Bioactive Compounds and Their Impact on Protein Modification in Human Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Bioactive Compounds
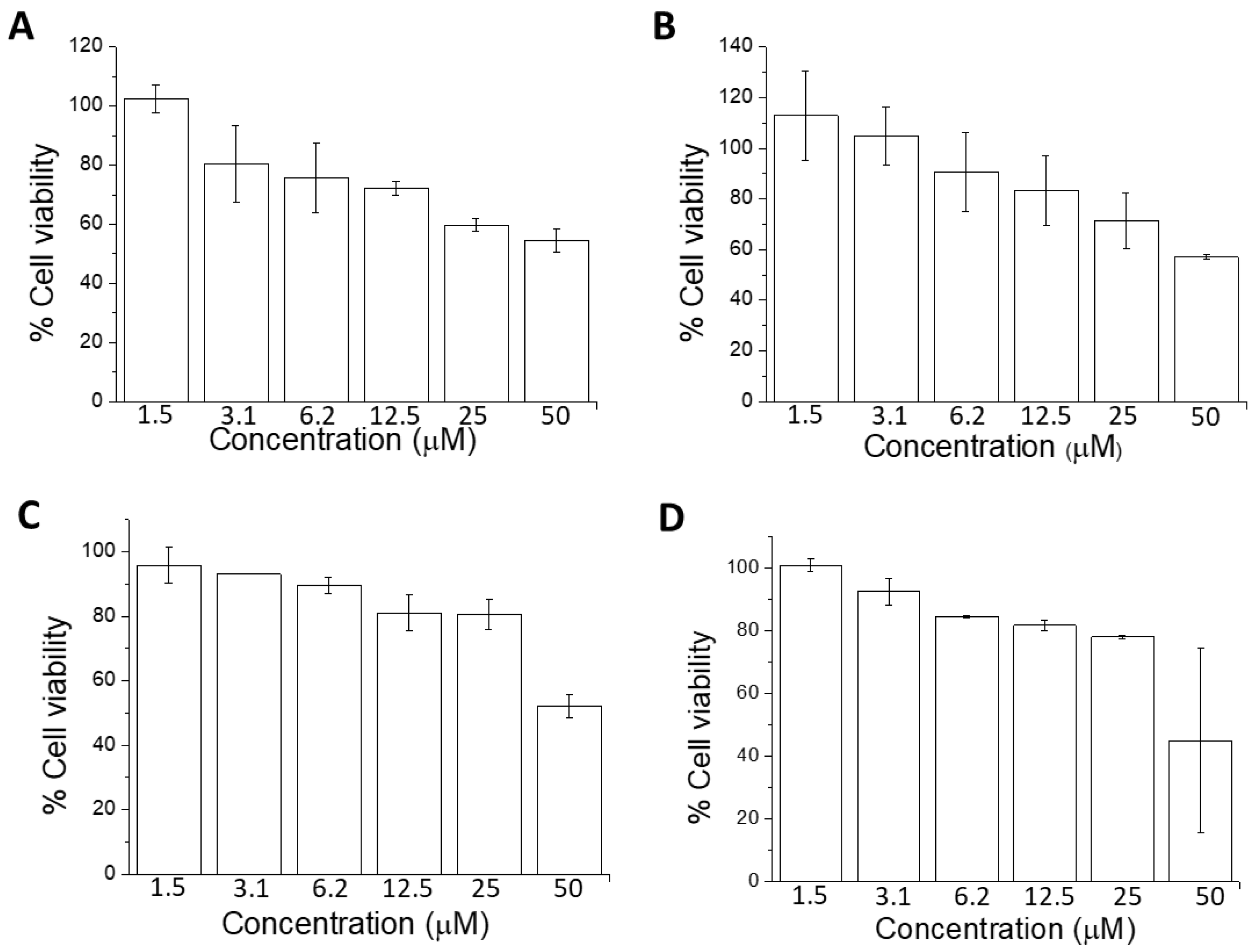
2.2. Cell Viability Using MTT Assay
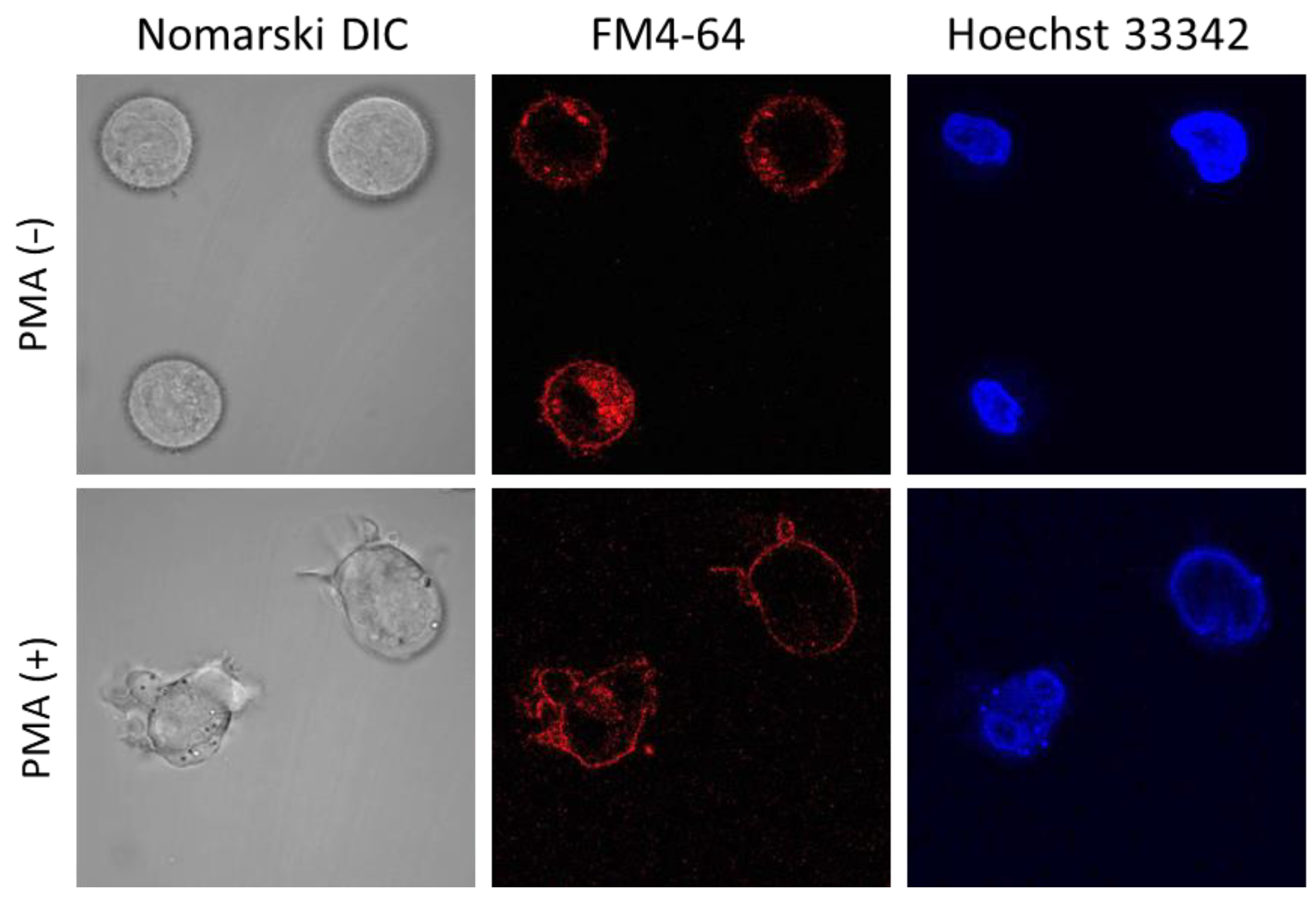
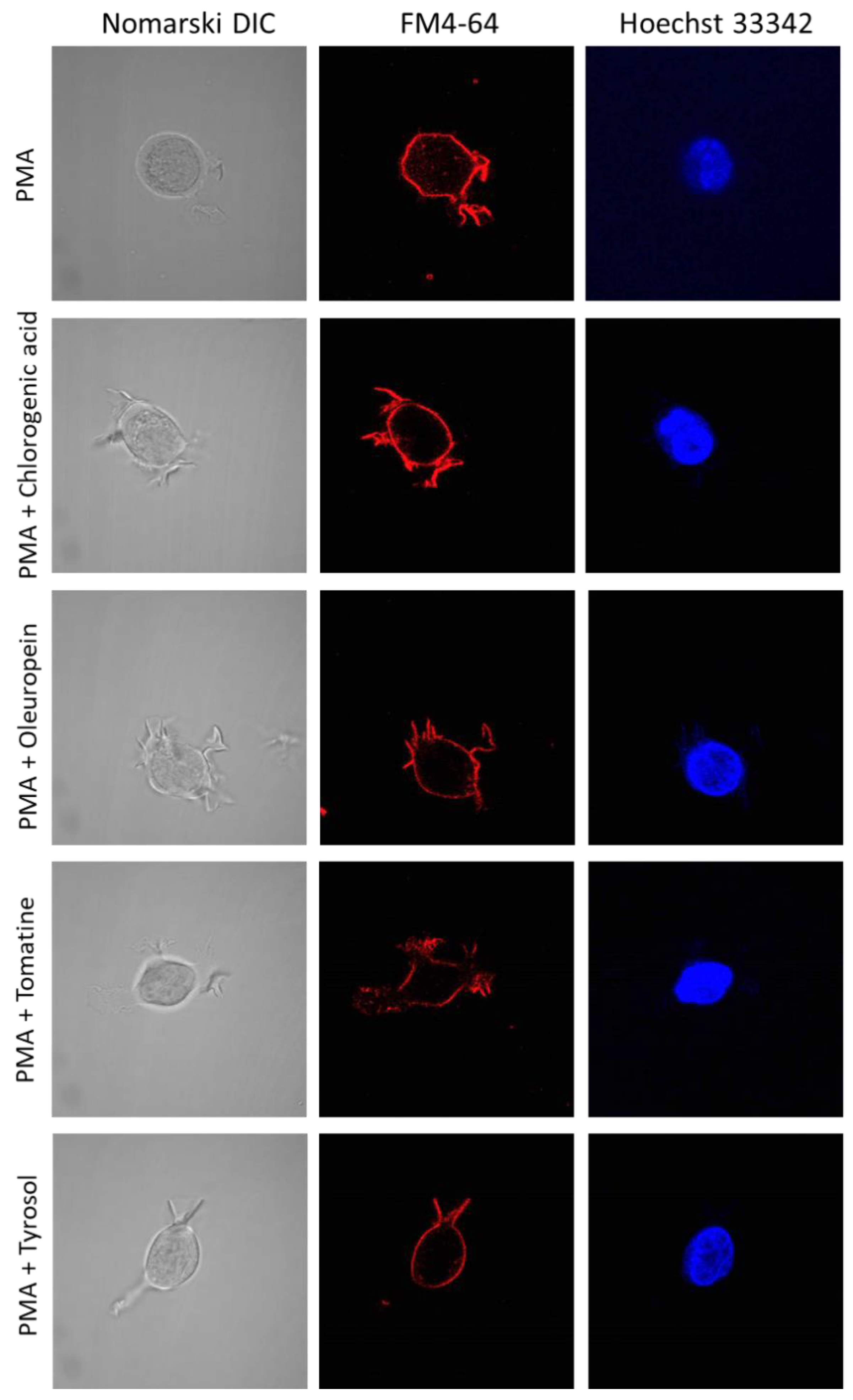
2.3. U-937 Cell Morphology
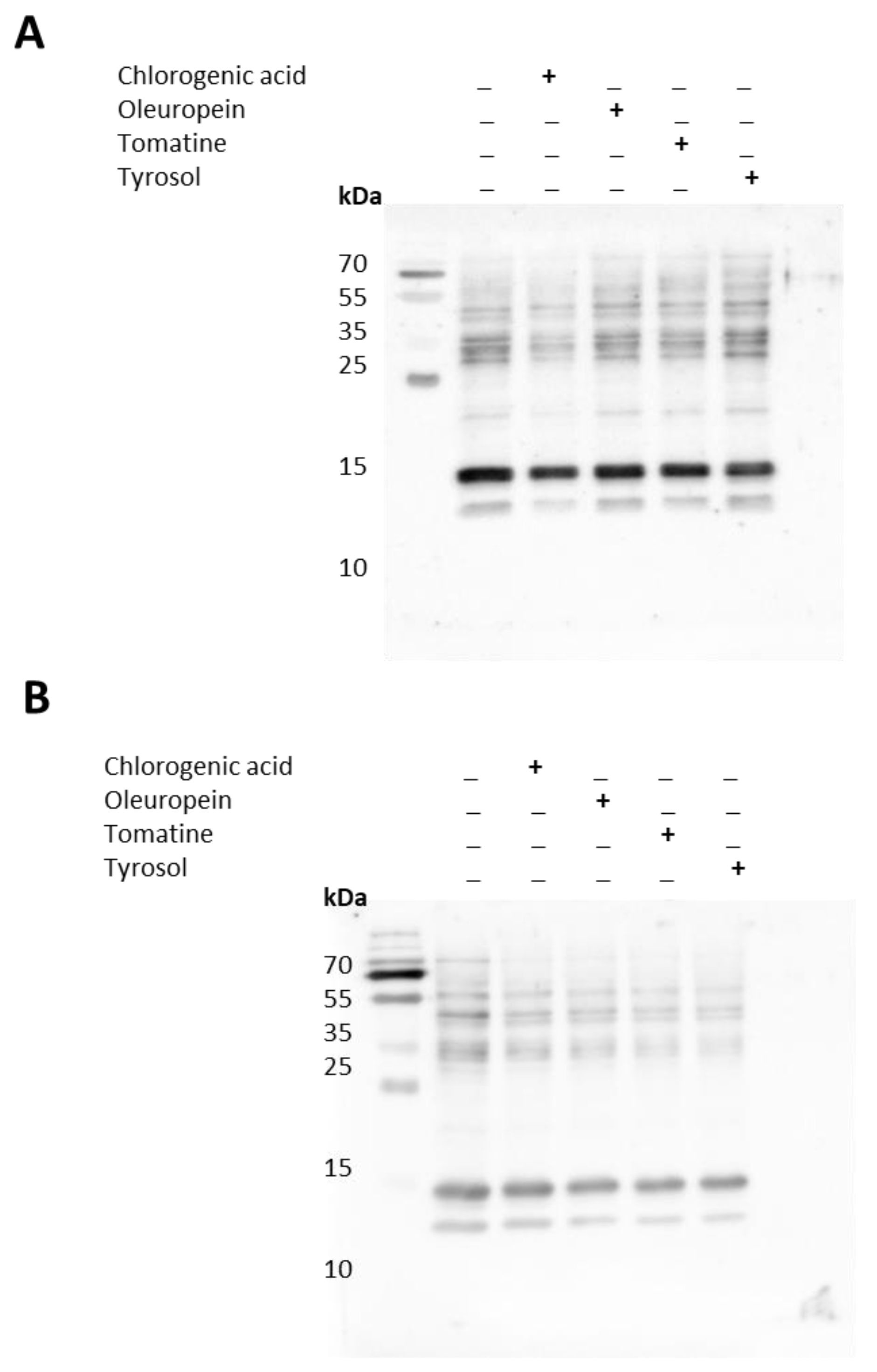
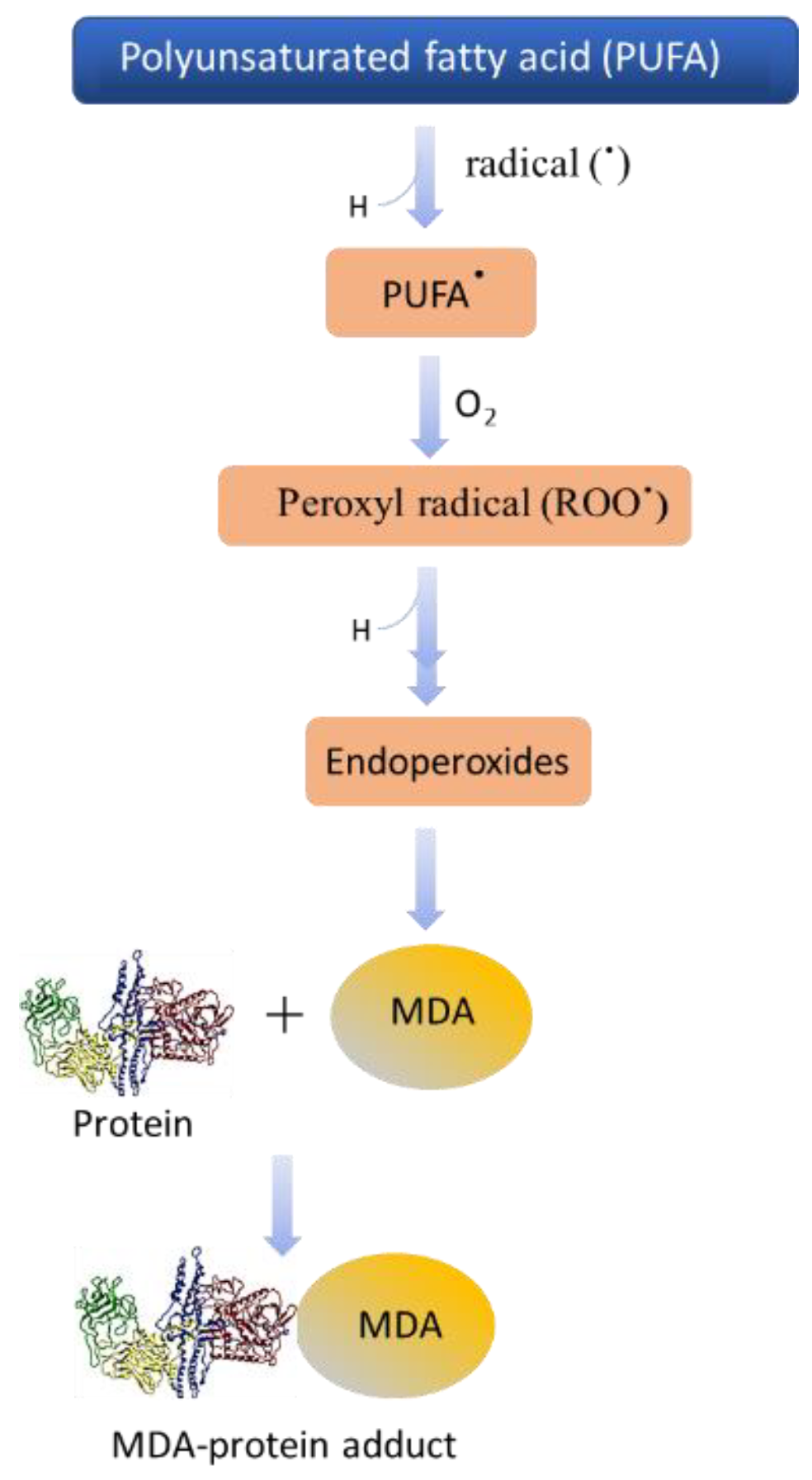
2.4. Protein Modification in Differentiating Cells and Impact of Bioactive Compounds
3. Methods and Materials
3.1. Extracts
3.2. Reagents and Antibodies
3.3. HPLC-MS Analysis for α-Tomatine Determination in Tomato Leaf Extracts
3.4. HPLC-UV Analysis for Oleuropein Determination in Olive Leaf Extracts
3.5. Cell Line Origin and Cultivation
3.6. Cell Line and Differentiation Condition
3.7. MTT Cell Proliferation Assay
3.8. Confocal Laser Scanning Microscopy
3.9. Protein Immunoblotting
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gutteridge, J.M.C.; Halliwell, B. Free radicals and antioxidants in the year 2000—A historical look to the future. Ann. N. Y. Acad. Sci. 2000, 899, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B.; Gutteridge, J. Free Radicals in Biology and Medicine, 4th ed.; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Wang, X.; Quinn, P.J. Vitamin E and its function in membranes. Prog. Lipid Res. 1999, 38, 309–336. [Google Scholar] [CrossRef]
- Young, A.J.; Lowe, G.M. Antioxidant and prooxidant properties of carotenoids. Arch. Biochem. Biophys. 2001, 385, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Munne-Bosch, S.; Alegre, L. The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 2002, 21, 31–57. [Google Scholar] [CrossRef]
