Abstract
Connexin (Cx) hemichannels (HCs) are large pore hexameric structures that allow the exchange of ions, metabolites and a variety of other molecules between the cell cytoplasm and extracellular milieu. HC inhibitors are attracting growing interest as drug candidates because deregulated fluxes through HCs have been implicated in a plethora of genetic conditions and other diseases. HC activity has been mainly investigated by electrophysiological methods and/or using HC-permeable dye uptake measurements. Here, we present an all-optical assay based on fluorometric measurements of ionized calcium (Ca2+) uptake with a Ca2+-selective genetically encoded indicator (GCaMP6s) that permits the optical tracking of cytosolic Ca2+ concentration ([Ca2+]cyt) changes with high sensitivity. We exemplify use of the assay in stable pools of HaCaT cells overexpressing human Cx26, Cx46, or the pathological mutant Cx26G45E, under control of a tetracycline (Tet) responsive element (TRE) promoter (Tet-on). We demonstrate the usefulness of the assay for the characterization of new monoclonal antibodies (mAbs) targeting the extracellular domain of the HCs. Although we developed the assay on a spinning disk confocal fluorescence microscope, the same methodology can be extended seamlessly to high-throughput high-content platforms to screen other kinds of inhibitors and/or to probe HCs expressed in primary cells and microtissues.
1. Introduction
Connexin (Cx) hemichannels (HCs) may function as large-pore plasma membrane channels or dock head-to-head with other HCs from an opposing cell, leading to the formation of intercellular gap junction channels (IGJCs) [1,2,3,4]. Both Cx HCs and IGJCs are permeable to a variety of molecular species, such as ions, second messengers, metabolites and even small noncoding RNA (siRNAs and miRNAs). Based on pore size estimates, the list of permeating molecules could potentially include >35,000 members of the Human Metabolome Database [5], including Ca2+ [6,7,8,9,10].
Not only Ca2+ can permeate through Cx HCs but, in addition, HC gating depends on Ca2+ concentration in the extracellular milieu ([Ca2+]ex). In particular, it is well known that lowering the [Ca2+]ex favors the opening of HCs formed by several different types of Cxs, whereas relatively high Ca2+ concentrations prevent opening [11,12,13,14,15,16,17]. Numerous other factors modulate the open probability of Cx HCs, including mechanical strain, changes in pH, transmembrane voltage, post translational modifications (especially protein phosphorylation), cellular redox state, metabolic inhibition, nitric oxide and linoleic acid; reviewed in [17,18,19,20].
The opening of Cx HCs promotes the diffusive release of paracrine messengers, including ATP [21,22,23,24,25,26], glutamate [27], prostaglandins [28], NAD(+) [29] and glutathione [30,31]; reviewed in [32]. In several instances, molecules released through open Cx HCs, most importantly ATP, may act as damage-associated molecular patterns (DAMPs) [33], activating inflammatory pathways and promoting cell death [34,35,36,37]. Not surprisingly, point mutations that render Cx HCs constitutively more active or abnormally active (referred to also as “leaky” HCs) have been implicated in numerous genetic diseases affecting the facial appearance, dentition (small and carious teeth), eyes (microphthalmia, microcornea), fingers (syndactyly), lens (cataracts), nervous system (myelination defects), heart (atrial fibrillation), inner ear (deafness) and skin (various disorders); reviewed in [38,39,40].
Deregulated Ca2+ flux through HCs has been implicated in several diseases linked to Cx mutations [18,38,39,40]. In addition, the opening of wild type (wt) Cx HCs is associated with an impressive list of pathological conditions that affect a major proportion of the world population, including ischemia/stroke, Alzheimer’s disease, epilepsy, liver fibrosis and cirrhosis, nonalcoholic steatohepatitis, inflammation and others [41,42,43,44,45,46]. Of note, inhibition of Cx HCs in animal models in vivo exerted anticonvulsant effects [47], lowered the degree of liver fibrosis induced by administration of thioacetamide [48] and improved recovery from spinal cord injury [49].
Cx HC activity has been probed indirectly by the release of ATP and other metabolites as mentioned above [21,22,23,24,25,26]. Other classical methods include electrophysiology (patch clamp) and dye uptake or leakage assays. The latter are based on passive diffusion of fluorescent molecules through open HCs in the cell plasma membrane [50]. Experiments of this type have been mainly conducted in model cell systems but occasionally also in native tissues [24,25] or in vivo [26]. The aim of this work was to establish a complementary, all-optical method to investigate Cx HC activity based on the uptake of Ca2+.
We focused on Cx26, which is expressed at extremely low, oftentimes undetectable levels in epidermal keratinocytes, yet, paradoxically, mutant Cx26 leaky/hyperactive HCs cause genodermatoses, including keratitis-ichthyosis-deafness syndrome (KID) and palmoplantar keratoderma with deafness (PPKDFN) that may drastically compromise quality of life and life-long health [40,51,52] (Table 1). To broaden the impact of this study and demonstrate the wider applicability of the methodology, we also investigated HCs composed of Cx46 [11,12], which is an important target for cancer therapy [53,54,55,56,57] and, when mutated, is involved in cataract formation [58].

Table 1.
Rare genodermatoses caused by leaky/hyperactive HCs. (P = prevalence; C = number of cases described in the literature; and E = estimated number of people with this disease).
The Ca2+ uptake assay we developed provides a highly sensitive readout of Cx HC activity, as detailed hereafter. Furthermore, the assay is also suitable to evaluate the efficacy of different HC inhibitors that are attracting a growing attention as potential therapeutics [41,42,43,44,45,46,47,48], including antibodies targeting the extracellular domain of HCs [49,59,60,61,62].
2. Results
2.1. Generation of Stable Cell Pools Overexpressing the Cx of Interest and GCaMP6s
HaCaT cells are spontaneously immortalized line of aneuploid human keratinocytes [63] widely adopted to model the epidermis in vitro [64,65]. We and others previously overexpressed Cx HCs in HaCaT cells [59,61,66]. Here, given that uncontrolled expression of leaky HCs causes toxicity and rapidly leads to cell death (see, e.g., [66,67,68,69,70,71,72,73]), we designed and produced lentiviruses (LVs) for Tet-on bicistronic expression of the Cx of interest and the genetically encoded indicator GCaMP6s that permits optical tracking of [Ca2+]cyt changes with high sensitivity [74] (see Materials and Methods).
We inserted the Cx coding sequence (CDS) upstream of an internal ribosome entry site (IRES) sequence, in turn, followed by the CDS of GCaMP6s. Next, by puromycin selection of virally transduced cells, we obtained stable cell pools expressing human Cx26 (referred to as HaCaT-Cx26-GCaMP6s cells) or the lethal KID-related mutant Cx26G45E that generates leaky HCs [68,70,75,76,77] (referred to as HaCaT-Cx26G45E-GCaMP6s cells). The expression levels of both IRES and GCaMP6s mRNAs increased over 350-fold within 24 h of doxycycline (dox) addition (2 µg/mL) to the culture medium (Figure 1). The non-zero levels of both mRNAs in the absence of dox can be attributed to the minimal basal leakiness of the TRE promoter in the lentiviral construct [78].
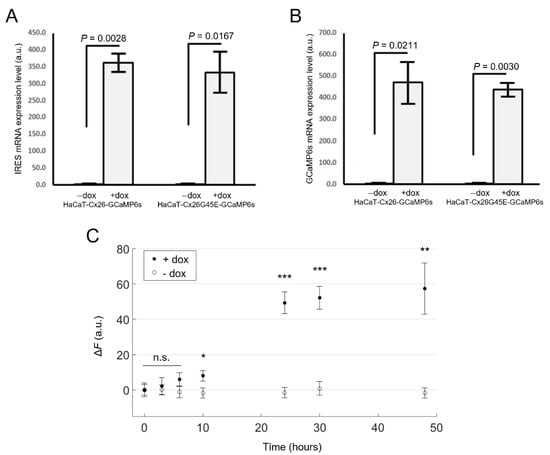
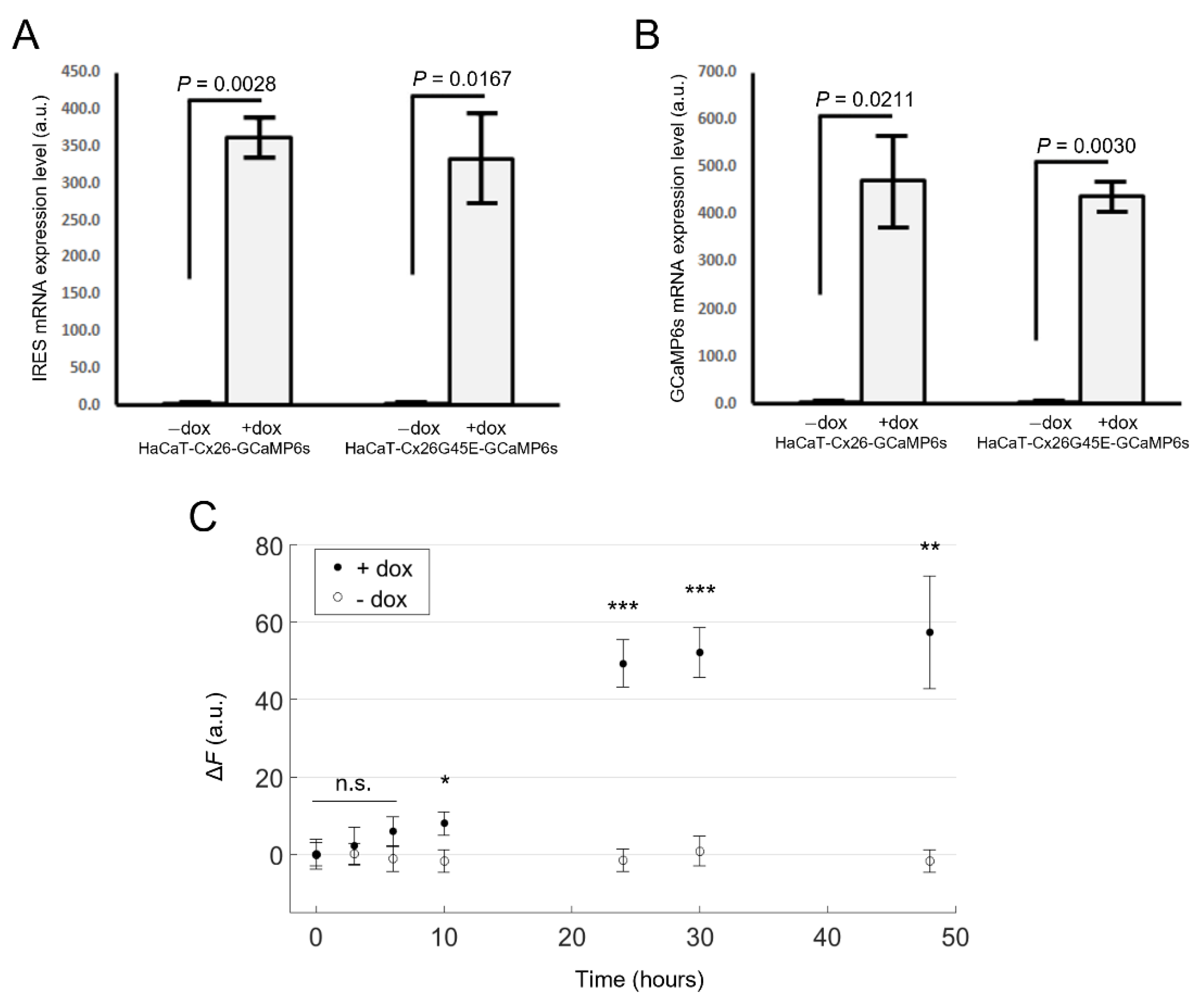
Figure 1.
Validation of Tet-on bicistronic lentiviral constructs used to generate stable cell pools. mRNA expression levels of IRES (A) or GCaMP6s (B) assayed by qPCR (mean values ± standard deviation) in HaCaT-Cx26-GCaMP6s and HaCaT-Cx26G45E-GCaMP6s cells exposed to doxycycline for 24 h (+dox), or not exposed to doxycycline (−dox). p-values (p) were calculated using the two-tailed t-test. (C) GCaMP6s fluorescence levels in live HaCaT-Cx26-GCaMP6s cells vs. time after addition of dox (2 µg/mL, filled symbols) or vehicle (empty symbols) to the culture medium. The mean values ± standard error of the mean (s.e.m.) for Δ = were computed for each condition as average fluorescence signal ( ) of pixels that exceeded an arbitrary threshold minus the average value obtained before dox administration (, t = 0). For each condition, values of n = 4–7 fields of view from one cell culture were averaged, and the statistical significance was calculated using a two-tailed t-test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and n.s., not significant.
Analyses of HaCaT-Cx26-GCaMP6s cells by live microscopy showed a monotonic growth of GCaMP6s fluorescence levels with time, reaching a steady state after ~30 h from the onset of dox exposure (Figure 1C). In HaCaT-Cx26-GCaMP6s cells pre-exposed to dox for 24 h, we confirmed overexpression of Cx26 by qPCR (Figure 2A) as well as by immunofluorescence staining with a commercial antibody that recognizes an intracellular epitope (Figure 2B). Note that Cx26 was detected at very low levels also in the absence of dox. Immunoreactivity against Cx26 produced fluorescent puncta at points of contacts between adjacent cells, corresponding to IGJCs (Figure 2C).
