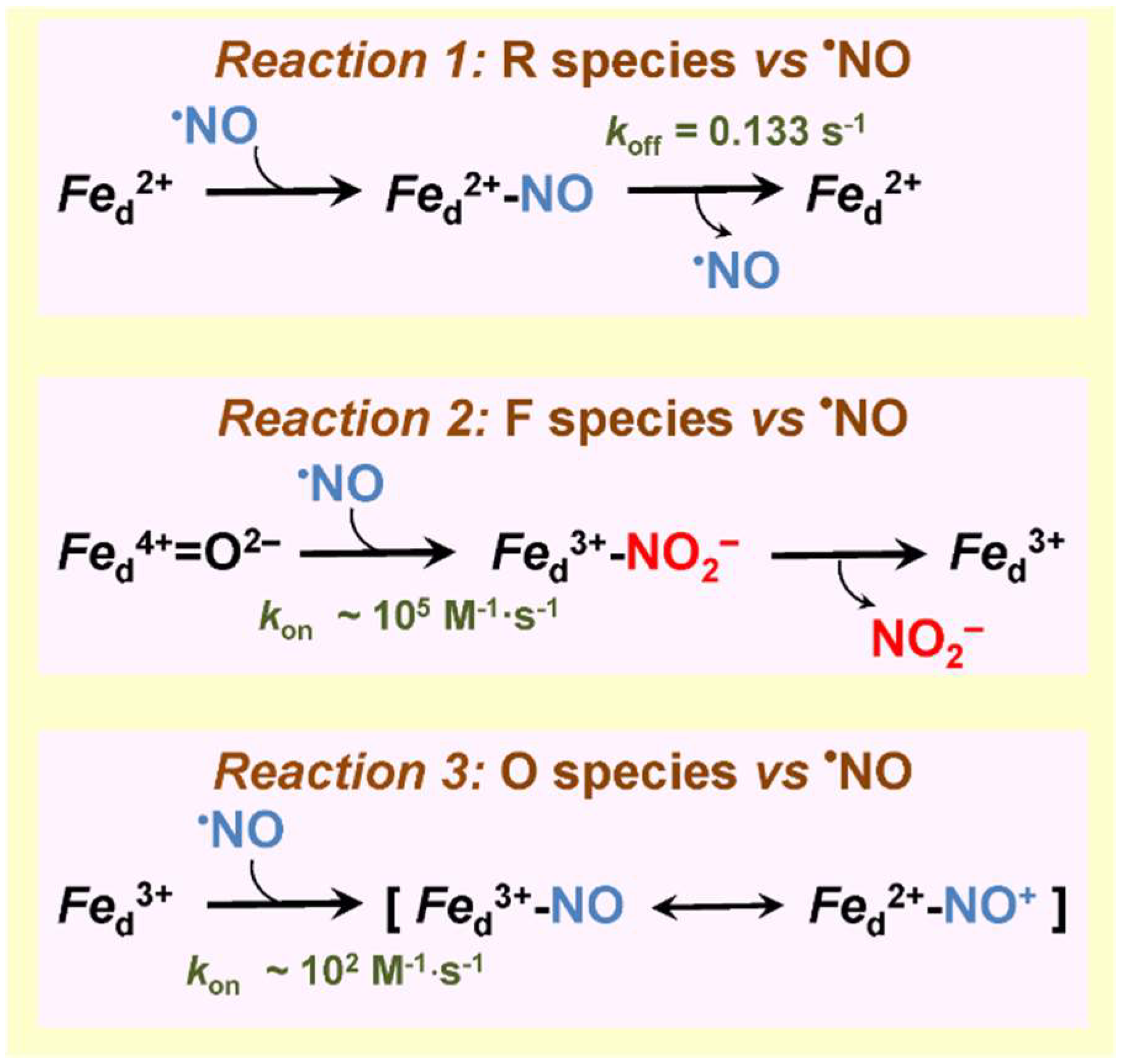
3.2. bd-Type Oxidases Confer Bacterial Resistance to •NO
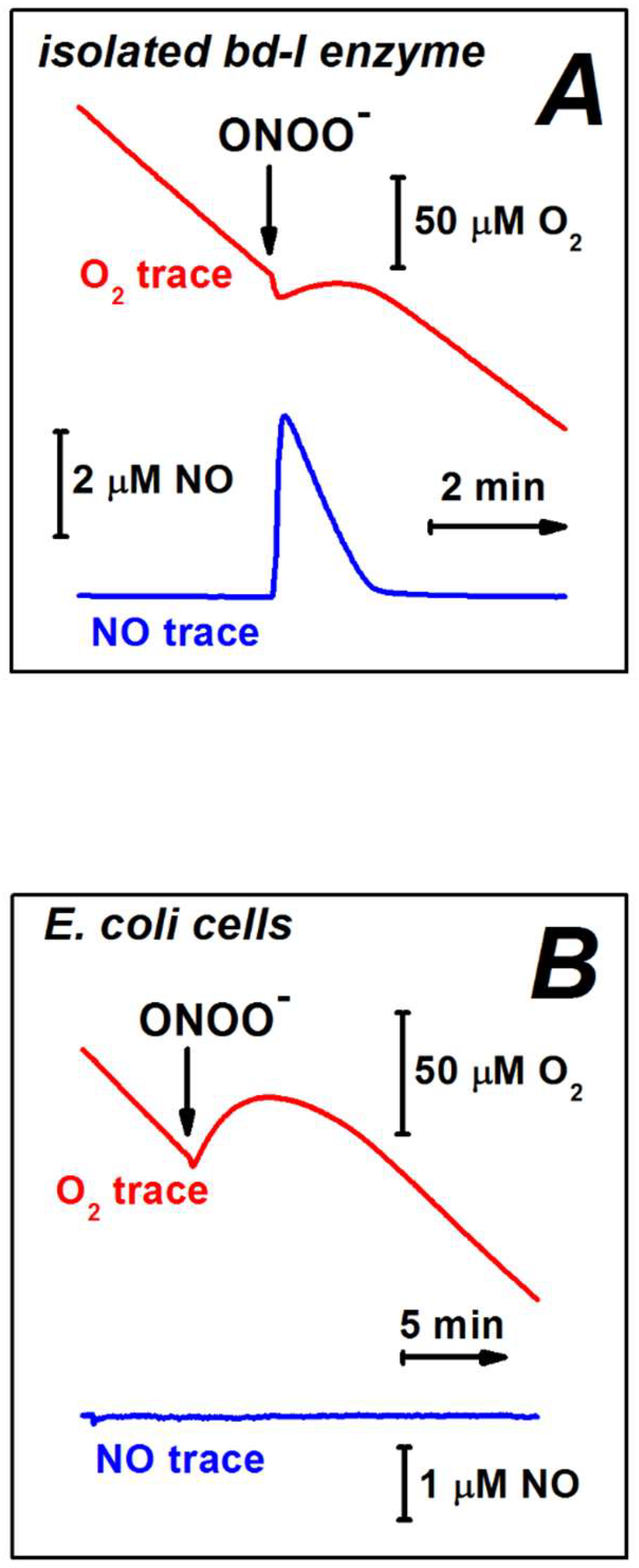
4. Peroxynitrite and Bacterial Terminal Oxidases
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martinez, M.C.; Andriantsitohaina, R. Reactive nitrogen species: Molecular mechanisms and potential significance in health and disease. Antioxid. Redox Signal. 2009, 11, 669–702. [Google Scholar] [CrossRef] [PubMed]
- Wareham, L.K.; Southam, H.M.; Poole, R.K. Do nitric oxide, carbon monoxide and hydrogen sulfide really qualify as ‘gasotransmitters’ in bacteria? Biochem. Soc. Trans. 2018, 46, 1107–1118. [Google Scholar] [CrossRef]
- Bryan, N.S.; Lefer, D.J. Update on gaseous signaling molecules nitric oxide and hydrogen sulfide: Strategies to capture their functional activity for human therapeutics. Mol. Pharmacol. 2019, 96, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Mendes, S.S.; Miranda, V.; Saraiva, L.M. Hydrogen sulfide and carbon monoxide tolerance in bacteria. Antioxidants 2021, 10, 729. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Forte, E. Impact of hydrogen sulfide on mitochondrial and bacterial bioenergetics. Int. J. Mol. Sci. 2021, 22, 12688. [Google Scholar] [CrossRef] [PubMed]
- Radi, R. Oxygen radicals, nitric oxide, and peroxynitrite: Redox pathways in molecular medicine. Proc. Natl. Acad. Sci. USA 2018, 115, 5839–5848. [Google Scholar] [CrossRef] [PubMed]
- Perez de la Lastra, J.M.; Juan, C.A.; Plou, F.J.; Perez-Lebena, E. The nitration of proteins, lipids and DNA by peroxynitrite derivatives-chemistry involved and biological relevance. Stresses 2022, 2, 3. [Google Scholar] [CrossRef]
- Fang, F.C.; Vazquez-Torres, A. Reactive nitrogen species in host-bacterial interactions. Curr. Opin. Immunol. 2019, 60, 96–102. [Google Scholar] [CrossRef]
- Carvalho, S.M.; Beas, J.Z.; Videira, M.A.M.; Saraiva, L.M. Defenses of multidrug resistant pathogens against reactive nitrogen species produced in infected hosts. Adv. Microb. Physiol. 2022, 80, 85–155. [Google Scholar] [CrossRef]
- Gusarov, I.; Nudler, E. Protein S-nitrosylation: Enzymatically controlled, but intrinsically unstable, post-translational modification. Mol. Cell 2018, 69, 351–353. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Mondal, A.; Venkataramaiah, M.; Chauhan, N.K.; Rajamohan, G. Role of oxyRKP, a novel LysR-family transcriptional regulator, in antimicrobial resistance and virulence in Klebsiella pneumoniae. Microbiology (Reading) 2013, 159, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Anes, J.; Dever, K.; Eshwar, A.; Nguyen, S.; Cao, Y.; Sivasankaran, S.K.; Sakalauskaite, S.; Lehner, A.; Devineau, S.; Daugelavicius, R.; et al. Analysis of the oxidative stress regulon identifies soxS as a genetic target for resistance reversal in multidrug-resistant Klebsiella pneumoniae. mBio 2021, 12, e0086721. [Google Scholar] [CrossRef] [PubMed]
- Way, S.S.; Goldberg, M.B. Clearance of Shigella flexneri infection occurs through a nitric oxide-independent mechanism. Infect. Immun. 1998, 66, 3012–3016. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A.; Dubchak, I.L.; Arkin, A.P.; Alm, E.J.; Gelfand, M.S. Dissimilatory metabolism of nitrogen oxides in bacteria: Comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 2005, 1, e55. [Google Scholar] [CrossRef] [PubMed]
- Kint, N.; Alves Feliciano, C.; Martins, M.C.; Morvan, C.; Fernandes, S.F.; Folgosa, F.; Dupuy, B.; Texeira, M.; Martin-Verstraete, I. How the anaerobic enteropathogen Clostridioides difficile tolerates low O2 tensions. mBio 2020, 11, e01559-01520. [Google Scholar] [CrossRef]
- Folgosa, F.; Martins, M.C.; Teixeira, M. The multidomain flavodiiron protein from Clostridium difficile 630 is an NADH:oxygen oxidoreductase. Sci. Rep. 2018, 8, 10164. [Google Scholar] [CrossRef]
- Kumar, M.; Adhikari, S.; Hurdle, J.G. Action of nitroheterocyclic drugs against Clostridium difficile. Int. J. Antimicrob. Agents 2014, 44, 314–319. [Google Scholar] [CrossRef]
- Caruana, N.J.; Stroud, D.A. The road to the structure of the mitochondrial respiratory chain supercomplex. Biochem. Soc. Trans. 2020, 48, 621–629. [Google Scholar] [CrossRef]
- Cogliati, S.; Cabrera-Alarcon, J.L.; Enriquez, J.A. Regulation and functional role of the electron transport chain supercomplexes. Biochem. Soc. Trans. 2021, 49, 2655–2668. [Google Scholar] [CrossRef]
- Sharma, P.; Maklashina, E.; Cecchini, G.; Iverson, T.M. Crystal structure of an assembly intermediate of respiratory Complex II. Nat. Commun. 2018, 9, 274. [Google Scholar] [CrossRef]
- Hederstedt, L. Molecular biology of Bacillus subtilis cytochromes anno 2020. Biochemistry 2021, 86, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.M.; Teixeira, M. Supramolecular organization of bacterial aerobic respiratory chains: From cells and back. Biochim. Biophys. Acta 2016, 1857, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Kaila, V.R.I.; Wikstrom, M. Architecture of bacterial respiratory chains. Nat. Rev. Microbiol. 2021, 19, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Nuber, F.; Merono, L.; Oppermann, S.; Schimpf, J.; Wohlwend, D.; Friedrich, T. A quinol anion as catalytic intermediate coupling proton translocation with electron transfer in E. coli respiratory complex I. Front. Chem. 2021, 9, 672969. [Google Scholar] [CrossRef]