- Liang, N.; Kitts, D.D. Role of chlorogenic acids in controlling oxidative and inflammatory stress conditions. Nutrients 2016, 8, 16. [Google Scholar] [CrossRef] [Green Version]
- Nabavi, S.F.; Tejada, S.; Setzer, W.N.; Gortzi, O.; Sureda, A.; Braidy, N.; Daglia, M.; Manayi, A. Chlorogenic acid and mental diseases: From chemistry to medicine. Curr. Neuropharmacol. 2017, 15, 471–479. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.J.; Tian, Z.M.; Cui, Y.Y.; Liu, Z.C.; Ma, X.Y. Chlorogenic acid: A comprehensive review of the dietary sources, processing effects, bioavailability, beneficial properties, mechanisms of action, and future directions. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3130–3158. [Google Scholar] [CrossRef]
- Lafay, S.; Gil-Izquierdo, A.; Manach, C.; Morand, C.; Besson, C.; Scalbert, A. Chlorogenic acid is absorbed in its intact form in the stomach of rats. J. Nutr. 2006, 136, 1192–1197. [Google Scholar] [CrossRef] [Green Version]
- Ekbatan, S.S.; Li, X.Q.; Ghorbani, M.; Azadi, B.; Kubow, S. Chlorogenic acid and its microbial metabolites exert anti-proliferative effects, s-phase cell-cycle arrest and apoptosis in human colon cancer caco-2 cells. Int. J. Mol. Sci. 2018, 19, 723. [Google Scholar] [CrossRef] [Green Version]
- Silva-Beltrán, N.P.; Ruiz-Cruz, S.; Cira-Chávez, L.A.; Estrada-Alvarado, M.I.; Ornelas-Paz, J.D.J.; López-Mata, M.A.; Del-Toro-Sánchez, C.L.; Zavala, J.F.A.; Márquez-Ríos, E. Total phenolic, flavonoid, tomatine, and tomatidine contents and antioxidant and antimicrobial activities of extracts of tomato plant. Int. J. Anal. Chem. 2015, 2015, 284071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcolongo, P.; Gamberucci, A.; Tamasi, G.; Pardini, A.; Bonechi, C.; Rossi, C.; Giunti, R.; Barone, V.; Borghini, A.; Fiorenzani, P.; et al. Chemical characterisation and antihypertensive effects of locular gel and serum of Lycopersicum esculentum L. var. “Camone” tomato in spontaneously hypertensive rats. Molecules 2020, 25, 3758. