Early Gonadal Development and Sex Determination in Mammal
Abstract
1. Introduction
2. Genital Ridge Formation
3. Differentiation of the Bipotential Gonads
3.1. Sex-Determining Region Y, SRY, and Sox9
3.2. WNT Family Member 4 (WNT4)
3.3. R-Spondin 1 (RSPO1)
3.4. Forkhead Box L2 (FOXL2)
4. Current Knowledge of PGCs
4.1. Formation of PGCs
4.2. In Vitro Derivation of PGCs
4.3. Migration of PGCs
4.4. Proliferation of PGCs and Gametogenesis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Witchel, S.F. Disorders of sex development. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 48, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Arboleda, V.A.; Sandberg, D.E.; Vilain, E. DSDs: Genetics, underlying pathologies and psychosexual differentiation. Nat. Rev. Endocrinol. 2014, 10, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Vander, B.M.; Wyns, C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018, 62, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Sex chromosomes and the evolution of sexual dimorphism. Evolution 1984, 38, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Herald, A.K.; Bashamboo, A. Gonadal development. Underst. Differ. Disord. Sex Dev. 2014, 27, 1–16. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, Z.; Wu, Z.; Hong, L. Sex Manipulation Technologies Progress in Livestock: A Review. Front. Vet. Sci. 2020, 7, 481. [Google Scholar] [CrossRef]
- She, Z.Y.; Yang, W.X. Sry and SoxE genes: How they participate in mammalian sex determination and gonadal development? Semin. Cell Dev. Biol. 2017, 63, 13–22. [Google Scholar] [CrossRef]
- Tanaka, S.S.; Nishinakamura, R. Regulation of male sex determination: Genital ridge formation and Sry activation in mice. Cell. Mol. Life Sci. 2014, 71, 4781–4802. [Google Scholar] [CrossRef] [PubMed]
- Biason-Lauber, A. WNT4, RSPO1, and FOXL2 in sex development. Semin. Reprod. Med. 2012, 30, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, A.; Volarevic, V.; Armstrong, L.; Lako, M.; Stojkovic, M. Primordial Germ Cells: Current Knowledge and Perspectives. Stem Cells Int. 2016, 2016, 1741072. [Google Scholar] [CrossRef] [PubMed]
- Marlow, F. Primordial Germ Cell Specification and Migration. F1000Research 2015, 4, Rev-1462. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.L.; DeFalco, T. Of Mice and Men: In Vivo and In Vitro Studies of Primordial Germ Cell Specification. Semin. Reprod. Med. 2017, 35, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Jost, A. A new look at the mechanisms controlling sex differentiation in mammals. Johns Hopkins Med. J. 1972, 1, 38–53. [Google Scholar]
- Wartenberg, H.; Kinsky, I.; Viebahn, C.; Schmolke, C. Fine structural characteristics of testicular cord formation in the developing rabbit gonad. J. Electron Microsc. Tech. 1991, 19, 133–157. [Google Scholar] [CrossRef] [PubMed]
- Pelliniemi, L.J. Ultrastructure of gonadal ridge in male and female pig embryos. Anat. Embryol. 1975, 147, 20–34. [Google Scholar] [CrossRef]
- Gropp, A.; Ohno, S. The presence of a common embryonic blastema for ovarian and testicular parenchymal (follicular, interstitial and tubular) cells in cattle Bos taurus. Z. Zellforsch. Mikrosk. Anat. 1966, 74, 505–528. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.; Renfree, M.B. Wolffian duct development. Sex. Dev. 2014, 8, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Hannema, S.E.; Hughes, I.A. Regulation of Wolffian duct development. Horm. Res. 2007, 67, 142–151. [Google Scholar] [CrossRef]
- Acien, P. Embryological observations on the female genital tract. Hum. Reprod. 1992, 7, 437–445. [Google Scholar] [CrossRef]
- Ross, A.J.; Capel, B. Signaling at the crossroads of gonad development. Trends Endocrinol. Metab. 2005, 16, 19–25. [Google Scholar] [CrossRef]
- Hirst, C.E.; Major, A.T.; Smith, C.A. Sex determination and gonadal sex differentiation in the chicken model. Int. J. Dev. Biol. 2018, 62, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Stevant, I.; Kuhne, F.; Greenfield, A.; Chaboissier, M.C.; Dermitzakis, E.T.; Nef, S. Dissecting Cell Lineage Specification and Sex Fate Determination in Gonadal Somatic Cells Using Single-Cell Transcriptomics. Cell Rep. 2019, 26, 3272–3283. [Google Scholar] [CrossRef]
- Guo, J.; Sosa, E.; Chitiashvili, T.; Nie, X.; Rojas, E.J.; Oliver, E.; Plath, K.; Hotaling, J.M.; Stukenborg, J.B.; Clark, A.T.; et al. Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell 2021, 28, 764–778. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Workman, S.; Wilson, M. The molecular pathways underlying early gonadal development. J. Mol. Endocrinol. 2018, 62, R47–R64. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Ikeda, Y.; Parker, K.L. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 1994, 77, 481–490. [Google Scholar] [CrossRef]
- Sadovsky, Y.; Crawford, P.A.; Woodson, K.G.; Polish, J.A.; Clements, M.A.; Tourtellotte, L.M.; Simburger, K.; Milbrandt, J. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc. Natl. Acad. Sci. USA 1995, 92, 10939–10943. [Google Scholar] [CrossRef]
- Shinoda, K.; Lei, H.; Yoshii, H.; Nomura, M.; Nagano, M.; Shiba, H.; Sasaki, H.; Osawa, Y.; Ninomiya, Y.; Niwa, O.; et al. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev. Dyn. 1995, 204, 22–29. [Google Scholar] [CrossRef]