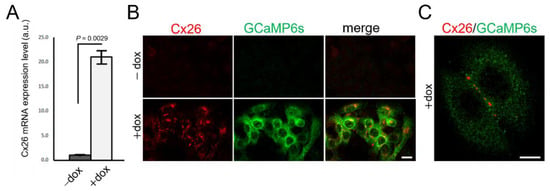
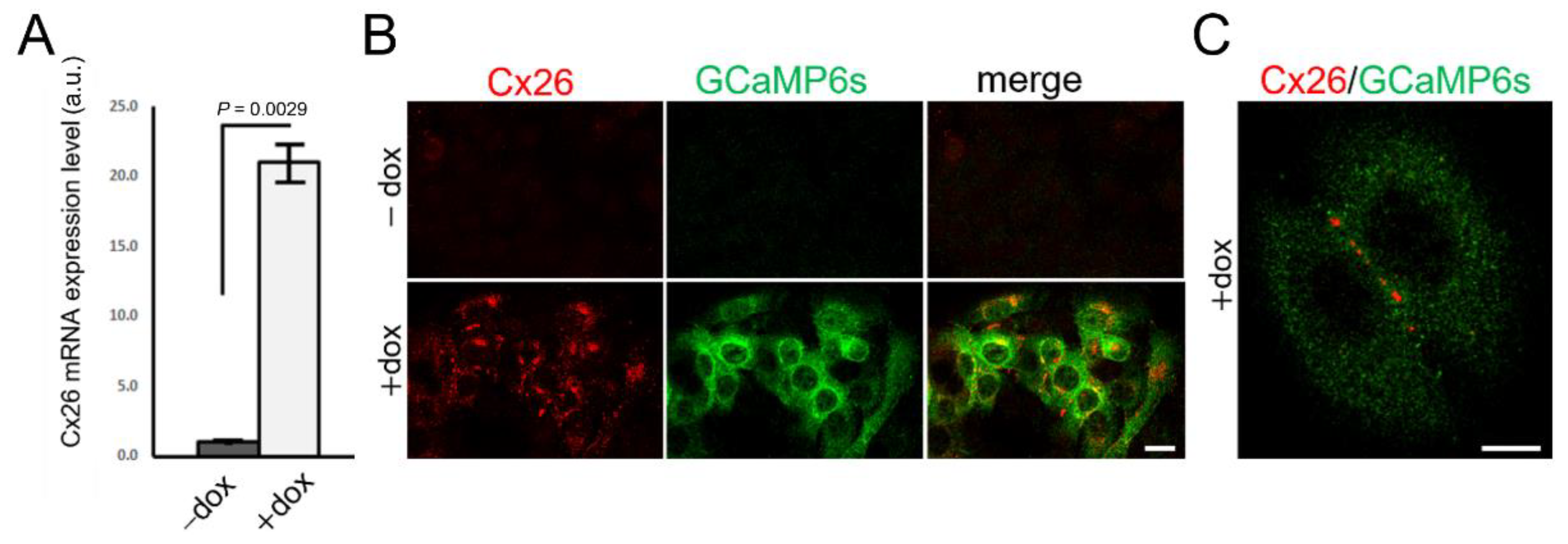
Figure 2.
Characterization of Cx26 expression in HaCaT-Cx26-GCaMP6s cell pools. (A) qPCR assay of Cx26 mRNA expression levels (mean values ± standard deviation) in HaCaT-Cx26-GCaMP6s cells not exposed (−dox) or exposed (+dox) to dox (2 µg/mL, 24 h). p-values (P) were calculated using a two-tailed t-test. (B) Representative confocal images of fixed and permeabilized cells labelled with anti-Cx26 (red) and anti-GFP (green) antibodies (the GFP epitope recognized by the antibody is also present in GCaMP6s); scale bar: 20 µm. (C) Higher magnification view showing fluorescent puncta at points of contacts between adjacent cells; scale bar: 10 µm.
To confirm that functional HCs were present in the plasma membrane of live HaCaT-Cx26-GCaMP6s cells, we performed a standard dye uptake assay (Figure 3) using 4′,6-diamidino-2-phenylindole (DAPI, 5 µM) that is non fluorescent in the medium but becomes fluorescent upon penetrating into the cell and binding to nucleic acids [50].
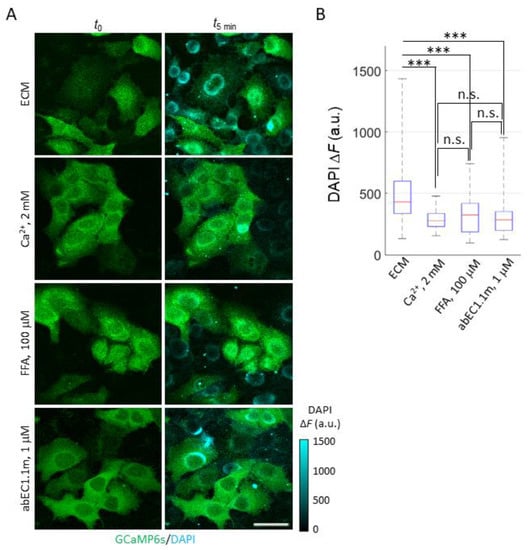
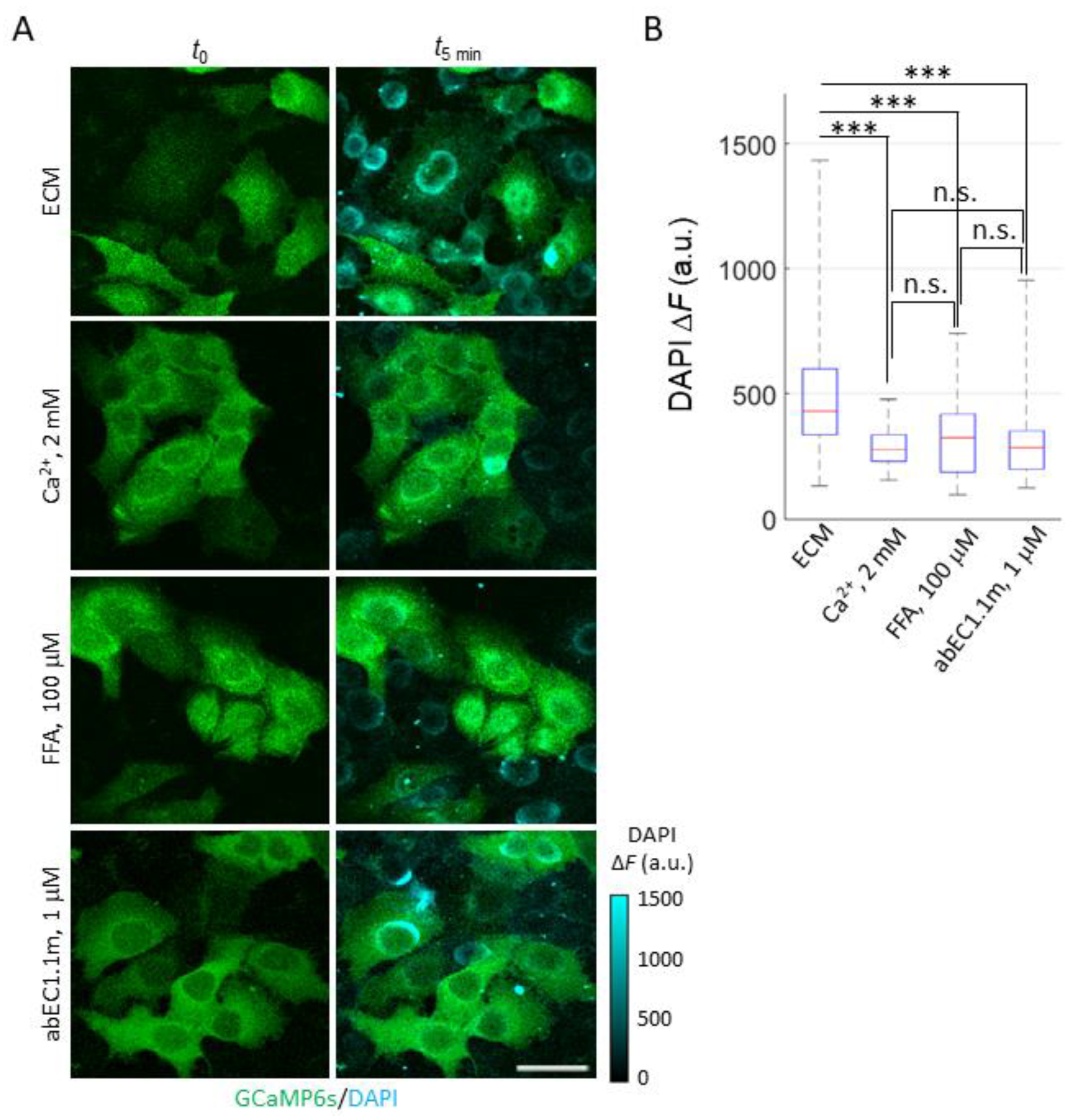
Figure 3.
DAPI uptake in HaCaT-Cx26-GCaMP6s cells. (A) Fluorescence images were acquired using the two-photon microscope described in [79] before () and after 5 min of incubation ( ) with DAPI (5 µM) in one of the following conditions (from top to bottom): ECM, ECM plus 2 mM Ca2+, ECM plus 100 µM FFA (denoted as FFA) or ECM plus 1 µM abEC1.1m (denoted as abEC1.1m). GCaMP6s signal (green) is shown as absolute fluorescence intensity (F). DAPI signal (cyan) is shown as Δ = . Scale bar: 40 µm. (B) Box plots of single cell Δ distributions for DAPI uptake experiments described in (A); for each condition, DAPI uptake was measured in 45 ≤ n ≤ 80 cells. Red horizontal bars represent the median. Statistical significance was calculated using the Dunn–Sidak post hoc test for pairwise comparisons after the Kruskal–Wallis’ test; ***, P < 0.001; and n.s., not significant.
Cells pre-exposed to dox for 24 h showed a significant increase of DAPI fluorescence emission when bathed in an extracellular medium (ECM) containing a low [Ca2+]ex (60 µM). HaCaT cells and isolated primary keratinocytes can be cultured in this and even lower [Ca2+]ex values [63,64,65,80,81,82] (which increase the open probability of HCs [11,12,13,14,15,16,17,18,19,20]). Indeed, the cell line “was designated HaCaT to denote its origin from human adult skin keratinocytes propagated under low Ca2+ conditions and elevated temperature” [63]. Supplementing ECM with 2 mM Ca2+, which reduces the open probability of HCs [11,12,13,14,15,16,17,18,19,20], inhibited DAPI uptake (Figure 3A). In contrast, in prior work with parental HaCaT cells that are known to express low levels of Cxs [59] and mainly Cx43 [83,84,85], DAPI uptake lacked the sensitivity required to discriminate between low and high [Ca2+]ex conditions [59].
In the experiments of Figure 3, DAPI uptake was also inhibited by flufenamic acid (FFA, 100 µM), a well-known and widely used aspecific blocker of HCs [77] (Figure 3B). To corroborate these findings, we used abEC1.1m, a previously characterized antagonist mAb, formatted as a single chain fragment variable (scFv)-fragment constant (Fc) diabody [59] with human scFv and mouse Fc domain, which specifically binds to the extracellular domain of Cx26, Cx30 and Cx32 HCs, however, is not effective on Cx43 HCs [60]. The effect of abEC1.1m (1 μM) was like that of FFA (Figure 3B). The simplest explanation for these results is that DAPI uptake occurred primarily through Cx26 HCs, whose contribution prevailed over that of HCs formed by other residual Cx isoforms expressed in dox-induced HaCaT-Cx26-GCaMP6s cells.
2.2. Development and Validation of the Ca2+ Uptake Assay
Based on the outcome of the experiments described above, we deemed it worthwhile to conduct Ca2+ uptake experiments in confluent cultures of HaCaT-Cx26-GCaMP6s cells using a custom-built spinning disk confocal fluoresce microscope (described in [86]) equipped with a 20× water-immersion objective. The wide field of view (FOV) achieved with this objective (630 µm ∅) allowed us to collect and average images of GCaMP6s fluorescence emission from more than 100 cells per culture. Experiments were carried out in accordance with the following procedure (Figure S1).
First, we pre-incubated the cells under the microscope in ECM for about 15 min to allow [Ca2+]cyt to equilibrate with the low [Ca2+]ex (60 µM) of the medium. Next, we excited GCaMP6s using a 488 nm laser and started to collect fluorescence images at 1 frame/s for 20 s of baseline (pre-stimulus) recording. Then, we added a bolus of CaCl2 (2 µL) to the medium in the recording chamber (1 mL volume) at different concentrations in different experiments, aiming to achieve final estimated [Ca2+]ex of 100 µM, 260 µM, 560 µM, 1 mM and 2 mM and continued to acquire images at 1 frame/s for a total of 10 min. By the end of each recording interval, we applied a bolus of the Ca2+ ionophore ionomycin (200 µM, 2 µL and 0.4 µM estimated final concentration) to confirm that GCaMP6s signals were not saturated (Figure S1). Thereafter, the experiment was terminated, and the culture was discarded.
For each experiment, GCaMP6s fluorescence emission was quantified pixel-by-pixel and frame-by-frame as ΔF = F − F0, where F represents emission at time t, and F0 represents emission averaged over all baseline (pre-stimulus) frames [87]. We then spatially averaged ΔF pixel signals over all cells in the FOV and produced a single ΔF trace for each experiment. In the first 20 s after CaCl2 addition, HaCaT-Cx26-GCaMP6s pre-exposed to dox for 24 h responded with a transient peak of [Ca2+]cyt followed by a delayed and broader secondary maximum the height of which depended on CaCl2 bolus concentration (see Discussion) (Figure 4A). Ionomycin generated a third response peak that largely exceeded the two previous ones (Figure S1).