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.; Levin, C.E.; Lee, S.-U.; Kim, H.-J.; Lee, I.-S.; Byun, J.-O.; Kozukue, N. Tomatine-containing green tomato extracts inhibit growth of human breast, colon, liver, and stomach cancer cells. J. Agric. Food Chem. 2009, 57, 5727–5733. [Google Scholar] [CrossRef] [PubMed]
- Serratì, S.; Porcelli, L.; Guida, S.; Ferretta, A.; Iacobazzi, R.M.; Cocco, T.; Maida, I.; Tamasi, G.; Rossi, C.; Manganelli, M.; et al. Tomatine displays antitumor potential in in vitro models of metastatic melanoma. Int. J. Mol. Sci. 2020, 21, 5243. [Google Scholar] [CrossRef]
- Toor, R.K.; Savage, G.P.; Lister, C.E. Release of antioxidant components from tomatoes determined by an in vitro digestion method. Int. J. Food Sci. Nutr. 2009, 60, 119–129. [Google Scholar] [CrossRef]
- Huang, S.-L.; He, H.-B.; Zou, K.; Bai, C.-H.; Xue, Y.-H.; Wang, J.-Z.; Chen, J.-F. Protective effect of tomatine against hydrogen peroxide-induced neurotoxicity in neuroblastoma (SH-SY5Y) cells. J. Pharm. Pharmacol. 2014, 66, 844–854. [Google Scholar] [CrossRef]
- Friedman, M. Anticarcinogenic, cardioprotective, and other health benefits of tomato compounds lycopene, alpha-tomatine, and tomatidine in pure form and in fresh and processed tomatoes. J. Agric. Food Chem. 2013, 61, 9534–9550. [Google Scholar] [CrossRef]
- Kúdelová, J.; Seifrtova, M.; Suchá, L.; Tomšík, P.; Havelek, R.; Řezáčová, M. Alpha-tomatine activates cell cycle checkpoints in the absence of DNA damage in human leukemic MOLT-4 cells. J. Appl. Biomed. 2013, 11, 93–103. [Google Scholar] [CrossRef]