- Bashamboo, A.; Ferraz-de-Souza, B.; Lourenco, D.; Lin, L.; Sebire, N.J.; Montjean, D.; Bignon-Topalovic, J.; Mandelbaum, J.; Siffroi, J.P.; Christin-Maitre, S.; et al. Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am. J. Hum. Genet. 2010, 87, 505–512. [Google Scholar] [CrossRef]
- Laan, M.; Kasak, L.; Timinskas, K.; Grigorova, M.; Venclovas, C.; Renaux, A.; Lenaerts, T.; Punab, M. NR5A1 c.991-1G > C splice-site variant causes familial 46,XY partial gonadal dysgenesis with incomplete penetrance. Clin. Endocrinol. 2021, 94, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Na, X.; Mao, Y.; Tang, Y.; Jiang, W.; Yu, J.; Cao, L.; Yang, J. Identification and functional analysis of fourteen NR5A1 variants in patients with the 46 XY disorders of sex development. Gene 2020, 760, 145004. [Google Scholar] [CrossRef]
- Yu, B.; Liu, Z.; Gao, Y.; Mao, J.; Wang, X.; Hao, M.; Ma, W.; Huang, Q.; Zhang, R.; Nie, M.; et al. Novel NR5A1 mutations found in Chinese patients with 46, XY disorders of sex development. Clin. Endocrinol. 2018, 89, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Achermann, J.C.; Ito, M.; Ito, M.; Hindmarsh, P.C.; Jameson, J.L. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat. Genet. 1999, 22, 125–126. [Google Scholar] [CrossRef] [PubMed]
- Knarston, I.M.; Robevska, G.; van den Bergen, J.A.; Eggers, S.; Croft, B.; Yates, J.; Hersmus, R.; Looijenga, L.; Cameron, F.J.; Monhike, K.; et al. NR5A1 gene variants repress the ovarian-specific WNT signaling pathway in 46,XX disorders of sex development patients. Hum. Mutat. 2019, 40, 207–216. [Google Scholar] [CrossRef]
- Jaillard, S.; Sreenivasan, R.; Beaumont, M.; Robevska, G.; Dubourg, C.; Knarston, I.M.; Akloul, L.; van den Bergen, J.; Odent, S.; Croft, B.; et al. Analysis of NR5A1 in 142 patients with premature ovarian insufficiency, diminished ovarian reserve, or unexplained infertility. Maturitas 2020, 131, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.C.; Okumura, L.M.; Page, D.C. Gata4 is required for formation of the genital ridge in mice. PLoS Genet. 2013, 9, e1003629. [Google Scholar] [CrossRef]
- Tevosian, S.G.; Albrecht, K.H.; Crispino, J.D.; Fujiwara, Y.; Eicher, E.M.; Orkin, S.H. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development 2002, 129, 4627–4634. [Google Scholar] [CrossRef]
- Lourenco, D.; Brauner, R.; Rybczynska, M.; Nihoul-Fekete, C.; McElreavey, K.; Bashamboo, A. Loss-of-function mutation in GATA4 causes anomalies of human testicular development. Proc. Natl. Acad. Sci. USA 2011, 108, 1597–1602. [Google Scholar] [CrossRef]
- Hammes, A.; Guo, J.K.; Lutsch, G.; Leheste, J.R.; Landrock, D.; Ziegler, U.; Gubler, M.C.; Schedl, A. Two splice variants of the Wilms’ tumor 1 gene have distinct functions during sex determination and nephron formation. Cell 2001, 106, 319–329. [Google Scholar] [CrossRef]
- Barbaux, S.; Niaudet, P.; Gubler, M.C.; Grunfeld, J.P.; Jaubert, F.; Kuttenn, F.; Fekete, C.N.; Souleyreau-Therville, N.; Thibaud, E.; Fellous, M.; et al. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat. Genet. 1997, 17, 467–470. [Google Scholar] [CrossRef]
- Klamt, B.; Koziell, A.; Poulat, F.; Wieacker, P.; Scambler, P.; Berta, P.; Gessler, M. Frasier syndrome is caused by defective alternative splicing of WT1 leading to an altered ratio of WT1 +/-KTS splice isoforms. Hum. Mol. Genet. 1998, 7, 709–714. [Google Scholar] [CrossRef]
- Birk, O.S.; Casiano, D.E.; Wassif, C.A.; Cogliati, T.; Zhao, L.; Zhao, Y.; Grinberg, A.; Huang, S.; Kreidberg, J.A.; Parker, K.L.; et al. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature 2000, 403, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Kusaka, M.; Katoh-Fukui, Y.; Ogawa, H.; Miyabayashi, K.; Baba, T.; Shima, Y.; Sugiyama, N.; Sugimoto, Y.; Okuno, Y.; Kodama, R.; et al. Abnormal epithelial cell polarity and ectopic epidermal growth factor receptor (EGFR) expression induced in Emx2 KO embryonic gonads. Endocrinology 2010, 151, 5893–5904. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Tanaka, S.S.; Yamaguchi, Y.L.; Kobayashi, H.; Kuroki, S.; Tachibana, M.; Shinomura, M.; Kanai, Y.; Morohashi, K.; Kawakami, K.; et al. Homeoproteins Six1 and Six4 regulate male sex determination and mouse gonadal development. Dev. Cell 2013, 26, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Ross, A.; Stallings, N.; Parker, K.L.; Capel, B.; Quaggin, S.E. Disrupted gonadogenesis and male-to-female sex reversal in Pod1 knockout mice. Development 2004, 131, 4095–4105. [Google Scholar] [CrossRef]
- Katoh-Fukui, Y.; Owaki, A.; Toyama, Y.; Kusaka, M.; Shinohara, Y.; Maekawa, M.; Toshimori, K.; Morohashi, K. Mouse Polycomb M33 is required for splenic vascular and adrenal gland formation through regulating Ad4BP/SF1 expression. Blood 2005, 106, 1612–1620. [Google Scholar] [CrossRef]
- Katoh-Fukui, Y.; Tsuchiya, R.; Shiroishi, T.; Nakahara, Y.; Hashimoto, N.; Noguchi, K.; Higashinakagawa, T. Male-to-female sex reversal in M33 mutant mice. Nature 1998, 393, 688–692. [Google Scholar] [CrossRef]
- Efstratiadis, A. Genetics of mouse growth. Int. J. Dev. Biol. 1998, 42, 955–976. [Google Scholar]
- Schnabel, C.A.; Selleri, L.; Cleary, M.L. Pbx1 is essential for adrenal development and urogenital differentiation. Genesis 2003, 37, 123–130. [Google Scholar] [CrossRef]