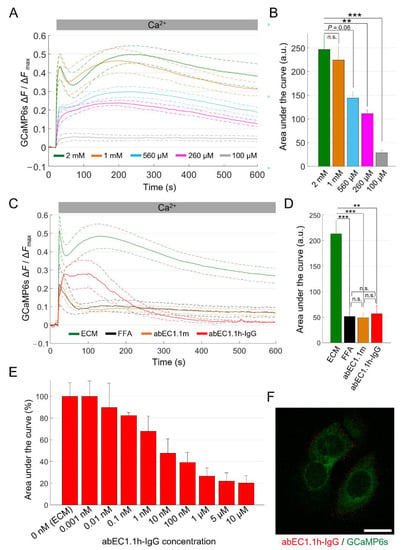
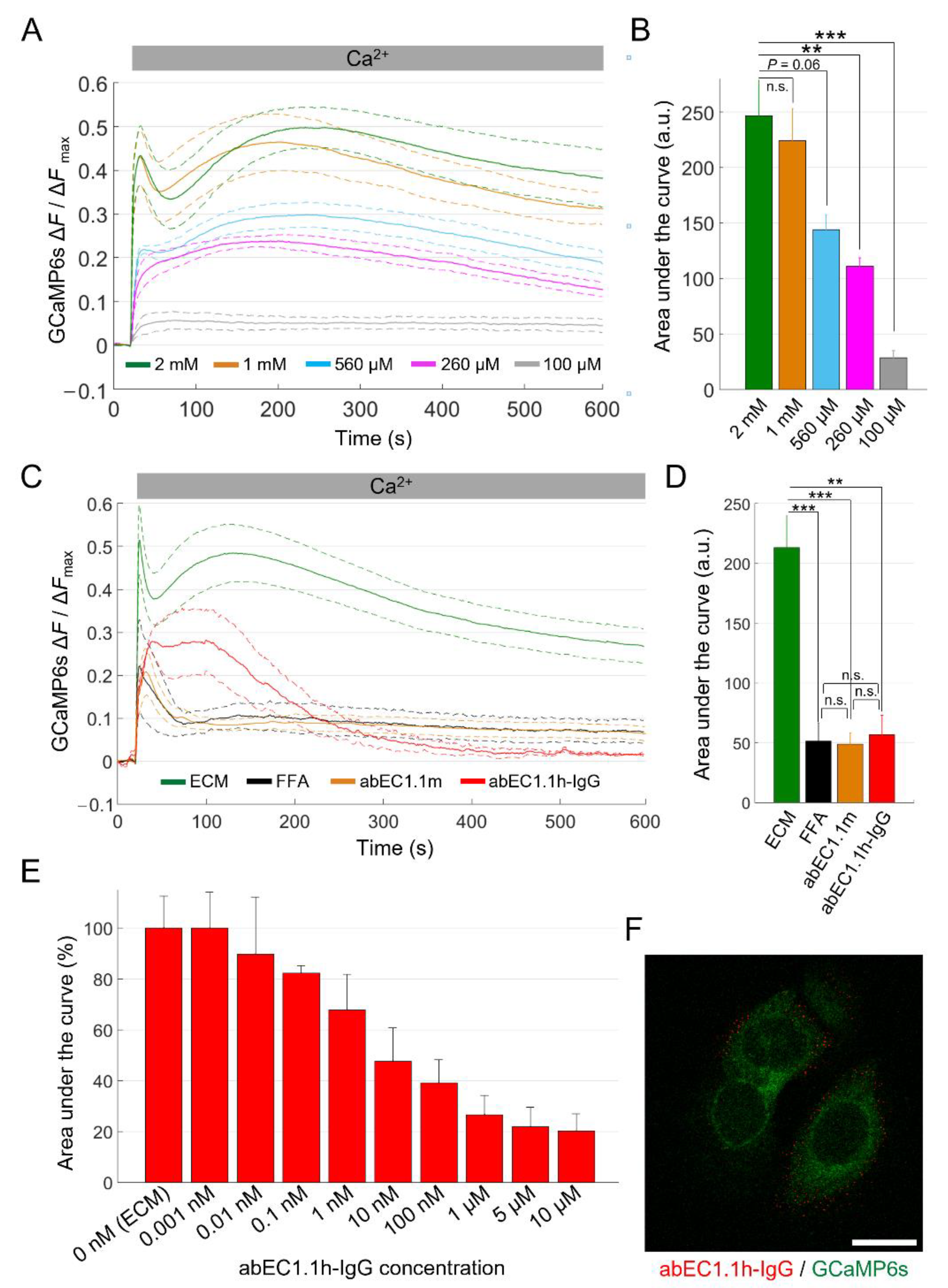
Figure 4.
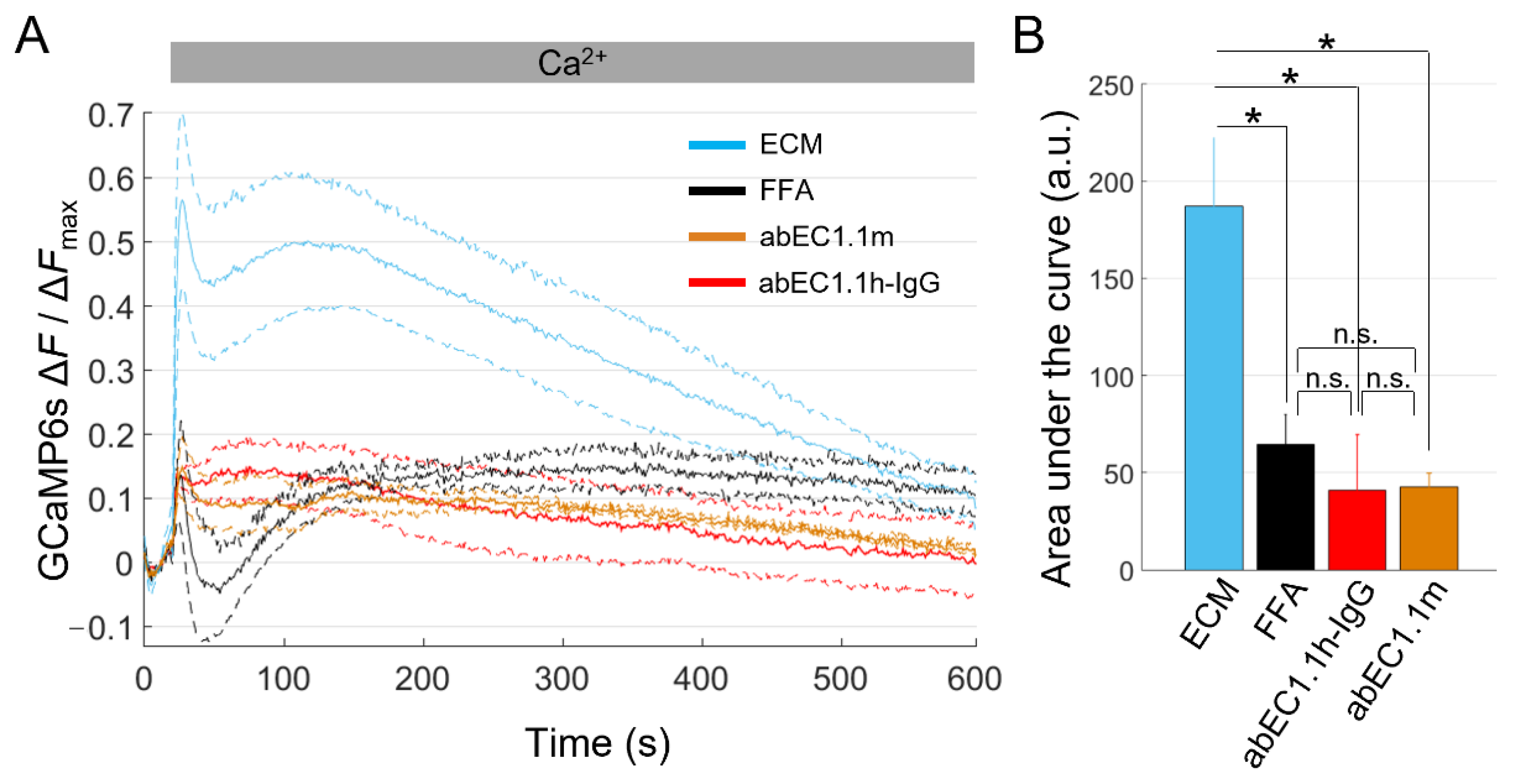
Ca2+ uptake in HaCaT-Cx26-GCaMP6s cells. (A) The mean Ca2+ uptake traces (solid lines) ± s.e.m. (dashed lines) obtained in HaCaT-Cx26-GCaMP6s cell cultures bathed in ECM and exposed to different CaCl2 bolus concentrations (BCs) to reach the following final [Ca2+]ex: 2 mM (green, n = 5, BC = 1 M), 1 mM (orange, n = 5, BC = 500 mM), 560 µM (light blue, n = 4, BC = 250 mM), 260 µM (magenta, n = 4, BC = 100 mM) and 100 µM (grey, n = 4, BC = 20 mM). Time t = 0 marks the onset of image acquisition. (B) Quantification of cytosolic Ca2+ load for the results in (A) computed as the areas subtended by the normalized Ca2+ uptake traces in the time interval between t = 20 s and t = 590 s (data are quoted as the mean ± s.e.m). Statistical significance was calculated using the Bonferroni post hoc test for pairwise comparisons after the ANOVA test; n.s., not significant; P, p-value; **, P < 0.01; ***, P < 0.001. (C) The mean Ca2+ uptake responses (solid lines) ± s.e.m. (dashed lines) to a 1 M CaCl2 bolus obtained from HaCaT-Cx26-GCaMP6s cell cultures bathed in the following extracellular media: ECM (green, n = 8), ECM plus FFA (100 µM, black, n = 4), ECM plus abEC1.1m (1 µM, orange, n = 7) and ECM plus abEC1.1h-IgG (1 µM, red, n = 3). (D) Quantification of cytosolic Ca2+ load for the results in (C) computed as described in (B). (E) Dose-dependent effect of the abEC1.1h-IgG for concentrations in the range 0.001 nM–10 µM (data are quoted as the mean ± s.e.m. for each condition). (F) Immunofluorescence labeling of HaCaT-Cx26-GCaMP6s cells with abEC1.1h-IgG; the antibody (15 nM) was applied to live cells that were then fixed but not permeabilized and counterstained with a red-fluorescent secondary antibody selective for the human Fc domain of the IgG. The green signal is due to GCaMP6s fluorescence. Scale bar: 20 μm.
Cells not exposed to dox showed minimal responses due to the basal leakiness of the TRE promoter (Figure S1). For statistical analyses, we normalized each ΔF trace for the peak response to ionomycin, ΔFmax (Figure S1). We computed average ΔF/ΔFmax traces from n independent experiments (see figure legends) in each condition, using a different culture for each experiment and displayed them as the mean (solid line) ± standard error of the mean (s.e.m., dashed lines, Figure 4A).
Finally, we quantified the [Ca2+]cyt load in terms of the area subtended by the ΔF/ΔFmax traces. The data analysis showed that the sensitivity of the assay plateaued between 1 mM and 2 mM final [Ca2+]ex (Figure 4B), therefore, we adopted 2 mM as the standard final [Ca2+]ex for all the other experiments. Reducing the initial [Ca2+]ex from 60 µM to 20 µM abolished the delayed and broader secondary maximum, resulting in a dramatically reduced assay sensitivity (Figure S2).
We next tested the effects of FFA and abEC1.1m on Ca2+ uptake quantified as explained above. Based on these analyses, abEC1.1m (1 µM, in ECM) exerted an inhibitory effect comparable to that of FFA (100 µM, in ECM; P = 1, ANOVA; Figure 4C and Figure S3), suggesting that Cx26 HCs provided the dominant uptake route for Ca2+. In this set of experiments, we tested also a new fully human immunoglobulin G (IgG) format of this mAb, henceforth referred as abEC1.1h-IgG (1 µM, in ECM). Although abEC1.1h-IgG modified the kinetics of Ca2+ uptake differently from abEC1.1m (Figure 4C), the overall blockade effect exerted by the two mAbs was undistinguishable (P = 1, ANOVA; Figure 4D).
To better characterize the new IgG, we constructed a dose–inhibition curve (Figure 4E and Figure S4) and determined that the half-maximal effective concentration (EC50) was comprised between 1 and 10 nM. At saturating concentrations (>1 µM), the inhibitory effect of abEC1.1h-IgG was about 80%, comparable to that the ScFv-Fc format previously estimated by the patch-clamp technique [59,61]. Immunolabeling with abEC1.1h-IgG produced fluorescent puncta at cell boundaries but not at points of contact between adjacent cells (Figure 4F; compare to Figure 2C) consistent with binding of abEC1.1h-IgG to unpaired HCs in the cell plasma membrane.
2.3. mAbs Targeting the Extracellular Domain of Cx26 Inhibit Ca2+ Uptake through Leaky HCs Implicated in KID Syndrome
Having successfully established working conditions to conduct Ca2+ uptake assays, we turned to cells overexpressing Cx26G45E. The percentage of pyknotic nuclei in HaCaT-Cx26G45E-GCaMP6s cells exposed to dox for 24 h (Table 2) was not significantly different from that of cultures not exposed to dox (Table 3). Therefore, we used cells pre-exposed to dox for 24 h to compare the effect of ECM alone or ECM supplemented with FFA (100 µM), abEC1.1m (1 µM) or abEC1.1h-IgG (1 µM). All three inhibitors significantly reduced Ca2+ uptake (Figure 5), suggesting that Ca2+ uptake was mediated by Cx26G45E HCs rather than by passive leakage through a compromised plasma membrane. Based on patch-clamp experiments conducted on Cx26G45E mutant HCs [59] and in vivo experiments on a mouse model of a related genodermatosis [61], these results suggest that the tested mAbs are candidate therapeutics for KID syndrome.

Table 2.
Percentage of pyknotic nuclei in HaCaT cell pools overexpressing Cx26 or Cx26G45E, exposed (+dox) or not exposed to dox (−dox) for 24 h.

Table 3.
Statistical comparisons for results presented in Table 2 calculated with the Bonferroni post hoc test for pairwise comparisons after the ANOVA test.
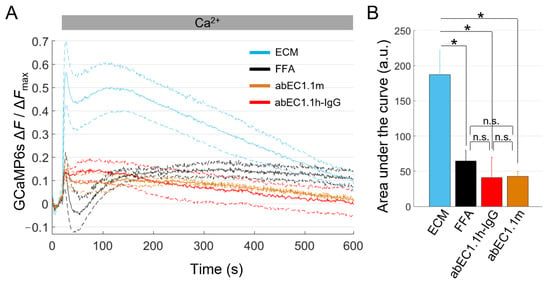
Figure 5.