- Chandra, H.M.; Ramalingam, S. Antioxidant potentials of skin, pulp, and seed fractions of commercially important tomato cultivars. Food Sci. Biotechnol. 2011, 20, 15–21. [Google Scholar] [CrossRef]
- Pardini, A.; Consumi, M.; Leone, G.; Bonechi, C.; Tamasi, G.; Sangiorgio, P.; Verardi, A.; Rossi, C.; Magnani, A. Effect of different post-harvest storage conditions and heat treatment on tomatine content in commercial varieties of green tomatoes. J. Food Compos. Anal. 2021, 96, 103735. [Google Scholar] [CrossRef]
- Motawea, M.H.; Ali, H.A.E.; Elharrif, M.G.; Desoky, A.A.E.; Ibrahimi, A. Evaluation of anti-inflammatory and antioxidant profile of oleuropein in experimentally induced ulcerative colitis. Int. J. Mol. Cell. Med. 2020, 9, 224–233. [Google Scholar] [PubMed]
- Burja, B.; Kuret, T.; Janko, T.; Topalović, D.; Živković, L.; Mrak-Poljšak, K.; Spremo-Potparević, B.; Žigon, P.; Distler, O.; Čučnik, S.; et al. Olive leaf extract attenuates inflammatory activation and DNA damage in human arterial endothelial cells. Front. Cardiovasc. Med. 2019, 6, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castejon, M.L.; Rosillo, M.A.; Montoya, T.; Gonzalez-Benjumea, A.; Fernandez-Bolanos, J.M.; Alarcon-de-la-Lastra, C. Oleuropein down-regulated IL-1 beta-induced inflammation and oxidative stress in human synovial fibroblast cell line SW982. Food Funct. 2017, 8, 1890–1898. [Google Scholar] [CrossRef]
- Ryu, S.-J.; Choi, H.-S.; Yoon, K.-Y.; Lee, O.-H.; Kim, K.-J.; Lee, B.-Y. Oleuropein suppresses LPS-induced inflammatory responses in RAW 264.7 cell and zebrafish. J. Agric. Food Chem. 2015, 63, 2098–2105. [Google Scholar] [CrossRef] [PubMed]