- Eozenou, C.; Bashamboo, A.; Bignon-Topalovic, J.; Merel, T.; Zwermann, O.; Lourenco, D.; Lottmann, H.; Lichtenauer, U.; Rojo, S.; Beuschlein, F.; et al. The TALE homeodomain of PBX1 is involved in human primary testis-determination. Hum. Mutat. 2019, 40, 1071–1076. [Google Scholar] [CrossRef]
- Wang, Q.; Lan, Y.; Cho, E.S.; Maltby, K.M.; Jiang, R. Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Dev. Biol. 2005, 288, 582–594. [Google Scholar] [CrossRef]
- McLaren, A. Somatic and germ-cell sex in mammals. Philos. Trans. R. Soc. London B Biol. Sci. 1988, 322, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Wyndham, N.R. A morphological study of testicular descent. J. Anat. 1943, 77, 179–188. [Google Scholar] [PubMed]
- Hacker, A.; Capel, B.; Goodfellow, P.; Lovell-Badge, R. Expression of Sry, the mouse sex determining gene. Development 1995, 121, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Schmahl, J.; Eicher, E.M.; Washburn, L.L.; Capel, B. Sry induces cell proliferation in the mouse gonad. Development 2000, 127, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Veitia, R.A. FOXL2 versus SOX9: A lifelong “battle of the sexes”. Bioessays 2010, 32, 375–380. [Google Scholar] [CrossRef]
- Minkina, A.; Matson, C.K.; Lindeman, R.E.; Ghyselinck, N.B.; Bardwell, V.J.; Zarkower, D. DMRT1 protects male gonadal cells from retinoid-dependent sexual transdifferentiation. Dev. Cell 2014, 29, 511–520. [Google Scholar] [CrossRef]
- Palmer, M.S.; Sinclair, A.H.; Berta, P.; Ellis, N.A.; Goodfellow, P.N.; Abbas, N.E.; Fellous, M. Genetic evidence that ZFY is not the testis-determining factor. Nature 1989, 342, 937–939. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.M.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef]
- Bowles, J.; Schepers, G.; Koopman, P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000, 227, 239–255. [Google Scholar] [CrossRef]
- Zhao, L.; Ng, E.T.; Davidson, T.L.; Longmuss, E.; Urschitz, J.; Elston, M.; Moisyadi, S.; Bowles, J.; Koopman, P. Structure-function analysis of mouse Sry reveals dual essential roles of the C-terminal polyglutamine tract in sex determination. Proc. Natl. Acad. Sci. USA 2014, 111, 11768–11773. [Google Scholar] [CrossRef]
- Harley, V.R.; Layfield, S.; Mitchell, C.L.; Forwood, J.K.; John, A.P.; Briggs, L.J.; McDowall, S.G.; Jans, D.A. Defective importin beta recognition and nuclear import of the sex-determining factor SRY are associated with XY sex-reversing mutations. Proc. Natl. Acad. Sci. USA 2003, 100, 7045–7050. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, W.; Chan, G.; Jancso-Radek, A.; Liu, S.; Weiss, M.A. Human sex reversal due to impaired nuclear localization of SRY. A clinical correlation. J. Biol. Chem. 2001, 276, 46480–46484. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Phillips, N.B.; Jancso-Radek, A.; Ittah, V.; Singh, R.; Jones, D.N.; Haas, E.; Weiss, M.A. SRY-directed DNA bending and human sex reversal: Reassessment of a clinical mutation uncovers a global coupling between the HMG box and its tail. J. Mol. Biol. 2006, 360, 310–328. [Google Scholar] [CrossRef] [PubMed]
- Battiloro, E.; Angeletti, B.; Tozzi, M.C.; Bruni, L.; Tondini, S.; Vignetti, P.; Verna, R.; D’Ambrosio, E. A novel double nucleotide substitution in the HMG box of the SRY gene associated with Swyer syndrome. Hum. Genet. 1997, 100, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.R. Mutational analysis of SRY in XY females. Hum. Mutat. 1993, 2, 347–350. [Google Scholar] [CrossRef]
- Kurtz, S.; Lucas-Hahn, A.; Schlegelberger, B.; Gohring, G.; Niemann, H.; Mettenleiter, T.C.; Petersen, B. Knockout of the HMG domain of the porcine SRY gene causes sex reversal in gene-edited pigs. Proc. Natl. Acad. Sci. USA 2021, 118, e2008743118. [Google Scholar] [CrossRef]
- Bergstrom, D.E.; Young, M.; Albrecht, K.H.; Eicher, E.M. Related function of mouse SOX3, SOX9, and SRY HMG domains assayed by male sex determination. Genesis 2000, 28, 111–124. [Google Scholar] [CrossRef]
- Miyawaki, S.; Kuroki, S.; Maeda, R.; Okashita, N.; Koopman, P.; Tachibana, M. The mouse Sry locus harbors a cryptic exon that is essential for male sex determination. Science 2020, 370, 121–124. [Google Scholar] [CrossRef]
- Hanley, N.A.; Hagan, D.M.; Clement-Jones, M.; Ball, S.G.; Strachan, T.; Salas-Cortes, L.; McElreavey, K.; Lindsay, S.; Robson, S.; Bullen, P.; et al. SRY, SOX9, and DAX1 expression patterns during human sex determination and gonadal development. Mech. Dev. 2000, 91, 403–407. [Google Scholar] [CrossRef]
- Mamsen, L.S.; Ernst, E.H.; Borup, R.; Larsen, A.; Olesen, R.H.; Ernst, E.; Anderson, R.A.; Kristensen, S.G.; Andersen, C.Y. Temporal expression pattern of genes during the period of sex differentiation in human embryonic gonads. Sci. Rep. 2017, 7, 15961. [Google Scholar] [CrossRef]
- Parma, P.; Radi, O. Molecular mechanisms of sexual development. Sex. Dev. 2012, 6, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Warr, N.; Carre, G.A.; Siggers, P.; Faleato, J.V.; Brixey, R.; Pope, M.; Bogani, D.; Childers, M.; Wells, S.; Scudamore, C.L.; et al. Gadd45gamma and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev. Cell 2012, 23, 1020–1031. [Google Scholar] [CrossRef] [PubMed]
- Bullejos, M.; Koopman, P. Spatially dynamic expression of Sry in mouse genital ridges. Dev. Dyn. 2001, 221, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Sekido, R.; Lovell-Badge, R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008, 453, 930–934. [Google Scholar] [CrossRef] [PubMed]