- Grba, D.N.; Blaza, J.N.; Bridges, H.R.; Agip, A.A.; Yin, Z.; Murai, M.; Miyoshi, H.; Hirst, J. Cryo-electron microscopy reveals how acetogenins inhibit mitochondrial respiratory complex I. J. Biol. Chem. 2022, 298, 101602. [Google Scholar] [CrossRef]
- Marreiros, B.C.; Sena, F.V.; Sousa, F.M.; Batista, A.P.; Pereira, M.M. Type II NADH:quinone oxidoreductase family: Phylogenetic distribution, structural diversity and evolutionary divergences. Environ. Microbiol. 2016, 18, 4697–4709. [Google Scholar] [CrossRef]
- Blaza, J.N.; Bridges, H.R.; Aragao, D.; Dunn, E.A.; Heikal, A.; Cook, G.M.; Nakatani, Y.; Hirst, J. The mechanism of catalysis by type-II NADH:quinone oxidoreductases. Sci. Rep. 2017, 7, 40165. [Google Scholar] [CrossRef]
- Bertsova, Y.V.; Baykov, A.A.; Bogachev, A.V. A simple strategy to differentiate between H+- and Na+-transporting NADH:quinone oxidoreductases. Arch. Biochem. Biophys. 2020, 681, 108266. [Google Scholar] [CrossRef]
- Liang, P.; Fang, X.; Hu, Y.; Yuan, M.; Raba, D.A.; Ding, J.; Bunn, D.C.; Sanjana, K.; Yang, J.; Rosas-Lemus, M.; et al. The aerobic respiratory chain of Pseudomonas aeruginosa cultured in artificial urine media: Role of NQR and terminal oxidases. PLoS ONE 2020, 15, e0231965. [Google Scholar] [CrossRef]
- Hreha, T.N.; Foreman, S.; Duran-Pinedo, A.; Morris, A.R.; Diaz-Rodriguez, P.; Jones, J.A.; Ferrara, K.; Bourges, A.; Rodriguez, L.; Koffas, M.A.G.; et al. The three NADH dehydrogenases of Pseudomonas aeruginosa: Their roles in energy metabolism and links to virulence. PLoS ONE 2021, 16, e0244142. [Google Scholar] [CrossRef]
- Kozlova, M.I.; Bushmakin, I.M.; Belyaeva, J.D.; Shalaeva, D.N.; Dibrova, D.V.; Cherepanov, D.A.; Mulkidjanian, A.Y. Expansion of the "Sodium World" through evolutionary time and taxonomic space. Biochemistry 2020, 85, 1518–1542. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.A.; Crofts, A.R. Dissecting the pattern of proton release from partial process involved in ubihydroquinone oxidation in the Q-cycle. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Francia, F.; Khalfaoui-Hassani, B.; Lanciano, P.; Musiani, F.; Noodleman, L.; Venturoli, G.; Daldal, F. The cytochrome b lysine 329 residue is critical for ubihydroquinone oxidation and proton release at the Qo site of bacterial cytochrome bc1. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Verkhovsky, M.I. Oxygen as Acceptor. EcoSal Plus 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, B.; Nitharwal, R.G.; Fedotovskaya, O.; Schafer, J.; Guo, H.; Kuang, Q.; Benlekbir, S.; Sjostrand, D.; Adelroth, P.; Rubinstein, J.L.; et al. Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis. Nat. Struct. Mol. Biol. 2018, 25, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Li, J.; Xu, A.; Tang, Y.; Ji, W.; Gao, R.; Wang, S.; Yu, L.; Tian, C.; Li, J.; et al. An electron transfer path connects subunits of a mycobacterial respiratory supercomplex. Science 2018, 362, eaat8923. [Google Scholar] [CrossRef]
- Kao, W.C.; Ortmann de Percin Northumberland, C.; Cheng, T.C.; Ortiz, J.; Durand, A.; von Loeffelholz, O.; Schilling, O.; Biniossek, M.L.; Klaholz, B.P.; Hunte, C. Structural basis for safe and efficient energy conversion in a respiratory supercomplex. Nat. Commun. 2022, 13, 545. [Google Scholar] [CrossRef]
- Fedotovskaya, O.; Albertsson, I.; Nordlund, G.; Hong, S.; Gennis, R.B.; Brzezinski, P.; Adelroth, P. Identification of a cytochrome bc1-aa3 supercomplex in Rhodobacter sphaeroides. Biochim. Biophys. Acta Bioenerg. 2021, 1862, 148433. [Google Scholar] [CrossRef]
- Friedrich, T.; Wohlwend, D.; Borisov, V.B. Recent advances in structural studies of cytochrome bd and its potential application as a drug target. Int. J. Mol. Sci. 2022, 23, 3166. [Google Scholar] [CrossRef]
- Vilcheze, C.; Weinrick, B.; Leung, L.W.; Jacobs, W.R., Jr. Plasticity of Mycobacterium tuberculosis NADH dehydrogenases and their role in virulence. Proc. Natl. Acad. Sci. USA 2018, 115, 1599–1604. [Google Scholar] [CrossRef]
- Siletsky, S.A.; Borisov, V.B. Proton pumping and non-pumping terminal respiratory oxidases: Active sites intermediates of these molecular machines and their derivatives. Int. J. Mol. Sci. 2021, 22, 10852. [Google Scholar] [CrossRef] [PubMed]
- Murali, R.; Gennis, R.B.; Hemp, J. Evolution of the cytochrome bd oxygen reductase superfamily and the function of CydAA’ in Archaea. ISME J. 2021, 15, 3534–3548. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Siletsky, S.A.; Paiardini, A.; Hoogewijs, D.; Forte, E.; Giuffre, A.; Poole, R.K. Bacterial oxidases of the cytochrome bd family: Redox enzymes of unique structure, function and utility as drug targets. Antioxid. Redox Signal. 2021, 34, 1280–1318. [Google Scholar] [CrossRef] [PubMed]
- Siletsky, S.A.; Borisov, V.B.; Mamedov, M.D. Photosystem II and terminal respiratory oxidases: Molecular machines operating in opposite directions. Front. Biosci. (Landmark Ed.) 2017, 22, 1379–1426. [Google Scholar] [CrossRef] [PubMed]
- Malatesta, F.; Antonini, G.; Sarti, P.; Brunori, M. Structure and function of a molecular machine: Cytochrome c oxidase. Biophys. Chem. 1995, 54, 1–33. [Google Scholar] [CrossRef]
- Pereira, M.M.; Sousa, F.L.; Verissimo, A.F.; Teixeira, M. Looking for the minimum common denominator in haem-copper oxygen reductases: Towards a unified catalytic mechanism. Biochim. Biophys. Acta 2008, 1777, 929–934. [Google Scholar] [CrossRef]
- Papa, S.; Capitanio, N.; Capitanio, G.; Palese, L.L. Protonmotive cooperativity in cytochrome c oxidase. Biochim. Biophys. Acta 2004, 1658, 95–105. [Google Scholar] [CrossRef]