- Ahamad, J.; Toufeeq, I.; Khan, M.A.; Ameen, M.S.M.; Anwer, E.T.; Uthirapathy, S.; Mir, S.R.; Ahmad, J. Oleuropein: A natural antioxidant molecule in the treatment of metabolic syndrome. Phytother. Res. 2019, 33, 3112–3128. [Google Scholar] [CrossRef] [PubMed]
- Lucas, R.; Comelles, F.; Alcántara, D.; Maldonado, O.S.; Curcuroze, M.; Parra, J.L.; Morales, J.C. Surface-active properties of lipophilic antioxidants tyrosol and hydroxytyrosol fatty acid esters: A potential explanation for the nonlinear hypothesis of the antioxidant activity in oil-in-water emulsions. J. Agric. Food Chem. 2010, 58, 8021–8026. [Google Scholar] [CrossRef]
- Giovannini, C.; Straface, E.; Modesti, D.; Coni, E.; Cantafora, A.; De Vincenzi, M.; Malorni, W.; Masella, R. Tyrosol, the major olive oil biophenol, protects against oxidized-LDL-induced injury in Caco-2 cells. J. Nutr. 1999, 129, 1269–1277. [Google Scholar] [CrossRef] [Green Version]
- Moreno, J.J. Effect of olive oil minor components on oxidative stress and arachidonic acid mobilization and metabolism by macrophages RAW 264.7. Free Radic. Biol. Med. 2003, 35, 1073–1081. [Google Scholar] [CrossRef]
- Muriana, F.J.G.; la Paz, S.M.-D.; Lucas, R.; Bermudez, B.; Jaramillo, S.; Morales, J.C.; Abia, R.; Lopez, S. Tyrosol and its metabolites as antioxidative and anti-inflammatory molecules in human endothelial cells. Food Funct. 2017, 8, 2905–2914. [Google Scholar] [CrossRef] [Green Version]
- Sundström, C.; Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 1976, 17, 565–577. [Google Scholar] [CrossRef]
- Prasad, A.; Sedlářová, M.; Balukova, A.; Rác, M.; Pospíšil, P. Reactive oxygen species as a response to wounding: In vivo imaging in Arabidopsis thaliana. Front. Plant Sci. 2020, 10, 1660. [Google Scholar] [CrossRef] [PubMed]