- Sekido, R.; Bar, I.; Narvaez, V.; Penny, G.; Lovell-Badge, R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev. Biol. 2004, 274, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Gubbay, J.; Collignon, J.; Koopman, P.; Capel, B.; Economou, A.; Munsterberg, A.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 1990, 346, 245–250. [Google Scholar] [CrossRef]
- Larney, C.; Bailey, T.L.; Koopman, P. Switching on sex: Transcriptional regulation of the testis-determining gene Sry. Development 2014, 141, 2195–2205. [Google Scholar] [CrossRef]
- Zangen, D.; Kaufman, Y.; Banne, E.; Weinberg-Shukron, A.; Abulibdeh, A.; Garfinkel, B.P.; Dweik, D.; Kanaan, M.; Camats, N.; Fluck, C.; et al. Testicular differentiation factor SF-1 is required for human spleen development. J. Clin. Investig. 2014, 124, 2071–2075. [Google Scholar] [CrossRef] [PubMed]
- Chaboissier, M.C.; Kobayashi, A.; Vidal, V.I.; Lutzkendorf, S.; van de Kant, H.J.; Wegner, M.; de Rooij, D.G.; Behringer, R.R.; Schedl, A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 2004, 131, 1891–1901. [Google Scholar] [CrossRef]
- Vidal, V.P.; Chaboissier, M.C.; de Rooij, D.G.; Schedl, A. Sox9 induces testis development in XX transgenic mice. Nat. Genet. 2001, 28, 216–217. [Google Scholar] [CrossRef]
- Qin, Y.; Bishop, C.E. Sox9 is sufficient for functional testis development producing fertile male mice in the absence of Sry. Hum. Mol. Genet. 2005, 14, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Wang, S.; Ning, Y.; Lamb, A.N.; Bartley, J. Autosomal XX sex reversal caused by duplication of SOX9. Am. J. Med. Genet. 1999, 87, 349–353. [Google Scholar] [CrossRef]
- Steinhart, Z.; Angers, S. Wnt signaling in development and tissue homeostasis. Development 2018, 145, dev146589. [Google Scholar] [CrossRef] [PubMed]
- Chassot, A.A.; Bradford, S.T.; Auguste, A.; Gregoire, E.P.; Pailhoux, E.; de Rooij, D.G.; Schedl, A.; Chaboissier, M.C. WNT4 and RSPO1 together are required for cell proliferation in the early mouse gonad. Development 2012, 139, 4461–4472. [Google Scholar] [CrossRef]
- Jeays-Ward, K.; Dandonneau, M.; Swain, A. Wnt4 is required for proper male as well as female sexual development. Dev. Biol. 2004, 276, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Vainio, S.; Heikkila, M.; Kispert, A.; Chin, N.; McMahon, A.P. Female development in mammals is regulated by Wnt-4 signalling. Nature 1999, 397, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Philibert, P.; Biason-Lauber, A.; Gueorguieva, I.; Stuckens, C.; Pienkowski, C.; Lebon-Labich, B.; Paris, F.; Sultan, C. Molecular analysis of WNT4 gene in four adolescent girls with mullerian duct abnormality and hyperandrogenism (atypical Mayer-Rokitansky-Kuster-Hauser syndrome). Fertil. Steril. 2011, 95, 2683–2686. [Google Scholar] [CrossRef] [PubMed]
- Biason-Lauber, A.; Konrad, D.; Navratil, F.; Schoenle, E.J. A WNT4 mutation associated with Mullerian-duct regression and virilization in a 46,XX woman. N. Engl. J. Med. 2004, 351, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.; Sim, H.; Knower, K.; Vilain, E.; Harley, V. Human SRY inhibits beta-catenin-mediated transcription. Int. J. Biochem. Cell Biol. 2008, 40, 2889–2900. [Google Scholar] [CrossRef]
- Jaaskelainen, M.; Prunskaite-Hyyrylainen, R.; Naillat, F.; Parviainen, H.; Anttonen, M.; Heikinheimo, M.; Liakka, A.; Ola, R.; Vainio, S.; Vaskivuo, T.E.; et al. WNT4 is expressed in human fetal and adult ovaries and its signaling contributes to ovarian cell survival. Mol. Cell. Endocrinol. 2010, 317, 106–111. [Google Scholar] [CrossRef][Green Version]
- Mandel, H.; Shemer, R.; Borochowitz, Z.U.; Okopnik, M.; Knopf, C.; Indelman, M.; Drugan, A.; Tiosano, D.; Gershoni-Baruch, R.; Choder, M.; et al. SERKAL syndrome: An autosomal-recessive disorder caused by a loss-of-function mutation in WNT4. Am. J. Hum. Genet. 2008, 82, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Heikkila, M.; Prunskaite, R.; Naillat, F.; Itaranta, P.; Vuoristo, J.; Leppaluoto, J.; Peltoketo, H.; Vainio, S. The partial female to male sex reversal in Wnt-4-deficient females involves induced expression of testosterone biosynthetic genes and testosterone production, and depends on androgen action. Endocrinology 2005, 146, 4016–4023. [Google Scholar] [CrossRef] [PubMed]
- Jeays-Ward, K.; Hoyle, C.; Brennan, J.; Dandonneau, M.; Alldus, G.; Capel, B.; Swain, A. Endothelial and steroidogenic cell migration are regulated by WNT4 in the developing mammalian gonad. Development 2003, 130, 3663–3670. [Google Scholar] [CrossRef] [PubMed]
- Heikkila, M.; Peltoketo, H.; Leppaluoto, J.; Ilves, M.; Vuolteenaho, O.; Vainio, S. Wnt-4 deficiency alters mouse adrenal cortex function, reducing aldosterone production. Endocrinology 2002, 143, 4358–4365. [Google Scholar] [CrossRef]
- Coveney, D.; Ross, A.J.; Slone, J.D.; Capel, B. A microarray analysis of the XX Wnt4 mutant gonad targeted at the identification of genes involved in testis vascular differentiation. Gene Expr. Patterns 2007, 7, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Naillat, F.; Prunskaite-Hyyrylainen, R.; Pietila, I.; Sormunen, R.; Jokela, T.; Shan, J.; Vainio, S.J. Wnt4/5a signalling coordinates cell adhesion and entry into meiosis during presumptive ovarian follicle development. Hum. Mol. Genet. 2010, 19, 1539–1550. [Google Scholar] [CrossRef]