Ca2+ uptake through leaky Cx26G45E HCs is blocked by antibodies targeting the extracellular domain of Cx26. (A) The mean normalized Ca2+ uptake traces (solid lines) ± s.e.m. (dashed lines) obtained from dox-induced HaCaT-Cx26G45E-GCaMP6s cell cultures maintained in the following extracellular media: ECM (light blue, n = 7), ECM plus FFA (100 µM, black, n = 5), ECM plus abEC1.1h-IgG (1 µM, red, n = 4) or ECM plus abEC1.1m (1 µM, orange, n = 4). (B) The mean ± s.e.m areas subtended by normalized Ca2+ uptake traces in the time interval between t = 20 s and t = 590 s. Statistical significance was calculated using the Bonferroni post hoc test for pairwise comparisons after the ANOVA test; *, P < 0.05; and n.s., not significant.
2.4. Effects of Uncharacterized mAbs, Targeting the Extracellular Domain of Cx46, on Ca2+ and DAPI Uptake
For the final set of experiments, we generated a stable pool of HaCaT-Cx46-GCaMP6s cells and used them to test new uncharacterized mAbs targeting the extracellular domain of Cx46 (Figure 6).
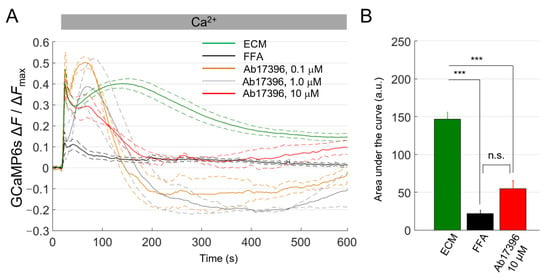
Figure 6.
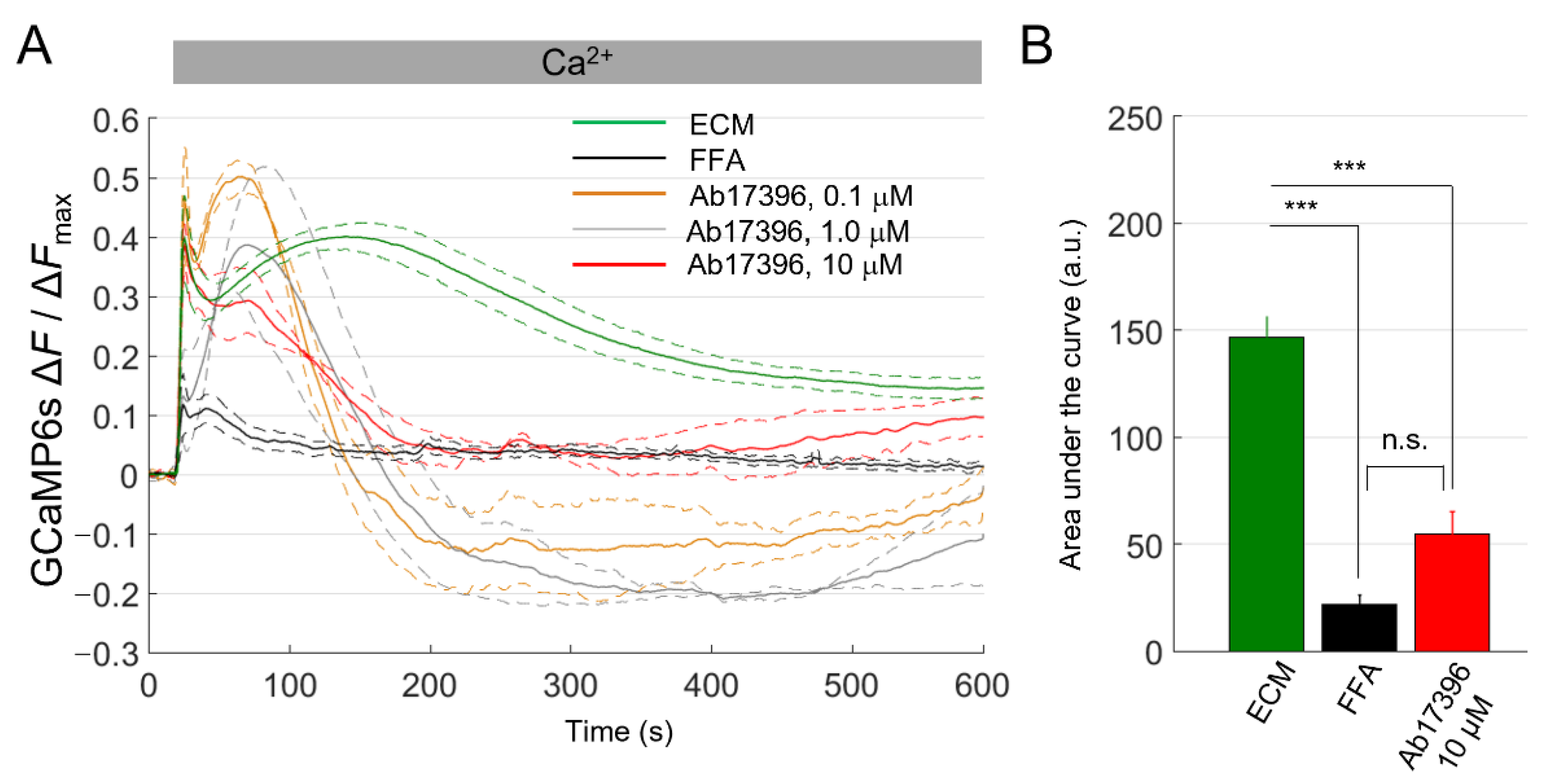
Ca2+ uptake assay in HaCaT-Cx46-GCaMP6s cells. (A) The mean normalized Ca2+ uptake traces (solid lines) ± s.e.m. (dashed lines) obtained from HaCaT-Cx46-GCaMP6s in ECM (green, n = 5), ECM plus FFA (100 µM, black, n = 6), ECM plus 0.1 µM anti-Cx46 mAb Ab17396 (orange, n = 3), ECM plus 1 µM Ab17396 (gray, n = 3) or ECM plus 10 µM Ab17396 (red, n = 3). (B) Areas subtended by normalized Ca2+ uptake traces (mean ± s.e.m.) in the time interval between t = 20 s and t = 590 s for some of the data shown on the left; 0.1 µM Ab17396 and 1 µM Ab17396 conditions were omitted due to the negative swing; this was never observed with other inhibitors or in control conditions. Statistical significance was calculated using the Bonferroni post hoc test for pairwise comparisons after the ANOVA test; ***, P < 0.001; n.s. not significant.
FFA (100 µM) strongly reduced Ca2+ uptake in HaCaT-Cx46-GCaMP6s cells pre-exposed to dox for 24 h. In contrast, we observed a significant, clear-cut inhibitory effect of about 63% only with Ab17396 at 10 µM concentration. At two lower concentrations (0.1 and 1.0 µM), we could not estimate the [Ca2+]cyt load from the area subtended by the ΔF/ΔFmax traces because, surprisingly, the traces dropped below the pre-stimulus levels after CaCl2 bolus delivery (see Discussion).
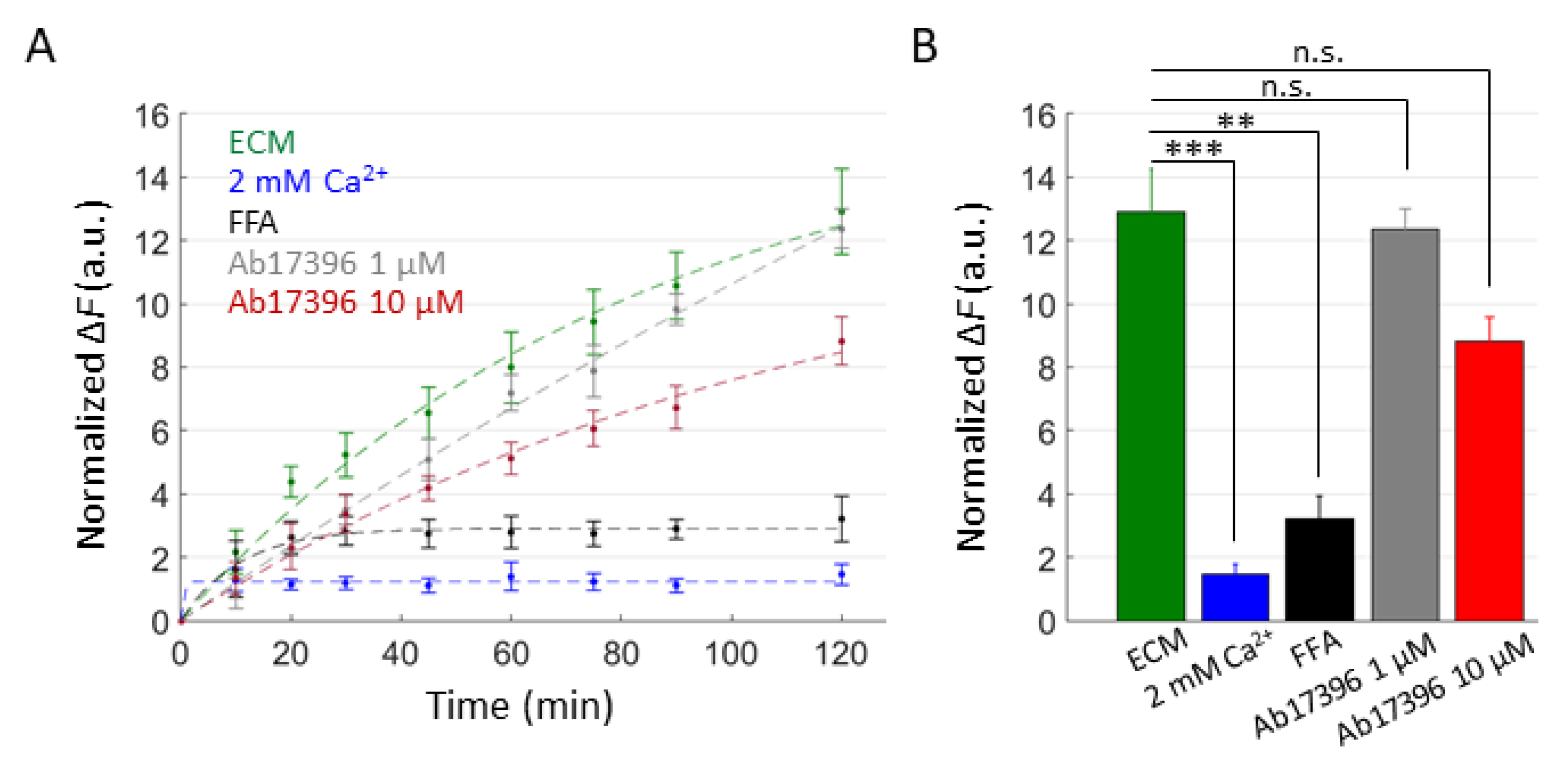
For a complementary estimate of Ab17396 activity, we performed DAPI uptake experiments in ECM alone or supplemented with Ca2+ (2 mM), FFA (100 µM) and Ab17396 (1.0 or 10 µM) (Figure 7). DAPI uptake was significantly inhibited by both Ca2+ and FFA, whereas the reductions provided by Ab17396 were not statistically significant (P = 0.085 between ECM and 10 µM Ab17396 conditions). However, these results were negatively affected by the limited number of experimental replicates that we were able to collect due to the shortage of Ab17396.
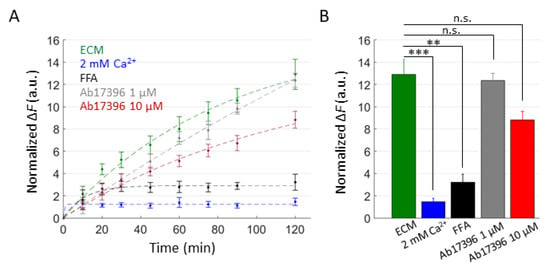
Figure 7.
DAPI uptake assay in HaCaT-Cx46-GCaMP6s cells. (A) Time course of DAPI uptake assay performed with a plate reader in dox-induced HaCaT-Cx46-GCaMP6s stable pools. Cells were exposed to DAPI (5 µM) dissolved in: ECM (green), ECM supplemented with 2 mM Ca2+ (blue), ECM plus FFA (100 µM, black), ECM plus Ab17396 1 µM (gray) and ECM plus Ab17396 10 µM (red). In each condition, DAPI uptake was measured as the average fluorescence signal variation Δ = of 3 ≤ n ≤ 6 cultures at different time points, from 5 min after the beginning of dye incubation (t = 0) up to 2 h later. The data are quoted as the mean values ± s.e.m. (B) The mean Δ values obtained for each condition shown in (A) at the end of the observation time window (2 h); statistical significance was calculated using the two-tailed t-test; **, P < 0.01; ***, P < 0.001; and n.s., not significant.