- Golubev, A.; Khrustalev, S.; Butov, A. An in silico investigation into the causes of telomere length heterogeneity and its implications for the Hayflick limit. J. Theor. Biol. 2003, 225, 153–170. [Google Scholar] [CrossRef]
- Gomez, D.E.; Armando, R.G.; Farina, H.G.; Lorenzano Menna, P.; Cerrudo, C.S.; Daniel Ghiringhelli, P.; Alonso, D.F. Telomere structure and telomerase in health and disease (Review). Int. J. Oncol. 2012, 41, 1561–1569. [Google Scholar] [CrossRef] [Green Version]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Droge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [Green Version]
- Prasad, A.; Manoharan, R.R.; Sedlářová, M.; Pospíšil, P. Free radical-mediated protein radical formation in differentiating monocytes. Int. J. Mol. Sci. 2021, 22, 9963. [Google Scholar] [CrossRef]
- Maeß, M.B.; Wittig, B.; Cignarella, A.; Lorkowski, S. Reduced PMA enhances the responsiveness of transfected THP-1 macrophages to polarizing stimuli. J. Immunol. Methods 2014, 402, 76–81. [Google Scholar] [CrossRef]
- Lund, M.E.; To, J.; O’Brien, B.A.; Donnelly, S. The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J. Immunol. Methods 2016, 430, 64–70. [Google Scholar] [CrossRef]
- STedesco, S.; De Majo, F.; Kim, J.; Trenti, A.; Trevisi, L.; Fadini, G.P.; Bolego, C.; Zandstra, P.W.; Cignarella, A.; Vitiello, L. Convenience versus biological significance: Are PMA-differentiated THP-1 cells a reliable substitute for blood-derived macrophages when studying in vitro polarization? Front. Pharmacol. 2018, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Giridhary, K.V.; Zhang, L.; Xu, S.; Wang, Q.J. A protein kinase C/protein kinase D pathway protects LNCaP prostate cancer cells from phorbol ester-induced apoptosis by promoting ERK1/2 and NF-kappa B activities. Carcinogenesis 2011, 32, 