- Boyer, A.; Lapointe, E.; Zheng, X.; Cowan, R.G.; Li, H.; Quirk, S.M.; DeMayo, F.J.; Richards, J.S.; Boerboom, D. WNT4 is required for normal ovarian follicle development and female fertility. FASEB J. 2010, 24, 3010–3025. [Google Scholar] [CrossRef]
- Kamata, T.; Katsube, K.; Michikawa, M.; Yamada, M.; Takada, S.; Mizusawa, H. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim. Biophys. Acta 2004, 1676, 51–62. [Google Scholar] [CrossRef]
- Kazanskaya, O.; Glinka, A.; Del, B.B.I.; Stannek, P.; Niehrs, C.; Wu, W. R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 2004, 7, 525–534. [Google Scholar] [CrossRef]
- Yoon, J.K.; Lee, J.S. Cellular signaling and biological functions of R-spondins. Cell. Signal. 2012, 24, 369–377. [Google Scholar] [CrossRef]
- Parma, P.; Radi, O.; Vidal, V.; Chaboissier, M.C.; Dellambra, E.; Valentini, S.; Guerra, L.; Schedl, A.; Camerino, G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet. 2006, 38, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Chassot, A.A.; Ranc, F.; Gregoire, E.P.; Roepers-Gajadien, H.L.; Taketo, M.M.; Camerino, G.; de Rooij, D.G.; Schedl, A.; Chaboissier, M.C. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum. Mol. Genet. 2008, 17, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- Chassot, A.A.; Gregoire, E.P.; Magliano, M.; Lavery, R.; Chaboissier, M.C. Genetics of ovarian differentiation: Rspo1, a major player. Sex. Dev. 2008, 2, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, S.; Megiorni, F.; De Bernardo, C.; Felici, A.; Marrocco, G.; Maggiulli, G.; Grammatico, B.; Remotti, D.; Saccucci, P.; Valentini, F.; et al. Syndromic true hermaphroditism due to an R-spondin1 (RSPO1) homozygous mutation. Hum. Mutat. 2008, 29, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Buscara, L.; Montazer-Torbati, F.; Chadi, S.; Auguste, A.; Laubier, J.; Chassot, A.A.; Renault, L.; Passet, B.; Costa, J.; Pannetier, M.; et al. Goat RSPO1 over-expression rescues sex-reversal in Rspo1-knockout XX mice but does not perturb testis differentiation in XY or sex-reversed XX mice. Transgenic Res. 2009, 18, 649–654. [Google Scholar] [CrossRef]
- Naasse, Y.; Bakhchane, A.; Charoute, H.; Jennane, F.; Bignon-Topalovic, J.; Malki, A.; Bashamboo, A.; Barakat, A.; Rouba, H.; McElreavey, K. A Novel Homozygous Missense Mutation in the FU-CRD2 Domain of the R-spondin1 Gene Associated with Familial 46,XX DSD. Sex. Dev. 2017, 11, 269–274. [Google Scholar] [CrossRef]
- Tomizuka, K.; Horikoshi, K.; Kitada, R.; Sugawara, Y.; Iba, Y.; Kojima, A.; Yoshitome, A.; Yamawaki, K.; Amagai, M.; Inoue, A.; et al. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum. Mol. Genet. 2008, 17, 1278–1291. [Google Scholar] [CrossRef]
- Tomaselli, S.; Megiorni, F.; Lin, L.; Mazzilli, M.C.; Gerrelli, D.; Majore, S.; Grammatico, P.; Achermann, J.C. Human RSPO1/R-spondin1 is expressed during early ovary development and augments beta-catenin signaling. PLoS ONE 2011, 6, e16366. [Google Scholar] [CrossRef]
- Geng, A.; Wu, T.; Cai, C.; Song, W.; Wang, J.; Yu, Q.C.; Zeng, Y.A. A novel function of R-spondin1 in regulating estrogen receptor expression independent of Wnt/beta-catenin signaling. eLife 2020, 9, e56434. [Google Scholar] [CrossRef]
- Chassot, A.A.; Gregoire, E.P.; Lavery, R.; Taketo, M.M.; de Rooij, D.G.; Adams, I.R.; Chaboissier, M.C. RSPO1/beta-catenin signaling pathway regulates oogonia differentiation and entry into meiosis in the mouse fetal ovary. PLoS ONE 2011, 6, e25641. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, Y.; Xing, H.; Li, L.; Li, R.; Dai, J.; Li, Q.; Fang, S. The role of R-spondin 1 through activating Wnt/beta-catenin in the growth, survival and migration of ovarian cancer cells. Gene 2019, 689, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.; Ovitt, C.E.; Anlag, K.; Fehsenfeld, S.; Gredsted, L.; Treier, A.C.; Treier, M. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 2004, 131, 933–942. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Jakob, S.; Anlag, K.; Eisenberger, T.; Sekido, R.; Kress, J.; Treier, A.C.; Klugmann, C.; Klasen, C.; Holter, N.I.; et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 2009, 139, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
- Takasawa, K.; Kashimada, K.; Pelosi, E.; Takagi, M.; Morio, T.; Asahara, H.; Schlessinger, D.; Mizutani, S.; Koopman, P. FOXL2 transcriptionally represses Sf1 expression by antagonizing WT1 during ovarian development in mice. FASEB J. 2014, 28, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Boulanger, L.; Pannetier, M.; Gall, L.; Allais-Bonnet, A.; Elzaiat, M.; Le Bourhis, D.; Daniel, N.; Richard, C.; Cotinot, C.; Ghyselinck, N.B.; et al. FOXL2 is a female sex-determining gene in the goat. Curr. Biol. 2014, 24, 404–408. [Google Scholar] [CrossRef]
- Ottolenghi, C.; Pelosi, E.; Tran, J.; Colombino, M.; Douglass, E.; Nedorezov, T.; Cao, A.; Forabosco, A.; Schlessinger, D. Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Hum. Mol. Genet. 2007, 16, 2795–2804. [Google Scholar] [CrossRef]
- Auguste, A.; Chassot, A.A.; Gregoire, E.P.; Renault, L.; Pannetier, M.; Treier, M.; Pailhoux, E.; Chaboissier, M.C. Loss of R-spondin1 and Foxl2 amplifies female-to-male sex reversal in XX mice. Sex. Dev. 2011, 5, 304–317. [Google Scholar] [CrossRef]