- Borisov, V.B.; Siletsky, S.A. Features of organization and mechanism of catalysis of two families of terminal oxidases: Heme-copper and bd-type. Biochemistry 2019, 84, 1390–1402. [Google Scholar] [CrossRef]
- Borisov, V.B.; Siletsky, S.A.; Nastasi, M.R.; Forte, E. ROS defense systems and terminal oxidases in bacteria. Antioxidants 2021, 10, 839. [Google Scholar] [CrossRef]
- Sousa, F.L.; Alves, R.J.; Ribeiro, M.A.; Pereira-Leal, J.B.; Teixeira, M.; Pereira, M.M. The superfamily of heme-copper oxygen reductases: Types and evolutionary considerations. Biochim. Biophys. Acta 2012, 1817, 629–637. [Google Scholar] [CrossRef]
- Wikstrom, M.; Krab, K.; Sharma, V. Oxygen activation and energy conservation by cytochrome c oxidase. Chem. Rev. 2018, 118, 2469–2490. [Google Scholar] [CrossRef] [PubMed]
- Capitanio, N.; Palese, L.L.; Capitanio, G.; Martino, P.L.; Richter, O.M.; Ludwig, B.; Papa, S. Allosteric interactions and proton conducting pathways in proton pumping aa3 oxidases: Heme a as a key coupling element. Biochim. Biophys. Acta 2012, 1817, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Maneg, O.; Malatesta, F.; Ludwig, B.; Drosou, V. Interaction of cytochrome c with cytochrome oxidase: Two different docking scenarios. Biochim. Biophys. Acta 2004, 1655, 274–281. [Google Scholar] [CrossRef] [PubMed]
- von Ballmoos, C.; Adelroth, P.; Gennis, R.B.; Brzezinski, P. Proton transfer in ba3 cytochrome c oxidase from Thermus thermophilus. Biochim. Biophys. Acta 2012, 1817, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Rich, P.R. Mitochondrial cytochrome c oxidase: Catalysis, coupling and controversies. Biochem. Soc. Trans. 2017, 45, 813–829. [Google Scholar] [CrossRef]
- Yoshikawa, S.; Shimada, A. Reaction mechanism of cytochrome c oxidase. Chem. Rev. 2015, 115, 1936–1989. [Google Scholar] [CrossRef]
- Forte, E.; Giuffre, A.; Huang, L.S.; Berry, E.A.; Borisov, V.B. Nitric oxide does not inhibit but is metabolized by the cytochrome bcc-aa3 supercomplex. Int. J. Mol. Sci. 2020, 21, 8521. [Google Scholar] [CrossRef]
- Borisov, V.B. Defects in mitochondrial respiratory complexes III and IV, and human pathologies. Mol. Aspects Med. 2002, 23, 385–412. [Google Scholar] [CrossRef]
- Borisov, V.B. Mutations in respiratory chain complexes and human diseases. Ital. J. Biochem. 2004, 53, 34–40. [Google Scholar]
- Pereira, M.M.; Santana, M.; Teixeira, M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta 2001, 1505, 185–208. [Google Scholar] [CrossRef]
- Pereira, M.M.; Gomes, C.M.; Teixeira, M. Plasticity of proton pathways in haem-copper oxygen reductases. FEBS Lett. 2002, 522, 14–18. [Google Scholar] [CrossRef][Green Version]
- Pereira, M.M.; Teixeira, M. Proton pathways, ligand binding and dynamics of the catalytic site in haem-copper oxygen reductases: A comparison between the three families. Biochim. Biophys. Acta 2004, 1655, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Safarian, S.; Rajendran, C.; Muller, H.; Preu, J.; Langer, J.D.; Ovchinnikov, S.; Hirose, T.; Kusumoto, T.; Sakamoto, J.; Michel, H. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science 2016, 352, 583–586. [Google Scholar] [CrossRef]
- Thesseling, A.; Rasmussen, T.; Burschel, S.; Wohlwend, D.; Kagi, J.; Muller, R.; Bottcher, B.; Friedrich, T. Homologous bd oxidases share the same architecture but differ in mechanism. Nat. Commun. 2019, 10, 5138. [Google Scholar] [CrossRef] [PubMed]
- Safarian, S.; Hahn, A.; Mills, D.J.; Radloff, M.; Eisinger, M.L.; Nikolaev, A.; Meier-Credo, J.; Melin, F.; Miyoshi, H.; Gennis, R.B.; et al. Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science 2019, 366, 100–104. [Google Scholar] [CrossRef]
- Wang, W.; Gao, Y.; Tang, Y.; Zhou, X.; Lai, Y.; Zhou, S.; Zhang, Y.; Yang, X.; Liu, F.; Guddat, L.W.; et al. Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels. Nat. Commun. 2021, 12, 4621. [Google Scholar] [CrossRef]
- Safarian, S.; Opel-Reading, H.K.; Wu, D.; Mehdipour, A.R.; Hards, K.; Harold, L.K.; Radloff, M.; Stewart, I.; Welsch, S.; Hummer, G.; et al. The cryo-EM structure of the bd oxidase from M. tuberculosis reveals a unique structural framework and enables rational drug design to combat TB. Nat. Commun. 2021, 12, 5236. [Google Scholar] [CrossRef]
- Grauel, A.; Kagi, J.; Rasmussen, T.; Makarchuk, I.; Oppermann, S.; Moumbock, A.F.A.; Wohlwend, D.; Muller, R.; Melin, F.; Gunther, S.; et al. Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D. Nat. Commun. 2021, 12, 6498. [Google Scholar] [CrossRef]
- Grund, T.N.; Radloff, M.; Wu, D.; Goojani, H.G.; Witte, L.F.; Josting, W.; Buschmann, S.; Muller, H.; Elamri, I.; Welsch, S.; et al. Mechanistic and structural diversity between cytochrome bd isoforms of Escherichia coli. Proc. Natl. Acad. Sci. USA 2021, 118, e2114013118. [Google Scholar] [CrossRef]
- Hill, J.J.; Alben, J.O.; Gennis, R.B. Spectroscopic evidence for a heme-heme binuclear center in the cytochrome bd ubiquinol oxidase from Escherichia coli. Proc. Natl. Acad. Sci. USA 1993, 90, 5863–5867. [Google Scholar] [CrossRef]