1198–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garg, R.; Caino, M.C.; Kazanietz, M.G. Regulation of transcriptional networks by PKC isozymes: Identification of c-rel as a key transcription factor for PKC-regulated genes. PLoS ONE 2013, 8, e67319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, A.; Kumar, A.; Suzuki, M.; Kikuchi, H.; Sugai, T.; Kobayashi, M.; Pospíšil, P.; Tada, M.; Kasai, S. Detection of hydrogen peroxide in Photosystem II (PSII) using catalytic amperometric biosensor. Front. Plant Sci. 2015, 6, 862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, H.; Prasad, A.; Matsuoka, R.; Aoyagi, S.; Matsue, T.; Kasai, S. Scanning electrochemical microscopy imaging during respiratory burst in human cell. Front. Physiol. 2016, 7, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pospíšil, P.; Prasad, A.; Rác, M. Mechanism of the formation of electronically excited species by oxidative metabolic processes: Role of reactive oxygen species. Biomolecules 2019, 9, 258. [Google Scholar] [CrossRef] [Green Version]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxidative Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Hauck, A.K.; Bernlohr, D.A. Oxidative stress and lipotoxicity. J. Lipid Res. 2016, 57, 1976–1986. [Google Scholar] [CrossRef] [Green Version]
- McDonagh, B. Detection of ROS induced proteomic signatures by mass spectrometry. Front. Physiol. 2017, 8, 470. [Google Scholar] [CrossRef]
- Grimsrud, P.A.; Xie, H.; Griffin, T.J.; Bernlohr, D.A. oxidative stress and covalent modification of protein with bioactive aldehydes. J. Biol. Chem. 2008, 283, 21837–21841. [Google Scholar] [CrossRef] [Green Version]