- Ghochani, Y.; Saini, J.K.; Mellon, P.L.; Thackray, V.G. FOXL2 is involved in the synergy between activin and progestins on the follicle-stimulating hormone beta-subunit promoter. Endocrinology 2012, 153, 2023–2033. [Google Scholar] [CrossRef]
- Corpuz, P.S.; Lindaman, L.L.; Mellon, P.L.; Coss, D. FoxL2 Is required for activin induction of the mouse and human follicle-stimulating hormone beta-subunit genes. Mol. Endocrinol. 2010, 24, 1037–1051. [Google Scholar] [CrossRef]
- Herndon, M.K.; Nilson, J.H. Maximal expression of Foxl2 in pituitary gonadotropes requires ovarian hormones. PLoS ONE 2015, 10, e126527. [Google Scholar] [CrossRef]
- Eozenou, C.; Lesage-Padilla, A.; Mauffre, V.; Healey, G.D.; Camous, S.; Bolifraud, P.; Giraud-Delville, C.; Vaiman, D.; Shimizu, T.; Miyamoto, A.; et al. FOXL2 is a Progesterone Target Gene in the Endometrium of Ruminants. Int. J. Mol. Sci. 2020, 21, 1478. [Google Scholar] [CrossRef] [PubMed]
- Pisarska, M.D.; Bae, J.; Klein, C.; Hsueh, A.J. Forkhead l2 is expressed in the ovary and represses the promoter activity of the steroidogenic acute regulatory gene. Endocrinology 2004, 145, 3424–3433. [Google Scholar] [CrossRef] [PubMed]
- Georges, A.; L’Hote, D.; Todeschini, A.L.; Auguste, A.; Legois, B.; Zider, A.; Veitia, R.A. The transcription factor FOXL2 mobilizes estrogen signaling to maintain the identity of ovarian granulosa cells. eLife 2014, 3, e04207. [Google Scholar] [CrossRef] [PubMed]
- Gustin, S.E.; Hogg, K.; Stringer, J.M.; Rastetter, R.H.; Pelosi, E.; Miles, D.C.; Sinclair, A.H.; Wilhelm, D.; Western, P.S. WNT/beta-catenin and p27/FOXL2 differentially regulate supporting cell proliferation in the developing ovary. Dev. Biol. 2016, 412, 250–260. [Google Scholar] [CrossRef]
- Bellessort, B.; Bachelot, A.; Heude, E.; Alfama, G.; Fontaine, A.; Le Cardinal, M.; Treier, M.; Levi, G. Role of Foxl2 in uterine maturation and function. Hum. Mol. Genet. 2015, 24, 3092–3103. [Google Scholar] [CrossRef]
- Crisponi, L.; Deiana, M.; Loi, A.; Chiappe, F.; Uda, M.; Amati, P.; Bisceglia, L.; Zelante, L.; Nagaraja, R.; Porcu, S.; et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat. Genet. 2001, 27, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Moumne, L.; Dipietromaria, A.; Batista, F.; Kocer, A.; Fellous, M.; Pailhoux, E.; Veitia, R.A. Differential aggregation and functional impairment induced by polyalanine expansions in FOXL2, a transcription factor involved in cranio-facial and ovarian development. Hum. Mol. Genet. 2008, 17, 1010–1019. [Google Scholar] [CrossRef]
- Dipietromaria, A.; Benayoun, B.A.; Todeschini, A.L.; Rivals, I.; Bazin, C.; Veitia, R.A. Towards a functional classification of pathogenic FOXL2 mutations using transactivation reporter systems. Hum. Mol. Genet. 2009, 18, 3324–3333. [Google Scholar] [CrossRef][Green Version]
- Shah, S.P.; Kobel, M.; Senz, J.; Morin, R.D.; Clarke, B.A.; Wiegand, K.C.; Leung, G.; Zayed, A.; Mehl, E.; Kalloger, S.E.; et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009, 360, 2719–2729. [Google Scholar] [CrossRef]
- Lima, J.F.; Jin, L.; de Araujo, A.R.; Erikson-Johnson, M.R.; Oliveira, A.M.; Sebo, T.J.; Keeney, G.L.; Medeiros, F. FOXL2 mutations in granulosa cell tumors occurring in males. Arch. Pathol. Lab. Med. 2012, 136, 825–828. [Google Scholar] [CrossRef]
- Hes, O.; Vanecek, T.; Petersson, F.; Grossmann, P.; Hora, M.; Perez, M.D.; Steiner, P.; Dvorak, M.; Michal, M. Mutational analysis (c.402C>G) of the FOXL2 gene and immunohistochemical expression of the FOXL2 protein in testicular adult type granulosa cell tumors and incompletely differentiated sex cord stromal tumors. Appl. Immunohistochem. Mol. Morphol. 2011, 19, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Karnezis, A.N.; Wang, Y.; Keul, J.; Tessier-Cloutier, B.; Magrill, J.; Kommoss, S.; Senz, J.; Yang, W.; Proctor, L.; Schmidt, D.; et al. DICER1 and FOXL2 Mutation Status Correlates With Clinicopathologic Features in Ovarian Sertoli-Leydig Cell Tumors. Am. J. Surg. Pathol. 2019, 43, 628–638. [Google Scholar] [CrossRef]
- Al-Agha, O.M.; Huwait, H.F.; Chow, C.; Yang, W.; Senz, J.; Kalloger, S.E.; Huntsman, D.G.; Young, R.H.; Gilks, C.B. FOXL2 is a sensitive and specific marker for sex cord-stromal tumors of the ovary. Am. J. Surg. Pathol. 2011, 35, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Gunes, S.O.; Metin, M.A.; Agarwal, A. Genetic and epigenetic effects in sex determination. Birth Defects Res. Part C Embryo Today Rev. 2016, 108, 321–336. [Google Scholar] [CrossRef]
- Kuroki, S.; Matoba, S.; Akiyoshi, M.; Matsumura, Y.; Miyachi, H.; Mise, N.; Abe, K.; Ogura, A.; Wilhelm, D.; Koopman, P.; et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science 2013, 341, 1106–1109. [Google Scholar] [CrossRef] [PubMed]
- Shendy, N.; Broadhurst, A.L.; Shoemaker, K.; Read, R.; Abell, A.N. MAP3K4 kinase activity dependent control of mouse gonadal sex determinationdagger. Biol. Reprod. 2021, 105, 491–502. [Google Scholar] [CrossRef]
- Johnen, H.; Gonzalez-Silva, L.; Carramolino, L.; Flores, J.M.; Torres, M.; Salvador, J.M. Gadd45g is essential for primary sex determination, male fertility and testis development. PLoS ONE 2013, 8, e58751. [Google Scholar] [CrossRef]
- Wen, Y.; Ma, X.; Wang, X.; Wang, F.; Dong, J.; Wu, Y.; Lv, C.; Liu, K.; Zhang, Y.; Zhang, Z.; et al. hnRNPU in Sertoli cells cooperates with WT1 and is essential for testicular development by modulating transcriptional factors Sox8/9. Theranostics 2021, 11, 10030–10046. [Google Scholar] [CrossRef] [PubMed]