- Tsubaki, M.; Hori, H.; Mogi, T.; Anraku, Y. Cyanide-binding site of bd-type ubiquinol oxidase from Escherichia coli. J. Biol. Chem. 1995, 270, 28565–28569. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.; Arutyunyan, A.M.; Osborne, J.P.; Gennis, R.B.; Konstantinov, A.A. Magnetic circular dichroism used to examine the interaction of Escherichia coli cytochrome bd with ligands. Biochemistry 1999, 38, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Vos, M.H.; Borisov, V.B.; Liebl, U.; Martin, J.L.; Konstantinov, A.A. Femtosecond resolution of ligand-heme interactions in the high-affinity quinol oxidase bd: A di-heme active site? Proc. Natl. Acad. Sci. USA 2000, 97, 1554–1559. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Sedelnikova, S.E.; Poole, R.K.; Konstantinov, A.A. Interaction of cytochrome bd with carbon monoxide at low and room temperatures: Evidence that only a small fraction of heme b595 reacts with CO. J. Biol. Chem. 2001, 276, 22095–22099. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Liebl, U.; Rappaport, F.; Martin, J.L.; Zhang, J.; Gennis, R.B.; Konstantinov, A.A.; Vos, M.H. Interactions between heme d and heme b595 in quinol oxidase bd from Escherichia coli: A photoselection study using femtosecond spectroscopy. Biochemistry 2002, 41, 1654–1662. [Google Scholar] [CrossRef]
- Arutyunyan, A.M.; Borisov, V.B.; Novoderezhkin, V.I.; Ghaim, J.; Zhang, J.; Gennis, R.B.; Konstantinov, A.A. Strong excitonic interactions in the oxygen-reducing site of bd-type oxidase: The Fe-to-Fe distance between hemes d and b595 is 10 A. Biochemistry 2008, 47, 1752–1759. [Google Scholar] [CrossRef]
- Borisov, V.B. Interaction of bd-type quinol oxidase from Escherichia coli and carbon monoxide: Heme d binds CO with high affinity. Biochemistry 2008, 73, 14–22. [Google Scholar] [CrossRef]
- Bloch, D.A.; Borisov, V.B.; Mogi, T.; Verkhovsky, M.I. Heme/heme redox interaction and resolution of individual optical absorption spectra of the hemes in cytochrome bd from Escherichia coli. Biochim. Biophys. Acta 2009, 1787, 1246–1253. [Google Scholar] [CrossRef]
- Rappaport, F.; Zhang, J.; Vos, M.H.; Gennis, R.B.; Borisov, V.B. Heme-heme and heme-ligand interactions in the di-heme oxygen-reducing site of cytochrome bd from Escherichia coli revealed by nanosecond absorption spectroscopy. Biochim. Biophys. Acta 2010, 1797, 1657–1664. [Google Scholar] [CrossRef]
- Borisov, V.B.; Verkhovsky, M.I. Accommodation of CO in the di-heme active site of cytochrome bd terminal oxidase from Escherichia coli. J. Inorg. Biochem. 2013, 118, 65–67. [Google Scholar] [CrossRef]
- Siletsky, S.A.; Zaspa, A.A.; Poole, R.K.; Borisov, V.B. Microsecond time-resolved absorption spectroscopy used to study CO compounds of cytochrome bd from Escherichia coli. PLoS ONE 2014, 9, e95617. [Google Scholar] [CrossRef] [PubMed]
- Siletsky, S.A.; Rappaport, F.; Poole, R.K.; Borisov, V.B. Evidence for fast electron transfer between the high-spin haems in cytochrome bd-I from Escherichia coli. PLoS ONE 2016, 11, e0155186. [Google Scholar] [CrossRef]
- Siletsky, S.A.; Dyuba, A.V.; Elkina, D.A.; Monakhova, M.V.; Borisov, V.B. Spectral-kinetic analysis of recombination reaction of heme centers of bd-type quinol oxidase from Escherichia coli with carbon monoxide. Biochemistry 2017, 82, 1354–1366. [Google Scholar] [CrossRef]
- Borisov, V.B. Effect of membrane environment on ligand-binding properties of the terminal oxidase cytochrome bd-I from Escherichia coli. Biochemistry 2020, 85, 1603–1612. [Google Scholar] [CrossRef] [PubMed]
- Gavrikova, E.V.; Grivennikova, V.G.; Borisov, V.B.; Cecchini, G.; Vinogradov, A.D. Assembly of a chimeric respiratory chain from bovine heart submitochondrial particles and cytochrome bd terminal oxidase of Escherichia coli. FEBS Lett. 2009, 583, 1287–1291. [Google Scholar] [CrossRef] [PubMed]
- Azarkina, N.; Borisov, V.; Konstantinov, A.A. Spontaneous spectral changes of the reduced cytochrome bd. FEBS Lett. 1997, 416, 171–174. [Google Scholar] [CrossRef]
- Puustinen, A.; Finel, M.; Haltia, T.; Gennis, R.B.; Wikstrom, M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry 1991, 30, 3936–3942. [Google Scholar] [CrossRef]
- Jasaitis, A.; Borisov, V.B.; Belevich, N.P.; Morgan, J.E.; Konstantinov, A.A.; Verkhovsky, M.I. Electrogenic reactions of cytochrome bd. Biochemistry 2000, 39, 13800–13809. [Google Scholar] [CrossRef]
- Belevich, I.; Borisov, V.B.; Zhang, J.; Yang, K.; Konstantinov, A.A.; Gennis, R.B.; Verkhovsky, M.I. Time-resolved electrometric and optical studies on cytochrome bd suggest a mechanism of electron-proton coupling in the di-heme active site. Proc. Natl. Acad. Sci. USA 2005, 102, 3657–3662. [Google Scholar] [CrossRef]
- Belevich, I.; Borisov, V.B.; Verkhovsky, M.I. Discovery of the true peroxy intermediate in the catalytic cycle of terminal oxidases by real-time measurement. J. Biol. Chem. 2007, 282, 28514–28519. [Google Scholar] [CrossRef]
- Borisov, V.B.; Belevich, I.; Bloch, D.A.; Mogi, T.; Verkhovsky, M.I. Glutamate 107 in subunit I of cytochrome bd from Escherichia coli is part of a transmembrane intraprotein pathway conducting protons from the cytoplasm to the heme b595/heme d active site. Biochemistry 2008, 47, 7907–7914. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B. Cytochrome bd: Structure and properties. Biochemistry 1996, 61, 565–574. [Google Scholar]
- Junemann, S. Cytochrome bd terminal oxidase. Biochim. Biophys. Acta 1997, 1321, 107–127. [Google Scholar] [CrossRef] [PubMed]
- Forte, E.; Borisov, V.B.; Vicente, J.B.; Giuffre, A. Cytochrome bd and gaseous ligands in bacterial physiology. Adv. Microb. Physiol. 2017, 71, 171–234. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Smirnova, I.A.; Krasnosel’skaya, I.A.; Konstantinov, A.A. Oxygenated cytochrome bd from Escherichia coli can be converted into the oxidized form by lipophilic electron acceptors. Biochemistry 1994, 59, 437–443. [Google Scholar]