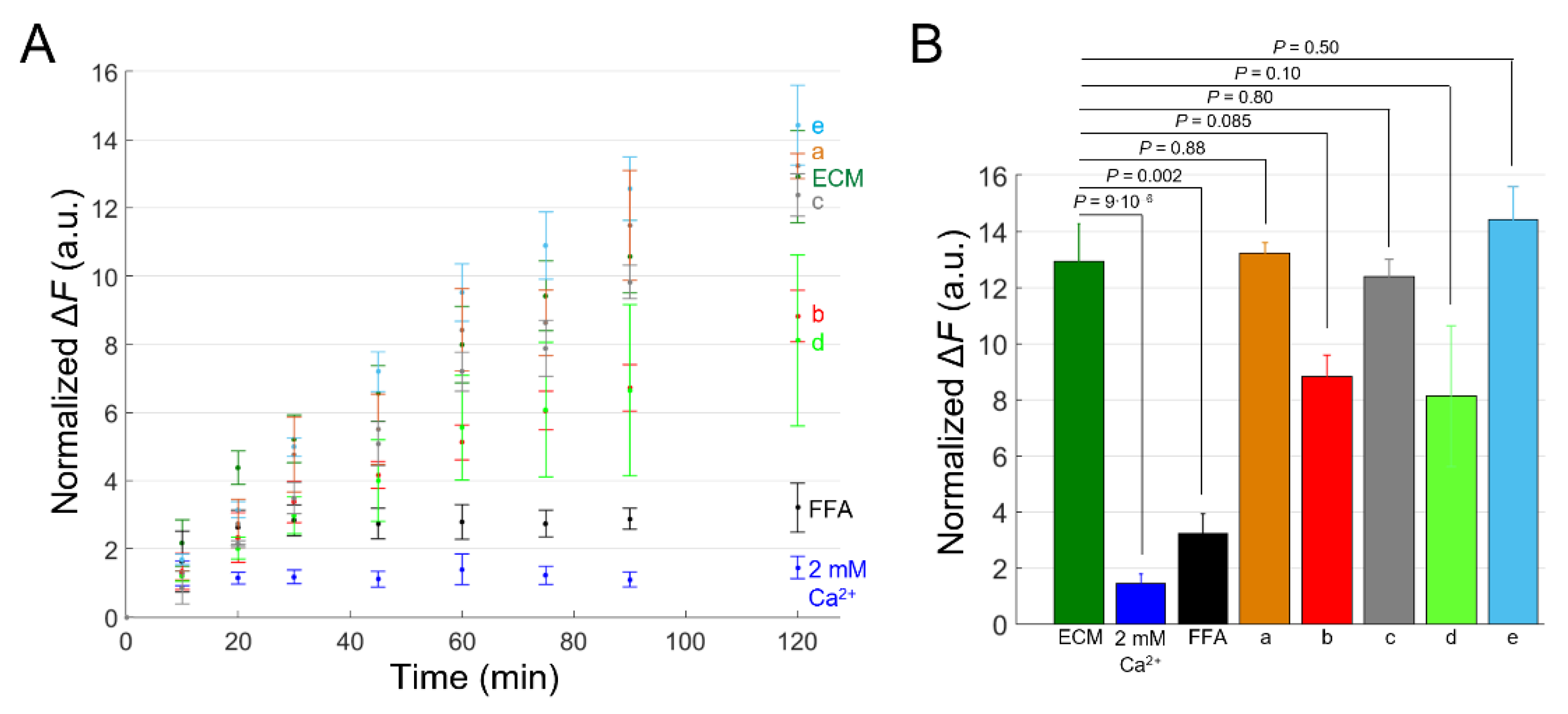
Finally, we also performed DAPI uptake experiments with other anti-Cx46 mAbs from the pool that led to the selection of Ab17396 (Figure 8). Together, based on the results of this preliminary screening, two of them, Ab17396 and Ab17398, appear worthy of further analyses to determine their potency, selectivity and usefulness as Cx46 HC inhibitors.
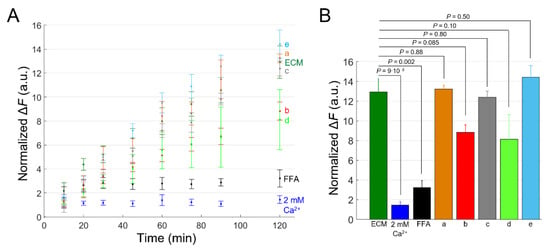
Figure 8.
Tests of different anti-Cx46 candidate mAbs by DAPI uptake assay in HaCaT-Cx46-GCaMP6s cells. (A) Time course of DAPI uptake assay performed with a plate reader in dox-induced HaCaT-Cx46-GCaMP6s stable pools. Cells were exposed to DAPI (5 µM) dissolved in: ECM (green), ECM supplemented with 2 mM Ca2+ (blue), ECM plus FFA (100 µM, black), ECM plus Ab17340 (a, 1 µM, orange), ECM plus Ab17396 (b, 10 µM, red), ECM plus Ab17397 (c, 1 µM, gray), ECM plus Ab17398 (d, 0.4 µM, light green) and ECM plus Ab17399 (e, 1 µM, light blue); for each condition, DAPI uptake was measured as average fluorescence signal variation Δ = of 3 ≤ n ≤ 6 cultures at different time points, from 5 min after the beginning of dye incubation (t = 0) up to 2 h later. The data are quoted as the mean values ± s.e.m. (B) The mean Δ values obtained for each condition shown in (A) at the end of the observation time window (2 h); p values (P) were calculated using the two-tailed t-test.
3. Discussion
In this work, we developed a novel all-optical assay to probe Ca2+ uptake through normal or pathological HCs, which are emerging drug targets for a variety of pathological conditions [41,42,43,44,45,46,47,48,49,61,62], using cytosolic GCaMP6s fluorescence intensity variations as a readout. This choice was dictated by different considerations. On the one hand, the study of Ca2+ dynamics in cells expressing pathological HCs is of utmost importance, given that deregulated Ca2+ influx through HCs has been implicated in different disorders, including but not limited to those affecting the skin, such as KID syndrome [40] and Clouston syndrome [61].
On the other hand, fluorometric assays based on intracellular Ca2+ variations are industry standards widely used in drug screening for membrane channels and G-protein coupled receptors (GPCRs) [88,89]. As shown here, the assay provided a sensitive indication for the effect of HC inhibitors, considering that it allowed us to detect changes in the uptake of Ca2+ with as little as 0.1 nM of the abEC1.1h-IgG mAb in HaCaT-Cx26-GCaMP6s cells.
In these experiments, GCaMP6s signals exhibited complex kinetics with an early peak in close temporal proximity to the CaCl2 bolus application, followed by a delayed response that generated a second relative maximum with a delay of 2 to 3 min. The reason for this biphasic response is unclear. We speculate that the sudden increase of the [Ca2+]ex had the contrasting effect of initiating Ca2+ uptake, due to the increased driving force for the cation, while also promoting HC closure. This would account for the short-duration first peak.
As for the delayed response producing the second peak (which was detectable at the final [Ca2+]ex > 500 µM, Figure 4A), it is worth considering that keratinocytes (and several other cell types) sense changes in the [Ca2+]ex via the G-protein coupled calcium-sensing receptor (CaSR) [90]. Activation of phospholipase C β isoforms downstream of CaSR-dependent G-protein stimulation results in the breakdown of phosphatidyl inositol-4,5-bisphosphate (PIP2), leading to the formation of inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). DAG remains associated with the plasma membrane and activates protein kinase C, whereas IP3 mediates Ca2+ release from the endoplasmic reticulum (ER), by activating IP3 receptors in a ligand-dependent manner [91]. Note that IP3 receptors are co-regulated by Ca2+ that promotes their opening at low [Ca2+]cyt and inhibits them at higher [Ca2+]cyt, depending on the IP3 level [92,93]. Therefore, the amplitude of this delayed, IP3-mediated response would depend strongly also on Ca2+ influx through HCs. In other words, the second peak that develops after CaCl2 bolus addition may be seen as and IP3-dependent (nonlinear) amplification mechanism for the signal generated by Ca2+ influx through HCs. This would explain its sensitivity to HC inhibitors in HaCaT-Cx26-GCaMP6s and HaCaT-Cx26G45E-GCaMP6s cells. The data in Figure 4A and Figure S2 support this conclusion. However, dedicated experiments, beyond the scope of this article, are required to validate this hypothesis (e.g., siRNA-mediated knock down of the CaSR and/or pre-empting of the intracellular Ca2+ stores with the SERCA pump inhibitor thapsigargin).
In the experiments with HaCaT-Cx46-GCaMP6s cells, Ca2+ uptake traces dropped below pre-stimulus levels in the recovery phase in the presence of Ab17396 at 0.1 µM and 1.0 µM but not with Ab17396 at 10 µM or with FFA at 100 µM. In contrast, in HaCaT-Cx26-GCaMP6s and HaCaT-Cx26G45E-GCaMP6s cells, the traces never dropped below baseline. Thus, we suspect that Ab17396 may affect, in a concentration-dependent manner, also other molecular players involved in the homeostasis of cytosolic Ca2+. In particular, low concentrations Ab17396 might facilitate the export of Ca2+ from the cytosol to the extracellular milieu, e.g., by potentiating the activity of plasma membrane Ca2+ ATP-ases or sodium–calcium exchangers. At higher concentrations, the prevailing effect of Ab17396 appears to be inhibition of Cx46 HCs. Further experiments are required to address this issue.
A stronghold of the Ca2+ uptake assay is the all-in-one Tet-on bicistronic system for the simultaneous, controlled expression of the Cx of interest and GCaMP6s in model cells. GCaMP6s provided not only an indirect fluorescent reporter for the successful expression of the Cx in HaCaT cells, but also a key element for the sensitive detection of [Ca2+]cyt variations at the core of the method. Although we used LVs, other systems can be contemplated, including the piggyBac transposon Tet-on vector system [94].
Furthermore, the methodology can be readily extended to different Cx isoforms or pathological variants in other popular cell lines that express low levels of endogenous Cxs, e.g., HeLa cells [95]. At the opposite extreme, the assay could be used to probe native HCs in cells, organoids, native tissues (e.g., slices), or even in vivo. In all these cases, the requirements are (i) achieving adequate levels of the Ca2+ indicator (e.g., by in vivo viral transduction or electroporation) and (ii) being able to rapidly modify the [Ca2+]ex. Indeed, biasing (overexpressed or native) HCs towards the open state in a low [Ca2+]ex medium prior to switching to a high [Ca2+]ex medium to promote Ca2+ uptake is key for these experiments.
The strengths of this approach for the study of HCs can be summarized as follows. The all-in-one Tet-on system allows for inducible bicistronic expression of both the Cx isoform/mutant of interest and the biosensor required to test HC functionality (GCaMP6s). After Cx-expressing stable cell pools are established, no further manipulations are required to run the assay, ensuring homogeneity across samples. The assay can be run on large cell populations to rapidly obtain statistically robust results. It is sensitive enough to construct dose–inhibition curves for drug candidates, including mAbs, without the need for electrophysiological recordings of HC-mediated currents, which may be challenging in the case of leaky HCs. Last but not least, the assay can be extended to high-throughput high-content platforms and/or primary cells and microtissues.
Whereas dye uptake assays provide a useful complement, as also demonstrated here, they may lack the required sensitivity to discriminate among different experimental conditions (e.g., concentrations and/or inhibitors), chiefly due to charge/size of permeating fluorescent molecule in relation to HC pore size and electrostatic profile [96,97]. On the other hand, the time course of the fluorescence changes caused by dye accumulation is typically monotonic and thus comparatively easier to analyze and interpret.
4. Materials and Methods
4.1. Viruses
For controlled expression of GCaMP6s and the Cx of interest, we modified the pCW57.1 vector, a mammalian expression, lentiviral, Tet-on, destination vector with a puromycin selection marker (a gift from David Root; Addgene plasmid #41393; http://n2t.net/addgene:41393, accessed on 26 June 2022; RRID: Addgene_41393). A multiple cloning site (MCS) linked to an IRES and GCaMP6s sequences (MCS-IRES-GCaMP6s) was gene-synthesized and inserted between the NheI (GCTAGC) and BamHI (GGATCC) restriction enzyme sites in the pCW57.1 vector to form a new lentiviral transfer vector, which we named pCW57-MSC-IRES-GCaMP6s.
Next, the MCS in this vector was used to subclone the CDS of human Cx26 (GJB2, NCBI CCDS ID: CCDS9290.1), its mutant Cx26G45E, or human Cx46 (GJA3, NCBI CCDS ID: CCDS9289.1). We thus obtained three corresponding inducible bicistronic lentiviral plasmids, named pCW57-Cx26-IRES-GCaMP6s, pCW57-Cx26G45E-IRES-GCaMP6s and pCW57-Cx46-IRES-GCaMP6s, respectively, for the simultaneous expression of the Cx of interest and cytosolic GCaMP6s. We deposited these plasmids in Addgene, where they have been respectively assigned the following IDs: 188236, 188237, 188238; https://www.addgene.org/Fabio_Mammano/, accessed on 26 June 2022.
4.2. Establishment of Stable Cell Pools
LVs were produced by transfection of HEK-293T cells (Cat. No. CRL-3216, American Type Culture Collection, ATCC, Manassas, VA, USA) using Lipofectamine 2000 (Cat. No. 11668-027, Thermo Fisher Scientific, TFS, Waltham, MA, USA) following a standard protocol [98]. LV-containing supernatant was collected, cleared by filtration and immediately used to infect HaCaT cells. The parental HaCaT cell line was maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Cat. No. 11995065, TFS) supplemented with heat-inactivated fetal bovine serum (FBS, 10% v/v, Cat. No. 10270-106, TFS) and penicillin/streptomycin solution (100 units/mL penicillin, 100 μg/mL streptomycin, Cat. No. 15070-063, TFS).
The day before infection, 2.5 × 105 cells were plated in a Petri dish (35 mm ∅) and cultured for 24 h. Then, the culture medium was removed and replaced with 0.5 mL of freshly collected undiluted LV-containing supernatant, supplemented with 8 µg/mL of hexadimethrine bromide (Cat. No. H9268, Merck KGaA, Darmstadt, Germany). The plate was incubated at 37 °C for 5 h with periodic gentle shaking every 5–10 min for the first 30 min; thereafter, 1.5 mL of normal culture medium was added, and incubation continued overnight.
Twenty-four hours post infection, the LV-containing medium was replaced with fresh medium. Forty-eight hours post infection, 1 µg/mL of puromycin (Cat. No. P9620, Merck KGaA) was added to the medium and kept until a selected homogeneous cell pool was established (typically 3 days). The cell pools thus obtained were denoted as HaCaT-Cx(…)-GCaMP6s. For precise gene regulation of Tet-on pools, we replaced the ordinary FBS (see above) with Tet-approved FBS (Cat. No. A4736401, TFS). All cell pools were routinely tested for mycoplasma contamination by staining with DAPI (Cat. No. D1306, TFS) followed by visual inspection at the fluorescence microscope.