- Tamasi, G.; Pardini, A.; Bonechi, C.; Donati, A.; Pessina, F.; Marcolongo, P.; Gamberucci, A.; Leone, G.; Consumi, M.; Magnani, A.; et al. Characterization of nutraceutical components in tomato pulp, skin and locular gel. Eur. Food Res. Technol. 2019, 245, 907–918. [Google Scholar] [CrossRef]
- Tamasi, G.; Baratto, M.C.; Bonechi, C.; Byelyakova, A.; Pardini, A.; Donati, A.; Leone, G.; Consumi, M.; Lamponi, S.; Magnani, A.; et al. Chemical characterization and antioxidant properties of products and by-products from Olea europaea L. Food Sci. Nutr. 2019, 7, 2907–2920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
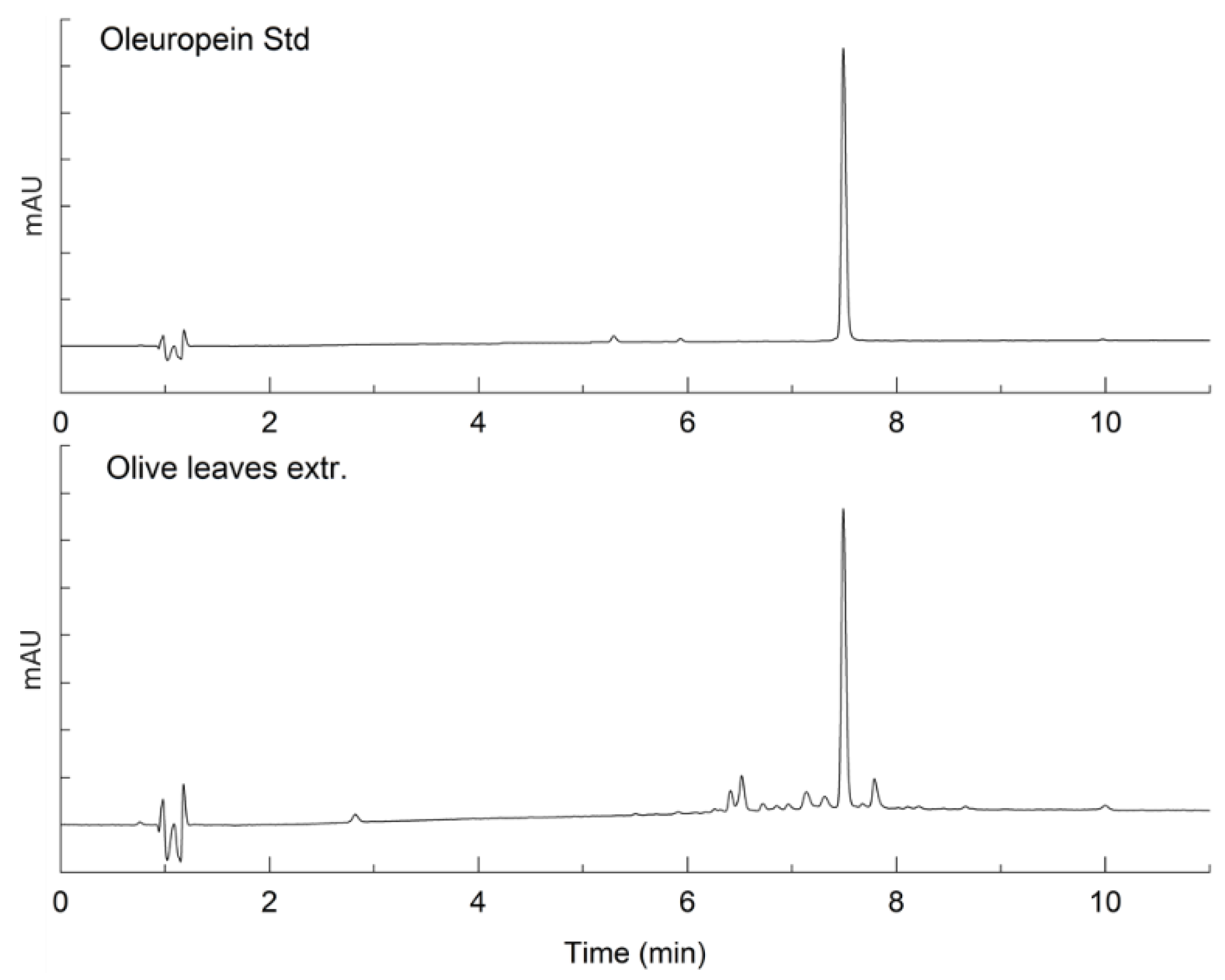
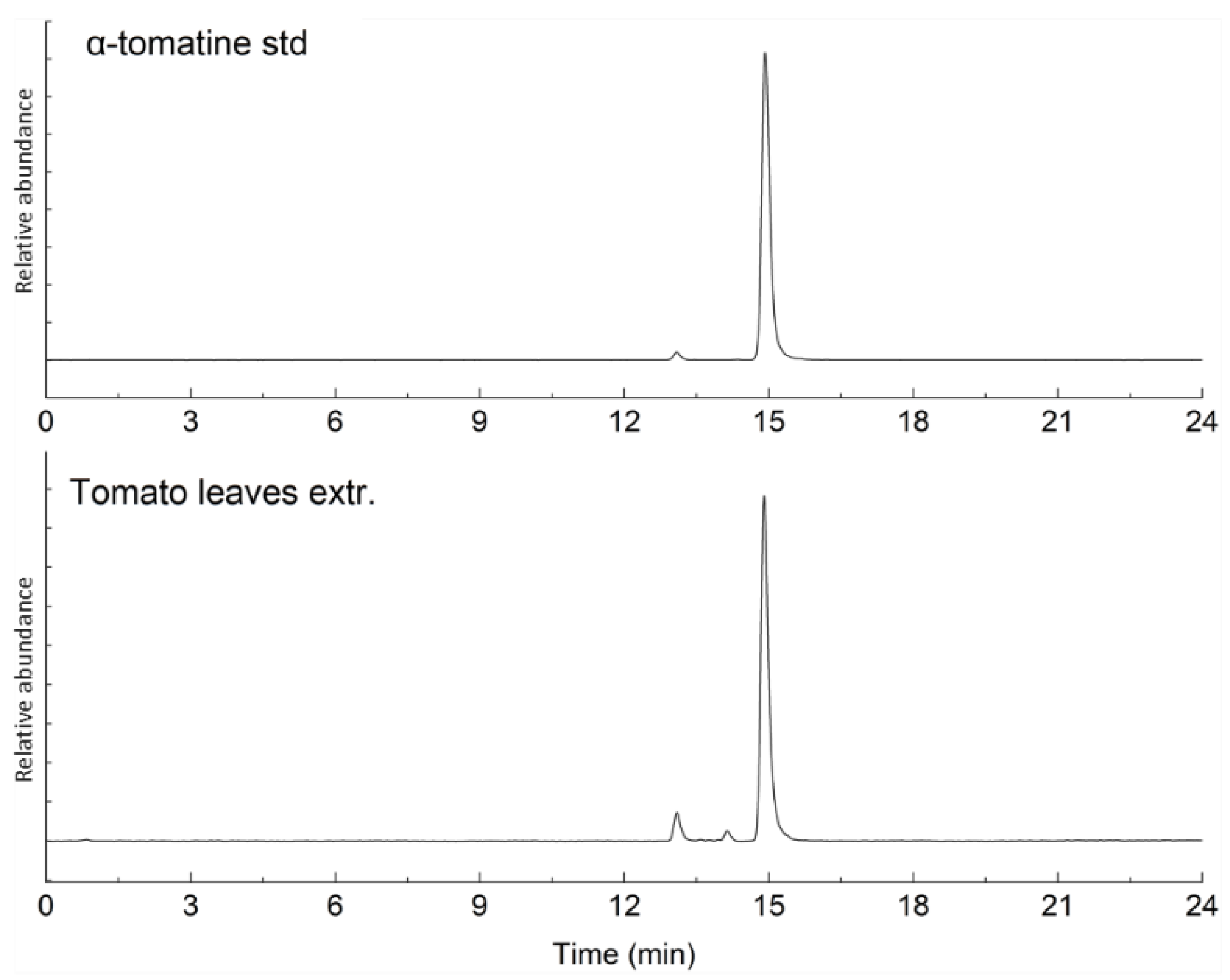

| Compound | Average Concentration (mg/g) | Standard Deviation | % RSD | % Purity (w/w) |
|---|---|---|---|---|
| Oleuropein | 0.602 | 0.010 | 0.01 | 60.8 ± 0.27 |
| α-Tomatine | 0.953 | 0.052 | 5.42 | 94.2 ± 0.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasad, A.; Rossi, C.; Manoharan, R.R.; Sedlářová, M.; Cangeloni, L.; Rathi, D.; Tamasi, G.; Pospíšil, P.; Consumi, M. Bioactive Compounds and Their Impact on Protein Modification in Human Cells. Int. J. Mol. Sci. 2022, 23, 7424. https://doi.org/10.3390/ijms23137424
Prasad A, Rossi C, Manoharan RR, Sedlářová M, Cangeloni L, Rathi D, Tamasi G, Pospíšil P, Consumi M. Bioactive Compounds and Their Impact on Protein Modification in Human Cells. International Journal of Molecular Sciences. 2022; 23(13):7424. https://doi.org/10.3390/ijms23137424
Chicago/Turabian StylePrasad, Ankush, Claudio Rossi, Renuka Ramalingam Manoharan, Michaela Sedlářová, Lorenzo Cangeloni, Deepak Rathi, Gabriella Tamasi, Pavel Pospíšil, and Marco Consumi. 2022. "Bioactive Compounds and Their Impact on Protein Modification in Human Cells" International Journal of Molecular Sciences 23, no. 13: 7424. https://doi.org/10.3390/ijms23137424