- D’mello, R.; Hill, S.; Poole, R.K. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two-oxygen-binding haems: Implicaitons for regulation of activity in vivo by oxygen inihibition. Microbiology 1996, 142, 755–763. [Google Scholar] [CrossRef]
- Belevich, I.; Borisov, V.B.; Konstantinov, A.A.; Verkhovsky, M.I. Oxygenated complex of cytochrome bd from Escherichia coli: Stability and photolability. FEBS Lett. 2005, 579, 4567–4570. [Google Scholar] [CrossRef]
- Belevich, I.; Borisov, V.B.; Bloch, D.A.; Konstantinov, A.A.; Verkhovsky, M.I. Cytochrome bd from Azotobacter vinelandii: Evidence for high-affinity oxygen binding. Biochemistry 2007, 46, 11177–11184. [Google Scholar] [CrossRef]
- Forte, E.; Borisov, V.B.; Siletsky, S.A.; Petrosino, M.; Giuffre, A. In the respiratory chain of Escherichia coli cytochromes bd-I and bd-II are more sensitive to carbon monoxide inhibition than cytochrome bo3. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 148088. [Google Scholar] [CrossRef]
- Azarkina, N.; Siletsky, S.; Borisov, V.; von Wachenfeldt, C.; Hederstedt, L.; Konstantinov, A.A. A cytochrome bb’-type quinol oxidase in Bacillus subtilis strain 168. J. Biol. Chem. 1999, 274, 32810–32817. [Google Scholar] [CrossRef]
- Borisov, V.B.; Gennis, R.B.; Hemp, J.; Verkhovsky, M.I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 2011, 1807, 1398–1413. [Google Scholar] [CrossRef] [PubMed]
- Arutyunyan, A.M.; Sakamoto, J.; Inadome, M.; Kabashima, Y.; Borisov, V.B. Optical and magneto-optical activity of cytochrome bd from Geobacillus thermodenitrificans. Biochim. Biophys. Acta 2012, 1817, 2087–2094. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Gennis, R.B.; Konstantinov, A.A. Interaction of cytochrome bd from Escherichia coli with hydrogen peroxide. Biochemistry 1995, 60, 231–239. [Google Scholar]
- Borisov, V.; Gennis, R.; Konstantinov, A.A. Peroxide complex of cytochrome bd: Kinetics of generation and stability. Biochem. Mol. Biol. Int. 1995, 37, 975–982. [Google Scholar]
- Borisov, V.B.; Forte, E.; Sarti, P.; Giuffre, A. Catalytic intermediates of cytochrome bd terminal oxidase at steady-state: Ferryl and oxy-ferrous species dominate. Biochim. Biophys. Acta 2011, 1807, 503–509. [Google Scholar] [CrossRef]
- Paulus, A.; Rossius, S.G.; Dijk, M.; de Vries, S. Oxoferryl-porphyrin radical catalytic intermediate in cytochrome bd oxidases protects cells from formation of reactive oxygen species. J. Biol. Chem. 2012, 287, 8830–8838. [Google Scholar] [CrossRef]
- Borisov, V.B.; Murali, R.; Verkhovskaya, M.L.; Bloch, D.A.; Han, H.; Gennis, R.B.; Verkhovsky, M.I. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc. Natl. Acad. Sci. USA 2011, 108, 17320–17324. [Google Scholar] [CrossRef]
- Forte, E.; Borisov, V.B.; Konstantinov, A.A.; Brunori, M.; Giuffre, A.; Sarti, P. Cytochrome bd, a key oxidase in bacterial survival and tolerance to nitrosative stress. Ital. J. Biochem. 2007, 56, 265–269. [Google Scholar]
- Borisov, V.B.; Forte, E.; Siletsky, S.A.; Arese, M.; Davletshin, A.I.; Sarti, P.; Giuffre, A. Cytochrome bd protects bacteria against oxidative and nitrosative stress: A potential target for next-generation antimicrobial agents. Biochemistry 2015, 80, 565–575. [Google Scholar] [CrossRef]
- Giuffre, A.; Borisov, V.B.; Mastronicola, D.; Sarti, P.; Forte, E. Cytochrome bd oxidase and nitric oxide: From reaction mechanisms to bacterial physiology. FEBS Lett. 2012, 586, 622–629. [Google Scholar] [CrossRef]
- Giuffre, A.; Borisov, V.B.; Arese, M.; Sarti, P.; Forte, E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim. Biophys. Acta 2014, 1837, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Davletshin, A.I.; Konstantinov, A.A. Peroxidase activity of cytochrome bd from Escherichia coli. Biochemistry 2010, 75, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Forte, E.; Davletshin, A.; Mastronicola, D.; Sarti, P.; Giuffre, A. Cytochrome bd oxidase from Escherichia coli displays high catalase activity: An additional defense against oxidative stress. FEBS Lett. 2013, 587, 2214–2218. [Google Scholar] [CrossRef] [PubMed]
- Forte, E.; Borisov, V.B.; Davletshin, A.; Mastronicola, D.; Sarti, P.; Giuffre, A. Cytochrome bd oxidase and hydrogen peroxide resistance in Mycobacterium tuberculosis. mBio 2013, 4, e01006-01013. [Google Scholar] [CrossRef]
- Al-Attar, S.; Yu, Y.; Pinkse, M.; Hoeser, J.; Friedrich, T.; Bald, D.; de Vries, S. Cytochrome bd displays significant quinol peroxidase activity. Sci. Rep. 2016, 6, 27631. [Google Scholar] [CrossRef] [PubMed]
- Forte, E.; Nastasi, M.R.; Borisov, V.B. Preparations of terminal oxidase cytochrome bd-II isolated from Escherichia coli reveal significant hydrogen peroxide scavenging activity. Biochemistry, 2022; in press. [Google Scholar]
- Forte, E.; Borisov, V.B.; Falabella, M.; Colaco, H.G.; Tinajero-Trejo, M.; Poole, R.K.; Vicente, J.B.; Sarti, P.; Giuffre, A. The terminal oxidase cytochrome bd promotes sulfide-resistant bacterial respiration and growth. Sci. Rep. 2016, 6, 23788. [Google Scholar] [CrossRef]
- Korshunov, S.; Imlay, K.R.; Imlay, J.A. The cytochrome bd oxidase of Escherichia coli prevents respiratory inhibition by endogenous and exogenous hydrogen sulfide. Mol. Microbiol. 2016, 101, 62–77. [Google Scholar] [CrossRef]