For imaging experiments or immunofluorescence analyses, 5 × 105 cells were plated onto round glass coverslips (12 mm ∅) in a 35 mm ∅ dish two days before the assay. The next day and at least 24 h before experiments, dox (2 µg/mL, Cat. No. D1822, Merck KGaA) was added to the culture medium to promote Cx and GCaMP6s expression. To evaluate time-dependent dox-induced GCaMP6s expression in HaCaT-Cx26-GCaMP6s cells (Figure 1C), samples were washed and imaged with a spinning disk fluorescence microscope adapted for live-cell imaging [86] at different time points after dox administration (3, 6, 10, 24, 30 and 48 h).
To estimate the percentage of pyknotic cells in HaCaT-Cx26-GCaMP6s and HaCaT-Cx26G45E-GCaMP6s pools (Table 2 and Table 3), cell-plated coverslips exposed to dox or vehicle for 24 h were fixed with 4% paraformaldehyde (PFA; Cat. No. 158127, Merck KGaA) for 15 min and permeabilized with 0.1% Triton X-100 (Cat. No. 1.08603, Merck KGaA) for 5 min. Next, samples were washed with PBS and nuclei were stained with DAPI (1 µM, 5 min). After visualization at the fluorescence microscope, cells that exhibited chromatin condensation and DNA fragmentation were counted in six different FOVs (n = 88–219 cells/FOV) for each condition, and the results were divided by the total number of cells in each FOV.
4.3. qPCR
The total RNA was extracted from HaCaT-Cx26-GCaMP6s and HaCaT-Cx26G45E-GCaMP6s cells exposed or not exposed to dox for 24 h using TRIzol Reagent (Cat. No. 15596018, TFS). 10 µg of the extracted total RNA was digested with DNase I (Cat. No. 18068015, TFS) and then purified using Rneasy mini Kit (Cat. No. 74104, Qiagen, Hilden, Germany) according to the manufacturer’s instructions. A NanoDrop spectrophotometer was used to quantify the RNA by measuring the absorbance and reverse transcription. (RT)-PCR was performed from 1μg of the purified RNA using high-capacity cDNA kit (Cat. No. 4368814, TFS), according to the manufacturer’s instructions but with a slight modification in which Oligo(dt) (Cat. No. 18418012, TFS) was combined with the cDNA kit random primers at a ratio of 1:5.
Quantitative PCR (qPCR) was performed on the cDNA with specific primers (see below). Samples were analyzed in triplicate and gene expression levels were estimated by the 2−∆∆CT method, using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal reference gene and Sybr green (SsoAdvanced Universal Sybr green Supermix, Cat. No. 1725274, BioRad, Hercules, CA, USA) on the ABI 7900HT sequence detection system equipped with the AB1 7900HT SDS software (Applied Biosystems, Waltham, MA, USA) applying the following amplification cycles:
50 °C: 2′,
95 °C: 10′
95 °C: 0.15′, 65 °C: 0.35′ (40 cycles).
- The primers used for these experiments are listed here.
- Cx26 (Cx26 wt and Cx26G45E) primers:
Forward 5′ATCGAAGGCTCCCTGTGGTG3′;
Reverse 5′ACAGTGTTGGGACAAGGCCA3′.
- EMCV-IRES primers:
Forward 5′AATGTGAGGGCCCGGAAACC3′;
Reverse 5′ACTCACAACGTGGCACTGGG3′.
- GCaMP6S primers:
Forward 5′ GGAGGACGGCAACATC 3′;
Reverse 5′GAAAGCCTCTTTAAATTCTGCG3′.
- GAPDH primers:
Forward 5′CACCATCTTCCAGGAGCGAG 3′;
Reverse 5′CCTTCTCCATGGTGGTGAAGAC3′.
4.4. Immunofluorescence
Cells from stable pools grown on glass coverslips and exposed to dox or vehicle for 24 h were fixed with 4% PFA for 15 min, permeabilized with 0.1% Triton X-100 for 5 min and incubated for 1 h in blocking buffer containing 0.5% bovine serum albumin (BSA; Cat. No. A4503, Merck KGaA) and 0.1% Triton. Samples were then incubated overnight at 4 °C with anti-Cx26 polyclonal antibody (1:200, Cat. No. 71-0500, TFS) diluted in blocking buffer.
The next day, samples were washed three times with PBS and incubated with cross-adsorbed Alexa Flour 555 secondary antibody (1:800, Cat. No. A-31572, TFS) for 3 h. For co-immunofluorescent staining, samples were then incubated overnight at 4 °C with anti-GFP mAb (1:50, Cat. No. A-11120, TFS) diluted in blocking buffer. The next day, samples were washed three times with PBS and incubated with cross-adsorbed Alexa Fluor 488 secondary antibody (1:800, Cat. No. A-21202, TFS) for 3 h.
For immunofluorescence labeling with abEC1.1h-IgG (Figure 4F), cells from stable pools, grown on glass coverslips and exposed to dox for 24 h, were incubated at 37 °C with ECM supplemented with 15 nM abEC1.1h-IgG for 1 h. Cells were then fixed with 4% PFA for 15 min and incubated for 1 h with a goat anti-human IgG Alexa Fluor 594 secondary antibody (1:800, Cat. No. 109-585-170, Jackson ImmunoReasearch, Ely, Cambridgeshire, UK). Samples were washed three times with PBS between all steps.
Fluorescence images (pixel size 96 nm) were acquired with a confocal microscope (TCS SP5 Leica, Wetzlar, Germany) equipped with a 63× oil immersion objective (HC PL Apo, UV optimized, NA 1.4, Leica). Alexa Fluor 488 and GCaMP6s fluorescence was excited by a 488-nm Argon laser and collected between 500 nm and 540 nm. Alexa Fluor 555 and Alexa Fluor 594 fluorescence was excited by a 543-nm HeNe laser and collected between 568 nm and 680 nm and between 600 and 670 nm, respectively. For visualization purposes, a σ = 0.8 Gaussian filter was applied on the green channel of fluorescence images.
4.5. Dye Uptake
DAPI (5 µM) was dissolved in ECM containing (in mM): 138 NaCl, 5 KCl, 0.06 CaCl2, 0.4 NaH2PO4, 6 D-Glucose, 10 HEPES (all from Merck KGaA), pH 7.3. For negative controls, ECM was supplemented with CaCl2 (2 mM) or flufenamic acid (FFA, 100 µM, Cat. No. F9005, Merck KGaA). Antibodies targeting the extracellular domain of HCs were dissolved in ECM at a concentration of 1 µM (abEC1.1m and abEC1.1h-IgG) or as stated in figure legends (anti-Cx46 antibodies).
For experiments with HaCaT-Cx26-GCaMP6s pools, coverslips with plated cells were washed with ECM and imaged using the two-photon microscope of Ref. [79] before and after 5 min incubation with one of the mentioned DAPI-containing solutions at room temperature. The microscope was equipped with a 25× water immersion multiphoton objective (XLPlanN25XWMP2, NA = 1.05, WD = 2 mm, Olympus Corporation, Tokyo, Japan). DAPI was excited at 780 nm and fluorescence emission signals were collected through a blue emission filter (460/50 nm, ET460/50m, Chroma Technology Corp, Bellow Falls, VT, USA). For each sample, we acquired multiple z-stacks (through-focus image sequences obtained with a 2 µm step size) of at least three different FOVs. FFA and abEC1.1m were pre-incubated at 37 °C for 30 min.
For experiments with HaCaT-Cx46-GCaMP6s pools, the day before the experiment cells were seeded into a black 96-well microplate for high content imaging (2 × 104 cells/well, #4580, Corning Inc., Corning, NY, USA) and dox (2 µg/mL) was added to the culture medium. The glass bottom of each well was pre-treated with laminin (2 µg/cm2 dissolved in water, 30 min at 37 °C; Cat. No. 11243217001, Merck KGaA).
On the day of the experiment, cells were washed in ECM supplemented by 2 mM CaCl2 to remove FBS and mean GCaMP6s fluorescence value was measured at the plate reader (Varioskan LUX, Cat. No. N16045, TFS) for future normalization of DAPI uptake results to the number of cells present in each well. Then, wells were pre-incubated with one of the mentioned media for 20 min at 37 °C without DAPI (n = 3–6 wells/condition) to allow the blockers to act on HCs. Next, DAPI-deprived media were replaced with analogous DAPI-containing solutions and the microplate was moved to the plate reader to evaluate DAPI uptake rate.
The mean DAPI fluorescence value for each well was measured every 10–15 min starting from 5 min after the onset of DAPI incubation (t = 0 in the graphs) and proceeded for 2 h. We ruled out any possible contribution of dead cells in the cultures (for which DAPI uptake is much faster than HC-dependent uptake in live cell) because for each well we treated the mean value of the first DAPI fluorescence reading (5 min after the onset of dye incubation) as a baseline and subtracted it from all the following measurements.
4.6. Ca2+ Uptake
On the day of the experiment, cell-plated coverslips with confluent cell population were transferred to the stage of the spinning disk microscope described in Ref. [86] and equipped with water immersion objective (20×, N.A. = 0.95, XLUMPlan Fl, Olympus Corporation). Cells were incubated at room temperature for 15 min in ECM, thereafter fluorescence images were acquired at 1 frame/s for 12 min. GCaMP6s fluorescence was excited by a 488 nm diode laser (Cat. No. COMPACT-150G-488-SM, World Star Tech, Markham, Ontario, Canada) and emission signals were collected though a 535/30 nm band-pass filter (Cat. No. ET535/30M, Chroma Technology Corp., Bellows Falls, VT, USA).
After 20 s of baseline recordings, a 2 µL bolus of 1 M CaCl2 solution was delivered through a manually operated pipette held at the edge of the FOV, and GCaMP6s fluorescence variations were monitored for 10 min. Finally, a 2 µL bolus of ionomycin solution (200 µM, Cat. No. I3909, Merck KGaA) was delivered to normalize the Ca2+ influx response to the maximum achievable fluorescence variation (i.e., to the level of biosensor expression throughout experiments or experimental sessions). We used each cell-plated coverslip only once. To assay the efficacy of HC inhibitors, coverslips were pre-incubated at 37 °C with ECM supplemented with one of the following HC compounds: 100 µM FFA (40 min), 1 µM abEC1.1m (30 min); abEC1.1h-IgG at a concentration varying from 0.001 nM to 10 µM for the construction of dose–inhibition curve (30 min).
4.7. Selection of Anti-Cx46 mAbs
A synthetic, phage-displayed antibody library [99] was selected for binding to Cx46 protein expressed on the surface of Virus-Like Particles (Cx46-VLPs). Briefly, Expi293 cells (Cat. No. A14527, TFS) were co-transfected with plasmids expressing either HA-tagged Cx46 together with HIV Gag (Cx46 VLPs) or HIV Gag alone (empty VLPs), and VLPs were isolated from supernatant 2 days post transfection. Cx46 VLPs (100 µL at 10 µg/mL) were immobilized on Maxisorp Immuno plates (Cat. No. 12-565-135, TFS) and used for positive binding selections with library phage pools that were first exposed to similarly immobilized empty VLPs to deplete non-specific binders. After four rounds of binding selections, clonal phage was prepared and evaluated by phage ELISA (using Cx46 VLP or empty VLP) and sequencing as previously described [99].
4.8. Statistics
Statistical analysis was performed with MATLAB (R2019a, The MathWorks, Inc. Natick, Massachusetts, USA). The normality of distributions was assessed using the Shapiro–Wilk test. For normal distributions, statistical comparisons of means were made by ANOVA and post hoc comparison was by the Bonferroni test (multiple groups) or by two-tailed t-test (two groups); for non-normal distributions, statistical comparisons of means were made using the Kruskal–Wallis and post hoc comparison by the Dunn–Sidak test. The mean values are quoted ± s.e.m. unless noted otherwise. Sample sizes (n) of experimental groups are indicated in figure legends. p = p-values < 0.05 indicate statistical significance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23137337/s1.