- Josso, N.; Belville, C.; di Clemente, N.; Picard, J.Y. AMH and AMH receptor defects in persistent Mullerian duct syndrome. Hum. Reprod. Update 2005, 11, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Ozisik, G.; Achermann, J.C.; Meeks, J.J.; Jameson, J.L. SF1 in the development of the adrenal gland and gonads. Horm. Res. 2003, 59 (Suppl. S1), 94–98. [Google Scholar] [CrossRef] [PubMed]
- Manuylov, N.L.; Fujiwara, Y.; Adameyko, I.I.; Poulat, F.; Tevosian, S.G. The regulation of Sox9 gene expression by the GATA4/FOG2 transcriptional complex in dominant XX sex reversal mouse models. Dev. Biol. 2007, 307, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, D.; Hiramatsu, R.; Mizusaki, H.; Widjaja, L.; Combes, A.N.; Kanai, Y.; Koopman, P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J. Biol. Chem. 2007, 282, 10553–10560. [Google Scholar] [CrossRef] [PubMed]
- Rossitto, M.; Ujjan, S.; Poulat, F.; Boizet-Bonhoure, B. Multiple roles of the prostaglandin D2 signaling pathway in reproduction. Reproduction 2015, 149, R49–R58. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, L.; Cui, X.; Lin, X.; Li, Y.; Wang, Y.; Wang, Y.; Qin, Y.; Chen, D.; Han, C.; et al. Wt1 directs the lineage specification of sertoli and granulosa cells by repressing Sf1 expression. Development 2017, 144, 44–53. [Google Scholar] [CrossRef]
- Gomes, N.L.; de Paula, L.; Silva, J.M.; Silva, T.E.; Lerario, A.M.; Nishi, M.Y.; Batista, R.L.; Faria, J.J.; Moraes, D.; Costa, E.; et al. A 46,XX testicular disorder of sex development caused by a Wilms’ tumour Factor-1 (WT1) pathogenic variant. Clin. Genet. 2019, 95, 172–176. [Google Scholar] [CrossRef]
- Eozenou, C.; Gonen, N.; Touzon, M.S.; Jorgensen, A.; Yatsenko, S.A.; Fusee, L.; Kamel, A.K.; Gellen, B.; Guercio, G.; Singh, P.; et al. Testis formation in XX individuals resulting from novel pathogenic variants in Wilms’ tumor 1 (WT1) gene. Proc. Natl. Acad. Sci. USA 2020, 117, 13680–13688. [Google Scholar] [CrossRef]
- Bhandari, R.K.; Sadler-Riggleman, I.; Clement, T.M.; Skinner, M.K. Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex determining gene SRY. PLoS ONE 2011, 6, e19935. [Google Scholar] [CrossRef]
- Katoh-Fukui, Y.; Miyabayashi, K.; Komatsu, T.; Owaki, A.; Baba, T.; Shima, Y.; Kidokoro, T.; Kanai, Y.; Schedl, A.; Wilhelm, D.; et al. Cbx2, a polycomb group gene, is required for Sry gene expression in mice. Endocrinology 2012, 153, 913–924. [Google Scholar] [CrossRef]
- Pitetti, J.L.; Calvel, P.; Romero, Y.; Conne, B.; Truong, V.; Papaioannou, M.D.; Schaad, O.; Docquier, M.; Herrera, P.L.; Wilhelm, D.; et al. Insulin and IGF1 receptors are essential for XX and XY gonadal differentiation and adrenal development in mice. PLoS Genet. 2013, 9, e1003160. [Google Scholar] [CrossRef]
- Jorgez, C.J.; Seth, A.; Wilken, N.; Bournat, J.C.; Chen, C.H.; Lamb, D.J. E2F1 regulates testicular descent and controls spermatogenesis by influencing WNT4 signaling. Development 2021, 148, dev191189. [Google Scholar] [CrossRef]
- Jameson, S.A.; Lin, Y.T.; Capel, B. Testis development requires the repression of Wnt4 by Fgf signaling. Dev. Biol. 2012, 370, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Bagheri-Fam, S.; Bird, A.D.; Zhao, L.; Ryan, J.M.; Yong, M.; Wilhelm, D.; Koopman, P.; Eswarakumar, V.P.; Harley, V.R. Testis Determination Requires a Specific FGFR2 Isoform to Repress FOXL2. Endocrinology 2017, 158, 3832–3843. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.H.; Matzuk, M.M.; Jorgez, C.J.; Menke, D.B.; Page, D.C.; Swain, A.; Capel, B. Follistatin operates downstream of Wnt4 in mammalian ovary organogenesis. Dev. Dyn. 2004, 230, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Jordan, B.K.; Mohammed, M.; Ching, S.T.; Delot, E.; Chen, X.N.; Dewing, P.; Swain, A.; Rao, P.N.; Elejalde, B.R.; Vilain, E. Up-regulation of WNT-4 signaling and dosage-sensitive sex reversal in humans. Am. J. Hum. Genet. 2001, 68, 1102–1109. [Google Scholar] [CrossRef]
- Kashimada, K.; Pelosi, E.; Chen, H.; Schlessinger, D.; Wilhelm, D.; Koopman, P. FOXL2 and BMP2 act cooperatively to regulate follistatin gene expression during ovarian development. Endocrinology 2011, 152, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, R.E.; Gearhart, M.D.; Minkina, A.; Krentz, A.D.; Bardwell, V.J.; Zarkower, D. Sexual cell-fate reprogramming in the ovary by DMRT1. Curr. Biol. 2015, 25, 764–771. [Google Scholar] [CrossRef]
- Lawson, K.A.; Hage, W.J. Clonal analysis of the origin of primordial germ cells in the mouse. Germline Dev. 1994, 165, 68–84. [Google Scholar] [CrossRef]
- Ginsburg, M.; Snow, M.H.; McLaren, A. Primordial germ cells in the mouse embryo during gastrulation. Development 1990, 110, 521–528. [Google Scholar] [CrossRef]
- Lawson, K.A.; Dunn, N.R.; Roelen, B.A.; Zeinstra, L.M.; Davis, A.M.; Wright, C.V.; Korving, J.P.; Hogan, B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999, 13, 424–436. [Google Scholar] [CrossRef]
- Ying, Y.; Qi, X.; Zhao, G.Q. Induction of primordial germ cells from murine epiblasts by synergistic action of BMP4 and BMP8B signaling pathways. Proc. Natl. Acad. Sci. USA 2001, 98, 7858–7862. [Google Scholar] [CrossRef]