- Forte, E.; Giuffre, A. How bacteria breathe in hydrogen sulphide-rich environments. Biochemist 2016, 38, 8–11. [Google Scholar] [CrossRef]
- Borisov, V.B.; Forte, E. Terminal oxidase cytochrome bd protects bacteria against hydrogen sulfide toxicity. Biochemistry 2021, 86, 22–32. [Google Scholar] [CrossRef]
- Forte, E.; Siletsky, S.A.; Borisov, V.B. In Escherichia coli ammonia inhibits cytochrome bo3 but activates cytochrome bd-I. Antioxidants 2021, 10, 13. [Google Scholar] [CrossRef]
- Xia, X.; Wu, S.; Li, L.; Xu, B.; Wang, G. The cytochrome bd complex is essential for chromate and sulfide resistance and is regulated by a GbsR-type regulator, CydE, in Alishewanella sp. WH16-1. Front. Microbiol. 2018, 9, 1849. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, J.; Koga, E.; Mizuta, T.; Sato, C.; Noguchi, S.; Sone, N. Gene structure and quinol oxidase activity of a cytochrome bd-type oxidase from Bacillus stearothermophilus. Biochim. Biophys. Acta 1999, 1411, 147–158. [Google Scholar] [CrossRef]
- Kalia, N.P.; Hasenoehrl, E.J.; Ab Rahman, N.B.; Koh, V.H.; Ang, M.L.T.; Sajorda, D.R.; Hards, K.; Gruber, G.; Alonso, S.; Cook, G.M.; et al. Exploiting the synthetic lethality between terminal respiratory oxidases to kill Mycobacterium tuberculosis and clear host infection. Proc. Natl. Acad. Sci. USA 2017, 114, 7426–7431. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.S.; Hards, K.; Engelhart, C.A.; Hasenoehrl, E.J.; Kalia, N.P.; Mackenzie, J.S.; Sviriaeva, E.; Chong, S.M.S.; Manimekalai, M.S.S.; Koh, V.H.; et al. Dual inhibition of the terminal oxidases eradicates antibiotic-tolerant Mycobacterium tuberculosis. EMBO Mol. Med. 2021, 13, e13207. [Google Scholar] [CrossRef] [PubMed]
- Meunier, B.; Madgwick, S.A.; Reil, E.; Oettmeier, W.; Rich, P.R. New inhibitors of the quinol oxidation sites of bacterial cytochromes bo and bd. Biochemistry 1995, 34, 1076–1083. [Google Scholar] [CrossRef]
- Radloff, M.; Elamri, I.; Grund, T.N.; Witte, L.F.; Hohmann, K.F.; Nakagaki, S.; Goojani, H.G.; Nasiri, H.; Hideto, M.; Bald, D.; et al. Short-chain aurachin D derivatives are selective inhibitors of E. coli cytochrome bd-I and bd-II oxidases. Sci. Rep. 2021, 11, 23852. [Google Scholar] [CrossRef]
- Miyoshi, H.; Takegami, K.; Sakamoto, K.; Mogi, T.; Iwamura, H. Characterization of the ubiquinol oxidation sites in cytochromes bo and bd from Escherichia coli using aurachin C analogues. J. Biochem. 1999, 125, 138–142. [Google Scholar] [CrossRef]
- Makarchuk, I.; Nikolaev, A.; Thesseling, A.; Dejon, L.; Lamberty, D.; Stief, L.; Speicher, A.; Friedrich, T.; Hellwig, P.; Nasiri, H.R.; et al. Identification and optimization of quinolone-based inhibitors against cytochrome bd oxidase using an electrochemical assay. Electrochim. Acta 2021, 381, 138293. [Google Scholar] [CrossRef]
- Lu, P.; Heineke, M.H.; Koul, A.; Andries, K.; Cook, G.M.; Lill, H.; van Spanning, R.; Bald, D. The cytochrome bd-type quinol oxidase is important for survival of Mycobacterium smegmatis under peroxide and antibiotic-induced stress. Sci. Rep. 2015, 5, 10333. [Google Scholar] [CrossRef]
- Harikishore, A.; Chong, S.S.M.; Ragunathan, P.; Bates, R.W.; Gruber, G. Targeting the menaquinol binding loop of mycobacterial cytochrome bd oxidase. Mol. Divers. 2021, 25, 517–524. [Google Scholar] [CrossRef]
- Hopfner, S.M.; Lee, B.S.; Kalia, N.P.; Miller, M.J.; Pethe, K.; Moraski, G.C. Structure guided generation of thieno[3,2-d]pyrimidin-4-amine Mycobacterium tuberculosis bd oxidase inhibitors. RSC Med. Chem. 2021, 12, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Akhter, Y. A review on enzyme complexes of electron transport chain from Mycobacterium tuberculosis as promising drug targets. Int. J. Biol. Macromol. 2022, 212, 474–494. [Google Scholar] [CrossRef] [PubMed]
- Hards, K.; Cheung, C.Y.; Waller, N.; Adolph, C.; Keighley, L.; Tee, Z.S.; Harold, L.K.; Menorca, A.; Bujaroski, R.S.; Buckley, B.J.; et al. An amiloride derivative is active against the F1Fo-ATP synthase and cytochrome bd oxidase of Mycobacterium tuberculosis. Commun. Biol. 2022, 5, 166. [Google Scholar] [CrossRef]
- Arjona, D.; Wikstrom, M.; Adelroth, P. Nitric oxide is a potent inhibitor of the cbb3-type heme-copper oxidases. FEBS Lett. 2015, 589, 1214–1218. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.C.; Cooper, C.E. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Lett. 1994, 356, 295–298. [Google Scholar] [CrossRef]
- Cooper, C.E.; Davies, N.A.; Psychoulis, M.; Canevari, L.; Bates, T.E.; Dobbie, M.S.; Casley, C.S.; Sharpe, M.A. Nitric oxide and peroxynitrite cause irreversible increases in the Km for oxygen of mitochondrial cytochrome oxidase: In Vitro and In Vivo studies. Biochim. Biophys. Acta 2003, 1607, 27–34. [Google Scholar] [CrossRef]
- Torres, J.; Cooper, C.E.; Wilson, M.T. A common mechanism for the interaction of nitric oxide with the oxidized binuclear centre and oxygen intermediates of cytochrome c oxidase. J. Biol. Chem. 1998, 273, 8756–8766. [Google Scholar] [CrossRef]
- Mason, M.G.; Nicholls, P.; Wilson, M.T.; Cooper, C.E. Nitric oxide inhibition of respiration involves both competitive (heme) and noncompetitive (copper) binding to cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2006, 103, 708–713. [Google Scholar] [CrossRef]
- Sarti, P.; Forte, E.; Mastronicola, D.; Giuffre, A.; Arese, M. Cytochrome c oxidase and nitric oxide in action: Molecular mechanisms and pathophysiological implications. Biochim. Biophys. Acta 2012, 1817, 610–619. [Google Scholar] [CrossRef]