Author Contributions
Conceptualization, F.M.; methodology, C.N., A.T.-M., M.G., S.S. and L.C.; formal analysis, C.N.; investigation, C.N., A.T.-M., V.D., C.P., D.M. and C.D.P.; resources, F.M., M.R., G.Y., F.Z. and S.S.S.; data curation, C.N., A.T.-M., V.D. and C.P.; writing—original draft preparation, C.N.; writing—review and editing, F.M.; visualization, C.N., C.P. and A.T.-M.; supervision, F.M., S.S.S. and G.Y.; project administration, F.M.; funding acquisition, F.M., M.R. and S.S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Padova, grant Prot. BIRD187130 to F.M., Fondazione Telethon, grant GGP19148 to F.M., the Italian Ministry of Education, Universities and Research (MUR) grants PRIN 20175C22WM to F.M. and PRIN 2017FTJ5ZE to M.R.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data were archived in a repository handled by the University of Padova and can be accessed at: http://researchdata.cab.unipd.it/645/, accessed on 26 June 2022. Lentiviral plasmids developed and used in this study have been deposited and will be made publicly available via the Addgene repository with the following IDs: 188236, 188237, 188238; https://www.addgene.org/Fabio_Mammano/, accessed on 26 June 2022.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Maeda, S.; Nakagawa, S.; Suga, M.; Yamashita, E.; Oshima, A.; Fujiyoshi, Y.; Tsukihara, T. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 2009, 458, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.B.; Haddad, B.G.; O’Neill, S.E.; Chorev, D.S.; Yoshioka, C.C.; Robinson, C.V.; Zuckerman, D.M.; Reichow, S.L. Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Nature 2018, 564, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jeong, H.; Hyun, J.; Ryu, B.; Park, K.; Lim, H.H.; Yoo, J.; Woo, J.S. Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel. Sci. Adv. 2020, 6, eaba4996. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.K.; Jagielnicki, M.; Bennett, B.C.; Purdy, M.D.; Yeager, M. Cryo-EM structure of an open conformation of a gap junction hemichannel in lipid bilayer nanodiscs. Structure 2021, 29, 1040–1047. [Google Scholar] [CrossRef] [PubMed]
- Esseltine, J.L.; Laird, D.W. Next-Generation Connexin and Pannexin Cell Biology. Trends Cell Biol. 2016, 26, 944–955. [Google Scholar] [CrossRef]
- Schalper, K.A.; Palacios-Prado, N.; Retamal, M.A.; Shoji, K.F.; Martinez, A.D.; Saez, J.C. Connexin hemichannel composition determines the FGF-1-induced membrane permeability and free [Ca2+]i responses. Mol. Biol. Cell 2008, 19, 3501–3513. [Google Scholar] [CrossRef]
- Schalper, K.A.; Sanchez, H.A.; Lee, S.C.; Altenberg, G.A.; Nathanson, M.H.; Saez, J.C. Connexin 43 hemichannels mediate the Ca2+ influx induced by extracellular alkalinization. Am. J. Physiology. Cell Physiol. 2010, 299, C1504–C1515. [Google Scholar] [CrossRef]
- Fiori, M.C.; Figueroa, V.; Zoghbi, M.E.; Saez, J.C.; Reuss, L.; Altenberg, G.A. Permeation of calcium through purified connexin 26 hemichannels. J. Biol. Chem. 2012, 287, 40826–40834. [Google Scholar] [CrossRef]
- Sanchez, H.A.; Orellana, J.A.; Verselis, V.K.; Saez, J.C. Metabolic inhibition increases activity of connexin-32 hemichannels permeable to Ca2+ in transfected HeLa cells. Am. J. Physiol. Cell Physiol. 2009, 297, C665–C678. [Google Scholar] [CrossRef]
- Mandal, A.; Shahidullah, M.; Delamere, N.A. Calcium entry via connexin hemichannels in lens epithelium. Exp. Eye Res. 2015, 132, 52–58. [Google Scholar] [CrossRef][Green Version]
- Paul, D.L.; Ebihara, L.; Takemoto, L.J.; Swenson, K.I.; Goodenough, D.A. Connexin46, a novel lens gap junction protein, induces voltage-gated currents in nonjunctional plasma membrane of Xenopus oocytes. J. Cell Biol. 1991, 115, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, L.; Steiner, E. Properties of a nonjunctional current expressed from a rat connexin46 cDNA in Xenopus oocytes. J. Gen. Physiol. 1993, 102, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Contreras, J.E.; Saez, J.C.; Bukauskas, F.F.; Bennett, M.V. Gating and regulation of connexin 43 (Cx43) hemichannels. Proc. Natl. Acad. Sci. USA 2003, 100, 11388–11393. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.V.; Contreras, J.E.; Bukauskas, F.F.; Saez, J.C. New roles for astrocytes: Gap junction hemichannels have something to communicate. Trends Neurosci. 2003, 26, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, D.A.; Paul, D.L. Beyond the gap: Functions of unpaired connexon channels. Nat. Rev. Mol. Cell Biol. 2003, 4, 285–294. [Google Scholar] [CrossRef]
- Saez, J.C.; Retamal, M.A.; Basilio, D.; Bukauskas, F.F.; Bennett, M.V. Connexin-based gap junction hemichannels: Gating mechanisms. Biochim. Biophys. Acta 2005, 1711, 215–224. [Google Scholar] [CrossRef]
- Saez, J.C.; Schalper, K.A.; Retamal, M.A.; Orellana, J.A.; Shoji, K.F.; Bennett, M.V. Cell membrane permeabilization via connexin hemichannels in living and dying cells. Exp. Cell Res. 2010, 316, 2377–2389. [Google Scholar] [CrossRef]
- Fasciani, I.; Temperan, A.; Perez-Atencio, L.F.; Escudero, A.; Martinez-Montero, P.; Molano, J.; Gomez-Hernandez, J.M.; Paino, C.L.; Gonzalez-Nieto, D.; Barrio, L.C. Regulation of connexin hemichannel activity by membrane potential and the extracellular calcium in health and disease. Neuropharmacology 2013, 75, 479–490. [Google Scholar] [CrossRef]
- Saez, J.C.; Leybaert, L. Hunting for connexin hemichannels. FEBS Lett. 2014, 588, 1205–1211. [Google Scholar] [CrossRef]
- Leybaert, L.; Lampe, P.D.; Dhein, S.; Kwak, B.R.; Ferdinandy, P.; Beyer, E.C.; Laird, D.W.; Naus, C.C.; Green, C.R.; Schulz, R. Connexins in Cardiovascular and Neurovascular Health and Disease: Pharmacological Implications. Pharmacol. Rev. 2017, 69, 396–478. [Google Scholar] [CrossRef]
- Cotrina, M.L.; Lin, J.H.; Alves-Rodrigues, A.; Liu, S.; Li, J.; Azmi-Ghadimi, H.; Kang, J.; Naus, C.C.; Nedergaard, M. Connexins regulate calcium signaling by controlling ATP release. Proc. Natl. Acad. Sci. USA 1998, 95, 15735–15740. [Google Scholar] [CrossRef] [PubMed]
- Stout, C.E.; Costantin, J.L.; Naus, C.C.; Charles, A.C. Intercellular calcium signaling in astrocytes via ATP release through connexin hemichannels. J. Biol. Chem. 2002, 277, 10482–10488. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.A.; Dale, N.; Llaudet, E.; Mobbs, P. ATP released via gap junction hemichannels from the pigment epithelium regulates neural retinal progenitor proliferation. Neuron 2005, 46, 731–744. [Google Scholar] [CrossRef]
- Anselmi, F.; Hernandez, V.H.; Crispino, G.; Seydel, A.; Ortolano, S.; Roper, S.D.; Kessaris, N.; Richardson, W.; Rickheit, G.; Filippov, M.A.; et al. ATP release through connexin hemichannels and gap junction transfer of second messengers propagate Ca2+ signals across the inner ear. Proc. Natl. Acad. Sci. USA 2008, 105, 18770–18775. [Google Scholar] [CrossRef]
- Mazzarda, F.; D’Elia, A.; Massari, R.; De Ninno, A.; Bertani, F.R.; Businaro, L.; Ziraldo, G.; Zorzi, V.; Nardin, C.; Peres, C.; et al. Organ-on-chip model shows that ATP release through connexin hemichannels drives spontaneous Ca(2+) signaling in non-sensory cells of the greater epithelial ridge in the developing cochlea. Lab A Chip 2020, 20, 3011–3023. [Google Scholar] [CrossRef] [PubMed]
- Nardin, C.; Peres, C.; Putti, S.; Orsini, T.; Colussi, C.; Mazzarda, F.; Raspa, M.; Scavizzi, F.; Salvatore, A.M.; Chiani, F.; et al. Connexin Hemichannel Activation by S-Nitrosoglutathione Synergizes Strongly with Photodynamic Therapy Potentiating Anti-Tumor Bystander Killing. Cancers 2021, 13, 5062. [Google Scholar] [CrossRef]
- Orellana, J.A.; Froger, N.; Ezan, P.; Jiang, J.X.; Bennett, M.V.; Naus, C.C.; Giaume, C.; Saez, J.C. ATP and glutamate released via astroglial connexin 43 hemichannels mediate neuronal death through activation of pannexin 1 hemichannels. J. Neurochem. 2011, 118, 826–840. [Google Scholar] [CrossRef]
- Cherian, P.P.; Siller-Jackson, A.J.; Gu, S.; Wang, X.; Bonewald, L.F.; Sprague, E.; Jiang, J.X. Mechanical strain opens connexin 43 hemichannels in osteocytes: A novel mechanism for the release of prostaglandin. Mol. Biol. Cell 2005, 16, 3100–3106. [Google Scholar] [CrossRef]
- De Flora, A.; Zocchi, E.; Guida, L.; Franco, L.; Bruzzone, S. Autocrine and paracrine calcium signaling by the CD38/NAD+/cyclic ADP-ribose system. Ann. N. Y. Acad. Sci. 2004, 1028, 176–191. [Google Scholar] [CrossRef]
- Rana, S.; Dringen, R. Gap junction hemichannel-mediated release of glutathione from cultured rat astrocytes. Neurosci. Lett. 2007, 415, 45–48. [Google Scholar] [CrossRef]
- Tong, X.; Lopez, W.; Ramachandran, J.; Ayad, W.A.; Liu, Y.; Lopez-Rodriguez, A.; Harris, A.L.; Contreras, J.E. Glutathione release through connexin hemichannels: Implications for chemical modification of pores permeable to large molecules. J. Gen. Physiol. 2015, 146, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; De Bock, M.; Decrock, E.; Bol, M.; Gadicherla, A.; Vinken, M.; Rogiers, V.; Bukauskas, F.F.; Bultynck, G.; Leybaert, L. Paracrine signaling through plasma membrane hemichannels. Biochim. Biophys. Acta 2013, 1828, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Vénéreau, E.; Ceriotti, C.; Bianchi, M.E. DAMPs from Cell Death to New Life. Front. Immunol. 2015, 6, 422. [Google Scholar] [CrossRef] [PubMed]
- Decrock, E.; Vinken, M.; De Vuyst, E.; Krysko, D.V.; D’Herde, K.; Vanhaecke, T.; Vandenabeele, P.; Rogiers, V.; Leybaert, L. Connexin-related signaling in cell death: To live or let die? Cell Death Differ. 2009, 16, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, A.; Bera, A.K. Hemichannels: Permeants and their effect on development, physiology and death. Cell Biochem. Funct. 2012, 30, 89–100. [Google Scholar] [CrossRef]
- Vinken, M. Connexin hemichannels: Novel mediators of toxicity. Arch. Toxicol. 2015, 89, 143–145. [Google Scholar] [CrossRef]
- Dosch, M.; Gerber, J.; Jebbawi, F.; Beldi, G. Mechanisms of ATP Release by Inflammatory Cells. Int. J. Mol. Sci. 2018, 19, 1222. [Google Scholar] [CrossRef]
- Retamal, M.A.; Reyes, E.P.; Garcia, I.E.; Pinto, B.; Martinez, A.D.; Gonzalez, C. Diseases associated with leaky hemichannels. Front. Cell. Neurosci. 2015, 9, 267. [Google Scholar] [CrossRef]
- Laird, D.W.; Lampe, P.D. Cellular mechanisms of connexin-based inherited diseases. Trends Cell Biol. 2022, 32, 58–69. [Google Scholar] [CrossRef]
- Garcia, I.E.; Bosen, F.; Mujica, P.; Pupo, A.; Flores-Munoz, C.; Jara, O.; Gonzalez, C.; Willecke, K.; Martinez, A.D. From Hyperactive Connexin26 Hemichannels to Impairments in Epidermal Calcium Gradient and Permeability Barrier in the Keratitis-Ichthyosis-Deafness Syndrome. J. Investig. Dermatol. 2016, 136, 574–583. [Google Scholar] [CrossRef]
- Willebrords, J.; Maes, M.; Crespo Yanguas, S.; Vinken, M. Inhibitors of connexin and pannexin channels as potential therapeutics. Pharmacol. Ther. 2017, 180, 144–160. [Google Scholar] [CrossRef] [PubMed]
- Delvaeye, T.; Vandenabeele, P.; Bultynck, G.; Leybaert, L.; Krysko, D.V. Therapeutic Targeting of Connexin Channels: New Views and Challenges. Trends Mol. Med. 2018, 24, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.W.; Lampe, P.D. Therapeutic strategies targeting connexins. Nat. Rev. Drug Discov. 2018, 17, 905–921. [Google Scholar] [CrossRef]
- Retamal, M.A.; Fernandez-Olivares, A.; Stehberg, J. Over-activated hemichannels: A possible therapeutic target for human diseases. Biochim. Biophys. Acta. Mol. Basis Dis. 2021, 1867, 166232. [Google Scholar] [CrossRef] [PubMed]
- Van Campenhout, R.; Gomes, A.R.; De Groof, T.W.M.; Muyldermans, S.; Devoogdt, N.; Vinken, M. Mechanisms Underlying Connexin Hemichannel Activation in Disease. Int. J. Mol. Sci. 2021, 22, 3503. [Google Scholar] [CrossRef]
- Giaume, C.; Naus, C.C.; Saez, J.C.; Leybaert, L. Glial Connexins and Pannexins in the Healthy and Diseased Brain. Physiol. Rev. 2021, 101, 93–145. [Google Scholar] [CrossRef]
- Walrave, L.; Pierre, A.; Albertini, G.; Aourz, N.; De Bundel, D.; Van Eeckhaut, A.; Vinken, M.; Giaume, C.; Leybaert, L.; Smolders, I. Inhibition of astroglial connexin43 hemichannels with TAT-Gap19 exerts anticonvulsant effects in rodents. Glia 2018, 66, 1788–1804. [Google Scholar] [CrossRef]