- Chen, J.; Xie, P.; Huang, Y.; Gao, H. Complex interplay of heme-copper oxidases with nitrite and nitric oxide. Int. J. Mol. Sci. 2022, 23, 979. [Google Scholar] [CrossRef]
- Borisov, V.B.; Forte, E.; Konstantinov, A.A.; Poole, R.K.; Sarti, P.; Giuffre, A. Interaction of the bacterial terminal oxidase cytochrome bd with nitric oxide. FEBS Lett. 2004, 576, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Forte, E.; Sarti, P.; Brunori, M.; Konstantinov, A.A.; Giuffre, A. Redox control of fast ligand dissociation from Escherichia coli cytochrome bd. Biochem. Biophys. Res. Commun. 2007, 355, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Carabet, L.A.; Guertin, M.; Lague, P.; Lamoureux, G. Mechanism of the nitric oxide dioxygenase reaction of Mycobacterium tuberculosis hemoglobin N. J. Phys. Chem. B 2017, 121, 8706–8718. [Google Scholar] [CrossRef] [PubMed]
- Giuffre, A.; Stubauer, G.; Sarti, P.; Brunori, M.; Zumft, W.G.; Buse, G.; Soulimane, T. The heme-copper oxidases of Thermus thermophilus catalyze the reduction of nitric oxide: Evolutionary implications. Proc. Natl. Acad. Sci. USA 1999, 96, 14718–14723. [Google Scholar] [CrossRef]
- Butler, C.; Forte, E.; Maria Scandurra, F.; Arese, M.; Giuffre, A.; Greenwood, C.; Sarti, P. Cytochrome bo3 from Escherichia coli: The binding and turnover of nitric oxide. Biochem. Biophys. Res. Commun. 2002, 296, 1272–1278. [Google Scholar] [CrossRef]
- Forte, E.; Urbani, A.; Saraste, M.; Sarti, P.; Brunori, M.; Giuffre, A. The cytochrome cbb3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur. J. Biochem. 2001, 268, 6486–6491. [Google Scholar] [CrossRef]
- Huang, Y.; Reimann, J.; Lepp, H.; Drici, N.; Adelroth, P. Vectorial proton transfer coupled to reduction of O2 and NO by a heme-copper oxidase. Proc. Natl. Acad. Sci. USA 2008, 105, 20257–20262. [Google Scholar] [CrossRef]
- Stubauer, G.; Giuffre, A.; Brunori, M.; Sarti, P. Cytochrome c oxidase does not catalyze the anaerobic reduction of NO. Biochem. Biophys. Res. Commun. 1998, 245, 459–465. [Google Scholar] [CrossRef]
- Ohta, T.; Soulimane, T.; Kitagawa, T.; Varotsis, C. Nitric oxide activation by caa3 oxidoreductase from Thermus thermophilus. Phys. Chem. Chem. Phys. 2015, 17, 10894–10898. [Google Scholar] [CrossRef]
- Blomberg, M.R.A. Activation of O2 and NO in heme-copper oxidases-mechanistic insights from computational modelling. Chem. Soc. Rev. 2020, 49, 7301–7330. [Google Scholar] [CrossRef]
- Jones-Carson, J.; Husain, M.; Liu, L.; Orlicky, D.J.; Vazquez-Torres, A. Cytochrome bd-dependent bioenergetics and antinitrosative defenses in Salmonella pathogenesis. mBio 2016, 7, e02052-02016. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, M.; Achard, M.E.; Idris, A.; Totsika, M.; Phan, M.D.; Peters, K.M.; Sarkar, S.; Ribeiro, C.A.; Holyoake, L.V.; Ladakis, D.; et al. The cytochrome bd-I respiratory oxidase augments survival of multidrug-resistant Escherichia coli during infection. Sci. Rep. 2016, 6, 35285. [Google Scholar] [CrossRef] [PubMed]
- Beebout, C.J.; Eberly, A.R.; Werby, S.H.; Reasoner, S.A.; Brannon, J.R.; De, S.; Fitzgerald, M.J.; Huggins, M.M.; Clayton, D.B.; Cegelski, L.; et al. Respiratory heterogeneity shapes biofilm formation and host colonization in uropathogenic Escherichia coli. mBio 2019, 10, e02400-18. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Shepherd, M.; Nicholls, P.; Dobbin, P.S.; Dodsworth, K.S.; Poole, R.K.; Cooper, C.E. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 2009, 5, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Yin, J.; Jin, M.; Gao, H. Distinct nitrite and nitric oxide physiologies in Escherichia coli and Shewanella oneidensis. Appl. Environ. Microbiol. 2018, 84, e00559-00518. [Google Scholar] [CrossRef] [PubMed]
- Pullan, S.T.; Gidley, M.D.; Jones, R.A.; Barrett, J.; Stevanin, T.M.; Read, R.C.; Green, J.; Poole, R.K. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: Unaltered methionine biosynthesis indicates lack of S nitrosation. J. Bacteriol. 2007, 189, 1845–1855. [Google Scholar] [CrossRef]
- Hyduke, D.R.; Jarboe, L.R.; Tran, L.M.; Chou, K.J.; Liao, J.C. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. USA 2007, 104, 8484–8489. [Google Scholar] [CrossRef]
- Richardson, A.R.; Dunman, P.M.; Fang, F.C. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 2006, 61, 927–939. [Google Scholar] [CrossRef]
- Moore, C.M.; Nakano, M.M.; Wang, T.; Ye, R.W.; Helmann, J.D. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J. Bacteriol. 2004, 186, 4655–4664. [Google Scholar] [CrossRef]
- Shi, L.; Sohaskey, C.D.; Kana, B.D.; Dawes, S.; North, R.J.; Mizrahi, V.; Gennaro, M.L. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under In Vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. USA 2005, 102, 15629–15634. [Google Scholar] [CrossRef]
- Cai, Y.; Jaecklein, E.; Mackenzie, J.S.; Papavinasasundaram, K.; Olive, A.J.; Chen, X.; Steyn, A.J.C.; Sassetti, C.M. Host immunity increases Mycobacterium tuberculosis reliance on cytochrome bd oxidase. PLoS Pathog. 2021, 17, e1008911. [Google Scholar] [CrossRef] [PubMed]
- Stevanin, T.M.; Ioannidis, N.; Mills, C.E.; Kim, S.O.; Hughes, M.N.; Poole, R.K. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo’ or bd, from nitric oxide. J. Biol. Chem. 2000, 275, 35868–35875. [Google Scholar] [CrossRef] [PubMed]
- Sarti, P.; Giuffre, A.; Forte, E.; Mastronicola, D.; Barone, M.C.; Brunori, M. Nitric oxide and cytochrome c oxidase: Mechanisms of inhibition and NO degradation. Biochem. Biophys. Res. Commun. 2000, 274, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Forte, E.; Sarti, P.; Brunori, M.; Konstantinov, A.A.; Giuffre, A. Nitric oxide reacts with the ferryl-oxo catalytic intermediate of the CuB-lacking cytochrome bd terminal oxidase. FEBS Lett. 2006, 580, 4823–4826. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Forte, E.; Giuffre, A.; Konstantinov, A.; Sarti, P. Reaction of nitric oxide with the oxidized di-heme and heme-copper oxygen-reducing centers of terminal oxidases: Different reaction pathways and end-products. J. Inorg. Biochem. 2009, 103, 1185–1187. [Google Scholar] [CrossRef]
- Giuffre, A.; Barone, M.C.; Mastronicola, D.; D’Itri, E.; Sarti, P.; Brunori, M. Reaction of nitric oxide with the turnover intermediates of cytochrome c oxidase: Reaction pathway and functional effects. Biochemistry 2000, 39, 15446–15453. [Google Scholar] [CrossRef]
- Yang, K.; Borisov, V.B.; Konstantinov, A.A.; Gennis, R.B. The fully oxidized form of the cytochrome bd quinol oxidase from E. coli does not participate in the catalytic cycle: Direct evidence from rapid kinetics studies. FEBS Lett. 2008, 582, 3705–3709. [Google Scholar] [CrossRef]
- Borisov, V.B.; Forte, E.; Siletsky, S.A.; Sarti, P.; Giuffre, A. Cytochrome bd from Escherichia coli catalyzes peroxynitrite decomposition. Biochim. Biophys. Acta 2015, 1847, 182–188. [Google Scholar] [CrossRef]
- Sharpe, M.A.; Cooper, C.E. Interaction of peroxynitrite with mitochondrial cytochrome oxidase. Catalytic production of nitric oxide and irreversible inhibition of enzyme activity. J. Biol. Chem. 1998, 273, 30961–30972. [Google Scholar] [CrossRef]
- Floris, R.; Piersma, S.R.; Yang, G.; Jones, P.; Wever, R. Interaction of myeloperoxidase with peroxynitrite. A comparison with lactoperoxidase, horseradish peroxidase and catalase. Eur. J. Biochem. 1993, 215, 767–775. [Google Scholar] [CrossRef]
- Van Zyl, J.M.; Van der Walt, B.J. Apparent hydroxyl radical generation without transition metal catalysis and tyrosine nitration during oxidation of the anti-tubercular drug, isonicotinic acid hydrazide. Biochem. Pharmacol. 1994, 48, 2033–2042. [Google Scholar] [CrossRef]
- Stern, M.K.; Jensen, M.P.; Kramer, K. Peroxynitrite decomposition catalysts. J. Am. Chem. Soc. 1996, 118, 8735–8736. [Google Scholar] [CrossRef]
- Bryk, R.; Griffin, P.; Nathan, C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 2000, 407, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Sueta, G.; Campolo, N.; Trujillo, M.; Bartesaghi, S.; Carballal, S.; Romero, N.; Alvarez, B.; Radi, R. Biochemistry of peroxynitrite and protein tyrosine nitration. Chem. Rev. 2018, 118, 1338–1408. [Google Scholar] [CrossRef] [PubMed]


| Enzyme Complex | Electron Donor | Electron Acceptor | Energy Currency Produced |
|---|---|---|---|
| Complex I | NADH | ubiquinone | proton motive force (PMF) |
| Complex II | succinate | ubiquinone | none |
| Complex III | ubiquinol | ferricytochrome c | PMF |
| Complex IV | ferrocytochrome c | O2 | PMF |
| Enzyme Complex | Electron Donor | Electron Acceptor | Energy Currency Produced |
|---|---|---|---|
| NDH-1 | NADH | quinone | PMF |
| NDH-2 | NADH | quinone | none |
| NQR | NADH | quinone | Na+ motive force |
| Complex II | succinate | quinone | none |
| Complex III | quinol | ferricytochrome c | PMF |
| Heme–copper oxidases (aa3, caa3, bo3, cbb3, ba3) | ferrocytochrome c or quinol | O2 | PMF |
| Cytochrome bcc-aa3 supercomplex | quinol | O2 | PMF |
| Cytochrome bd (bd-I, bd-II) | quinol | O2 | PMF |
| Cyanide insensitive bd-type oxidase (CIO) | quinol | O2 | n.d. |
| Enzyme Complex | Inhibition by •NO | •NO Degradation in Turnover | Anaerobic •NO Degradation | •NO off-Rate | NO2– off-Rate | Reference |
|---|---|---|---|---|---|---|
| Mycobacterial cytochrome bcc-aa3 supercomplex | No | Yes (~300 mol •NO × (mol bcc-aa3)−1 × min−1) | Yes (~3 mol •NO × (mol bcc-aa3)−1 × min−1) | n.d. | n.d. | [57] |
| E. coli cytochrome bd-I | Yes (IC50 = 100 nM •NO at 70 μM O2) | No | No | 0.133 s−1 | n.d. | [142,143] |
| Enzyme Complex | Inhibition by ONOO− | •NO Production after ONOO− Addition | Short-Term Generation of O2 just after ONOO− Addition | Direct Observation of ONOO− Degradation in Turnover | Reference |
|---|---|---|---|---|---|
| Purified bovine heart aa3-type cytochrome c oxidase | Yes (irreversible damage to enzyme complex) | Yes | No | No | [170] |
| Purified E. coli cytochrome bd-I | No (up to 0.1 mM ONOO−) | Yes | Yes | Yes (~600 mol ONOO− × (mol bd-I)−1 × min−1) | [169] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borisov, V.B.; Forte, E. Bioenergetics and Reactive Nitrogen Species in Bacteria. Int. J. Mol. Sci. 2022, 23, 7321. https://doi.org/10.3390/ijms23137321
Borisov VB, Forte E. Bioenergetics and Reactive Nitrogen Species in Bacteria. International Journal of Molecular Sciences. 2022; 23(13):7321. https://doi.org/10.3390/ijms23137321
Chicago/Turabian StyleBorisov, Vitaliy B., and Elena Forte. 2022. "Bioenergetics and Reactive Nitrogen Species in Bacteria" International Journal of Molecular Sciences 23, no. 13: 7321. https://doi.org/10.3390/ijms23137321
APA StyleBorisov, V. B., & Forte, E. (2022). Bioenergetics and Reactive Nitrogen Species in Bacteria. International Journal of Molecular Sciences, 23(13), 7321. https://doi.org/10.3390/ijms23137321