- Crespo Yanguas, S.; da Silva, T.C.; Pereira, I.V.A.; Willebrords, J.; Maes, M.; Sayuri Nogueira, M.; Alves de Castro, I.; Leclercq, I.; Romualdo, G.R.; Barbisan, L.F.; et al. TAT-Gap19 and Carbenoxolone Alleviate Liver Fibrosis in Mice. Int. J. Mol. Sci. 2018, 19, 817. [Google Scholar] [CrossRef]
- Zhang, C.; Yan, Z.; Maknojia, A.; Riquelme, M.A.; Gu, S.; Booher, G.; Wallace, D.J.; Bartanusz, V.; Goswami, A.; Xiong, W.; et al. Inhibition of astrocyte hemichannel improves recovery from spinal cord injury. JCI Insight 2021, 6, e134611. [Google Scholar] [CrossRef]
- Johnson, R.G.; Le, H.C.; Evenson, K.; Loberg, S.W.; Myslajek, T.M.; Prabhu, A.; Manley, A.M.; O’Shea, C.; Grunenwald, H.; Haddican, M.; et al. Connexin Hemichannels: Methods for Dye Uptake and Leakage. J. Membr. Biol. 2016, 249, 713–741. [Google Scholar] [CrossRef]
- Lilly, E.; Sellitto, C.; Milstone, L.M.; White, T.W. Connexin channels in congenital skin disorders. Semin. Cell Dev. Biol. 2016, 50, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Sybert, V.P. Genetic Skin Disorders; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Hitomi, M.; Deleyrolle, L.P.; Mulkearns-Hubert, E.E.; Jarrar, A.; Li, M.; Sinyuk, M.; Otvos, B.; Brunet, S.; Flavahan, W.A.; Hubert, C.G.; et al. Differential connexin function enhances self-renewal in glioblastoma. Cell Rep. 2015, 11, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Mulkearns-Hubert, E.E.; Torre-Healy, L.A.; Silver, D.J.; Eurich, J.T.; Bayik, D.; Serbinowski, E.; Hitomi, M.; Zhou, J.; Przychodzen, B.; Zhang, R.; et al. Development of a Cx46 Targeting Strategy for Cancer Stem Cells. Cell Rep. 2019, 27, 1062–1072.e1065. [Google Scholar] [CrossRef] [PubMed]
- Acuna, R.A.; Varas-Godoy, M.; Berthoud, V.M.; Alfaro, I.E.; Retamal, M.A. Connexin-46 Contained in Extracellular Vesicles Enhance Malignancy Features in Breast Cancer Cells. Biomolecules 2020, 10, 676. [Google Scholar] [CrossRef] [PubMed]
- Orellana, V.P.; Tittarelli, A.; Retamal, M.A. Connexins in melanoma: Potential role of Cx46 in its aggressiveness. Pigment. Cell Melanoma Res. 2021, 34, 853–868. [Google Scholar] [CrossRef] [PubMed]
- Acuna, R.A.; Varas-Godoy, M.; Herrera-Sepulveda, D.; Retamal, M.A. Connexin46 Expression Enhances Cancer Stem Cell and Epithelial-to-Mesenchymal Transition Characteristics of Human Breast Cancer MCF-7 Cells. Int. J. Mol. Sci. 2021, 22, 12604. [Google Scholar] [CrossRef]
- Shi, Y.; Li, X.; Yang, J. Mutations of CX46/CX50 and Cataract Development. Front. Mol. Biosci. 2022, 9, 842399. [Google Scholar] [CrossRef]
- Xu, L.; Carrer, A.; Zonta, F.; Qu, Z.; Ma, P.; Li, S.; Ceriani, F.; Buratto, D.; Crispino, G.; Zorzi, V.; et al. Design and Characterization of a Human Monoclonal Antibody that Modulates Mutant Connexin 26 Hemichannels Implicated in Deafness and Skin Disorders. Front. Mol. Neurosci. 2017, 10, 298. [Google Scholar] [CrossRef]
- Ziraldo, G.; Buratto, D.; Kuang, Y.; Xu, L.; Carrer, A.; Nardin, C.; Chiani, F.; Salvatore, A.M.; Paludetti, G.; Lerner, R.A.; et al. A Human-Derived Monoclonal Antibody Targeting Extracellular Connexin Domain Selectively Modulates Hemichannel Function. Front. Physiol. 2019, 10, 392. [Google Scholar] [CrossRef]
- Kuang, Y.; Zorzi, V.; Buratto, D.; Ziraldo, G.; Mazzarda, F.; Peres, C.; Nardin, C.; Salvatore, A.M.; Chiani, F.; Scavizzi, F.; et al. A potent antagonist antibody targeting connexin hemichannels alleviates Clouston syndrome symptoms in mutant mice. EBioMedicine 2020, 57, 102825. [Google Scholar] [CrossRef]
- Buratto, D.; Donati, V.; Zonta, F.; Mammano, F. Harnessing the therapeutic potential of antibodies targeting connexin hemichannels. Biochim. Biophys. Acta. Mol. Basis Dis. 2021, 1867, 166047. [Google Scholar] [CrossRef] [PubMed]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [PubMed]
- Schoop, V.M.; Mirancea, N.; Fusenig, N.E. Epidermal organization and differentiation of HaCaT keratinocytes in organotypic coculture with human dermal fibroblasts. J. Investig. Dermatol. 1999, 112, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Maas-Szabowski, N.; Starker, A.; Fusenig, N.E. Epidermal tissue regeneration and stromal interaction in HaCaT cells is initiated by TGF-alpha. J. Cell Sci. 2003, 116, 2937–2948. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, R.; Wang, Z.; Zhou, S.; Song, Y.; Chen, L.; Chen, N.; Liu, W.; Ji, C.; Wu, W.; et al. Mechanistic effect of the human GJB6 gene and its mutations in HaCaT cell proliferation and apoptosis. Braz. J. Med. Biol. Res. 2018, 51, e7560. [Google Scholar] [CrossRef]
- Essenfelder, G.M.; Bruzzone, R.; Lamartine, J.; Charollais, A.; Blanchet-Bardon, C.; Barbe, M.T.; Meda, P.; Waksman, G. Connexin30 mutations responsible for hidrotic ectodermal dysplasia cause abnormal hemichannel activity. Hum. Mol. Genet. 2004, 13, 1703–1714. [Google Scholar]
- Stong, B.C.; Chang, Q.; Ahmad, S.; Lin, X. A novel mechanism for connexin 26 mutation linked deafness: Cell death caused by leaky gap junction hemichannels. Laryngoscope 2006, 116, 2205–2210. [Google Scholar] [CrossRef]
- Lee, J.R.; Derosa, A.M.; White, T.W. Connexin mutations causing skin disease and deafness increase hemichannel activity and cell death when expressed in Xenopus oocytes. J. Investig. Dermatol. 2009, 129, 870–878. [Google Scholar] [CrossRef]
- Sanchez, H.A.; Mese, G.; Srinivas, M.; White, T.W.; Verselis, V.K. Differentially altered Ca2+ regulation and Ca2+ permeability in Cx26 hemichannels formed by the A40V and G45E mutations that cause keratitis ichthyosis deafness syndrome. J. Gen. Physiol. 2010, 136, 47–62. [Google Scholar] [CrossRef]
- Terrinoni, A.; Codispoti, A.; Serra, V.; Didona, B.; Bruno, E.; Nistico, R.; Giustizieri, M.; Alessandrini, M.; Campione, E.; Melino, G. Connexin 26 (GJB2) mutations, causing KID Syndrome, are associated with cell death due to calcium gating deregulation. Biochem. Biophys. Res. Commun. 2010, 394, 909–914. [Google Scholar] [CrossRef]
- Sanchez, H.A.; Villone, K.; Srinivas, M.; Verselis, V.K. The D50N mutation and syndromic deafness: Altered Cx26 hemichannel properties caused by effects on the pore and intersubunit interactions. J. Gen. Physiol. 2013, 142, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.C.; Kelly, J.J.; Lajoie, P.; Shao, Q.; Laird, D.W. Mutations in Cx30 that are linked to skin disease and non-syndromic hearing loss exhibit several distinct cellular pathologies. J. Cell Sci. 2014, 127, 1751–1764. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef]
- Gerido, D.A.; DeRosa, A.M.; Richard, G.; White, T.W. Aberrant hemichannel properties of Cx26 mutations causing skin disease and deafness. Am. J. Physiol. Cell Physiol. 2007, 293, C337–C345. [Google Scholar] [CrossRef] [PubMed]
- Mese, G.; Sellitto, C.; Li, L.; Wang, H.Z.; Valiunas, V.; Richard, G.; Brink, P.R.; White, T.W. The Cx26-G45E mutation displays increased hemichannel activity in a mouse model of the lethal form of keratitis-ichthyosis-deafness syndrome. Mol. Biol. Cell 2011, 22, 4776–4786. [Google Scholar] [CrossRef] [PubMed]
- Sellitto, C.; Li, L.; White, T.W. Connexin hemichannel inhibition ameliorates epidermal pathology in a mouse model of keratitis ichthyosis deafness syndrome. Sci. Rep. 2021, 11, 24118. [Google Scholar] [CrossRef]
- Agha-Mohammadi, S.; O’Malley, M.; Etemad, A.; Wang, Z.; Xiao, X.; Lotze, M.T. Second-generation tetracycline-regulatable promoter: Repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J. Gene Med. 2004, 6, 817–828. [Google Scholar] [CrossRef]
- Donati, V.; Peres, C.; Nardin, C.; Scavizzi, F.; Raspa, M.; Ciubotaru, C.D.; Bortolozzi, M.; Pedersen, M.G.; Mammano, F. Calcium Signaling in the Photodamaged Skin: In Vivo Experiments and Mathematical Modeling. Function 2022, 3, zqab064. [Google Scholar] [CrossRef]
- Hennings, H.; Michael, D.; Cheng, C.; Steinert, P.; Holbrook, K.; Yuspa, S.H. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell 1980, 19, 245–254. [Google Scholar] [CrossRef]
- Watt, F.M.; Green, H. Stratification and terminal differentiation of cultured epidermal cells. Nature 1982, 295, 434–436. [Google Scholar] [CrossRef]
- Bikle, D.D.; Xie, Z.; Tu, C.L. Calcium regulation of keratinocyte differentiation. Expert Rev. Endocrinol. Metab. 2012, 7, 461–472. [Google Scholar] [CrossRef]
- Garcia-Vega, L.; O’Shaughnessy, E.M.; Jan, A.; Bartholomew, C.; Martin, P.E. Connexin 26 and 43 play a role in regulating proinflammatory events in the epidermis. J. Cell. Physiol. 2019, 234, 15594–15606. [Google Scholar] [CrossRef]
- Fitzgerald, D.J.; Fusenig, N.E.; Boukamp, P.; Piccoli, C.; Mesnil, M.; Yamasaki, H. Expression and function of connexin in normal and transformed human keratinocytes in culture. Carcinogenesis 1994, 15, 1859–1865. [Google Scholar] [CrossRef]
- Nugent, L.; Ofori-Frimpong, B.; Martin, P.E.; Green, C.R.; Wright, C.S. Cx31.1 expression is modulated in HaCaT cells exposed to UV-induced damage and scrape-wounding. J. Cell. Physiol. 2021, 236, 911–920. [Google Scholar] [CrossRef]
- Ceriani, F.; Ciubotaru, C.D.; Bortolozzi, M.; Mammano, F. Design and Construction of a Cost-Effective Spinning Disk System for Live Imaging of Inner Ear Tissue. Methods Mol. Biol. 2016, 1427, 223–241. [Google Scholar] [CrossRef]
- Mammano, F.; Bortolozzi, M. Ca2+ imaging: Principles of analysis and enhancement. In Calcium Measurement Methods; Verkhratsky, A., Petersen, O., Eds.; Humana Press: New York, NY, USA, 2010; Volume 43, pp. 57–80. [Google Scholar]
- Woszczek, G.; Fuerst, E.; Maguire, T.J.A. FLIPR Calcium Mobilization Assays in GPCR Drug Discovery. Methods Mol. Biol. 2021, 2268, 193–205. [Google Scholar] [CrossRef]
- Vetter, I.; Carter, D.; Bassett, J.; Deuis, J.R.; Tay, B.; Jami, S.; Robinson, S.D. High-Throughput Fluorescence Assays for Ion Channels and GPCRs. Adv. Exp. Med. Biol. 2020, 1131, 27–72. [Google Scholar] [CrossRef]
- Bikle, D.D.; Ratnam, A.; Mauro, T.; Harris, J.; Pillai, S. Changes in calcium responsiveness and handling during keratinocyte differentiation. Potential role of the calcium receptor. J. Clin. Investig. 1996, 97, 1085–1093. [Google Scholar] [CrossRef]
- Elsholz, F.; Harteneck, C.; Muller, W.; Friedland, K. Calcium—A central regulator of keratinocyte differentiation in health and disease. Eur. J. Dermatol. EJD 2014, 24, 650–661. [Google Scholar] [CrossRef]
- Bezprozvanny, I.; Watras, J.; Ehrlich, B.E. Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature 1991, 351, 751–754. [Google Scholar] [CrossRef]
- Woll, K.A.; Van Petegem, F. Calcium-release channels: Structure and function of IP3 receptors and ryanodine receptors. Physiol. Rev. 2022, 102, 209–268. [Google Scholar] [CrossRef]
- Glover, J.D.; Taylor, L.; Sherman, A.; Zeiger-Poli, C.; Sang, H.M.; McGrew, M.J. A novel piggyBac transposon inducible expression system identifies a role for AKT signalling in primordial germ cell migration. PLoS ONE 2013, 8, e77222. [Google Scholar] [CrossRef]
- Choi, E.J.; Palacios-Prado, N.; Saez, J.C.; Lee, J. Identification of Cx45 as a Major Component of GJs in HeLa Cells. Biomolecules 2020, 10, 1389. [Google Scholar] [CrossRef]
- Zonta, F.; Polles, G.; Zanotti, G.; Mammano, F. Permeation pathway of homomeric connexin 26 and connexin 30 channels investigated by molecular dynamics. J. Biomol. Struct. Dyn. 2012, 29, 985–998. [Google Scholar] [CrossRef]
- Zonta, F.; Polles, G.; Sanasi, M.F.; Bortolozzi, M.; Mammano, F. The 3.5 angstrom X-ray structure of the human connexin26 gap junction channel is unlikely that of a fully open channel. Cell Commun. Signal. CCS 2013, 11, 15. [Google Scholar] [CrossRef]
- Cribbs, A.P.; Kennedy, A.; Gregory, B.; Brennan, F.M. Simplified production and concentration of lentiviral vectors to achieve high transduction in primary human T cells. BMC Biotechnol. 2013, 13, 98. [Google Scholar] [CrossRef]
- Persson, H.; Ye, W.; Wernimont, A.; Adams, J.J.; Koide, A.; Koide, S.; Lam, R.; Sidhu, S.S. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. J. Mol. Biol. 2013, 425, 803–811. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).