- Ying, Y.; Zhao, G.Q. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev. Biol. 2001, 232, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Avilion, A.A.; Nicolis, S.K.; Pevny, L.H.; Perez, L.; Vivian, N.; Lovell-Badge, R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003, 17, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Campolo, F.; Gori, M.; Favaro, R.; Nicolis, S.; Pellegrini, M.; Botti, F.; Rossi, P.; Jannini, E.A.; Dolci, S. Essential role of Sox2 for the establishment and maintenance of the germ cell line. Stem Cells 2013, 31, 1408–1421. [Google Scholar] [CrossRef]
- Irie, N.; Weinberger, L.; Tang, W.W.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef]
- Guo, F.; Yan, L.; Guo, H.; Li, L.; Hu, B.; Zhao, Y.; Yong, J.; Hu, Y.; Wang, X.; Wei, Y.; et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell 2015, 161, 1437–1452. [Google Scholar] [CrossRef]
- Zuo, Q.; Jin, J.; Jin, K.; Zhou, J.; Sun, C.; Song, J.; Chen, G.; Zhang, Y.; Li, B. P53 and H3K4me2 activate N6-methylated LncPGCAT-1 to regulate primordial germ cell formation via MAPK signaling. J. Cell. Physiol. 2020, 235, 9895–9909. [Google Scholar] [CrossRef]
- Soto, D.A.; Ross, P.J. Similarities between bovine and human germline development revealed by single-cell RNA sequencing. Reproduction 2021, 161, 239–253. [Google Scholar] [CrossRef]
- Ohinata, Y.; Ohta, H.; Shigeta, M.; Yamanaka, K.; Wakayama, T.; Saitou, M. A signaling principle for the specification of the germ cell lineage in mice. Cell 2009, 137, 571–584. [Google Scholar] [CrossRef]
- Ohinata, Y.; Payer, B.; O’Carroll, D.; Ancelin, K.; Ono, Y.; Sano, M.; Barton, S.C.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436, 207–213. [Google Scholar] [CrossRef]
- Mochizuki, K.; Hayashi, Y.; Sekinaka, T.; Otsuka, K.; Ito-Matsuoka, Y.; Kobayashi, H.; Oki, S.; Takehara, A.; Kono, T.; Osumi, N.; et al. Repression of Somatic Genes by Selective Recruitment of HDAC3 by BLIMP1 Is Essential for Mouse Primordial Germ Cell Fate Determination. Cell Rep. 2018, 24, 2682–2693. [Google Scholar] [CrossRef]
- Yamaguchi, Y.L.; Tanaka, S.S.; Kumagai, M.; Fujimoto, Y.; Terabayashi, T.; Matsui, Y.; Nishinakamura, R. Sall4 is essential for mouse primordial germ cell specification by suppressing somatic cell program genes. Stem Cells 2015, 33, 289–300. [Google Scholar] [CrossRef] [PubMed]
- West, J.A.; Viswanathan, S.R.; Yabuuchi, A.; Cunniff, K.; Takeuchi, A.; Park, I.H.; Sero, J.E.; Zhu, H.; Perez-Atayde, A.; Frazier, A.L.; et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 2009, 460, 909–913. [Google Scholar] [CrossRef]
- Yamaji, M.; Seki, Y.; Kurimoto, K.; Yabuta, Y.; Yuasa, M.; Shigeta, M.; Yamanaka, K.; Ohinata, Y.; Saitou, M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008, 40, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Sybirna, A.; Tang, W.; Pierson, S.M.; Dietmann, S.; Gruhn, W.H.; Brosh, R.; Surani, M.A. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat. Commun. 2020, 11, 1282. [Google Scholar] [CrossRef]
- Aramaki, S.; Hayashi, K.; Kurimoto, K.; Ohta, H.; Yabuta, Y.; Iwanari, H.; Mochizuki, Y.; Hamakubo, T.; Kato, Y.; Shirahige, K.; et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell 2013, 27, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Eckert, D.; Nettersheim, D.; Gillis, A.J.; Schafer, S.; Kuckenberg, P.; Ehlermann, J.; Werling, U.; Biermann, K.; Looijenga, L.H.; et al. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol. Reprod. 2010, 82, 214–223. [Google Scholar] [CrossRef]
- Tremblay, K.D.; Dunn, N.R.; Robertson, E.J. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 2001, 128, 3609–3621. [Google Scholar] [CrossRef]
- Chang, H.; Matzuk, M.M. Smad5 is required for mouse primordial germ cell development. Mech. Dev. 2001, 104, 61–67. [Google Scholar] [CrossRef]
- Mochizuki, K.; Tando, Y.; Sekinaka, T.; Otsuka, K.; Hayashi, Y.; Kobayashi, H.; Kamio, A.; Ito-Matsuoka, Y.; Takehara, A.; Kono, T.; et al. SETDB1 is essential for mouse primordial germ cell fate determination by ensuring BMP signaling. Development 2018, 145, dev164160. [Google Scholar] [CrossRef]
- Nady, N.; Gupta, A.; Ma, Z.; Swigut, T.; Koide, A.; Koide, S.; Wysocka, J. ETO family protein Mtgr1 mediates Prdm14 functions in stem cell maintenance and primordial germ cell formation. eLife 2015, 4, e10150. [Google Scholar] [CrossRef]
- Okamura, E.; Tam, O.H.; Posfai, E.; Li, L.; Cockburn, K.; Lee, C.; Garner, J.; Rossant, J. Esrrb function is required for proper primordial germ cell development in presomite stage mouse embryos. Dev. Biol. 2019, 455, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Angulo, B.; Xia, N.; Sukhwani, M.; Wang, Z.; Carey, C.C.; Mazurie, A.; Cui, J.; Wilkinson, R.; Wiedenheft, B.; et al. A PAX5-OCT4-PRDM1 developmental switch specifies human primordial germ cells. Nat. Cell Biol. 2018, 20, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Jin, K.; Song, J.; Zhang, Y.; Chen, G.; Li, B. Interaction of the primordial germ cell-specific protein C2EIP with PTCH2 directs differentiation of embryonic stem cells via HH signaling activation. Cell Death Dis. 2018, 9, 497. [Google Scholar] [CrossRef] [PubMed]
- De Felici, M.; McLaren, A. Isolation of mouse primordial germ cells. Exp. Cell Res. 1982, 142, 476–482. [Google Scholar] [CrossRef]
- Kuhholzer, B.; Baguisi, A.; Overstrom, E.W. Long-term culture and characterization of goat primordial germ cells. Theriogenology 2000, 53, 1071–1079. [Google Scholar] [CrossRef]
- Kakegawa, R.; Teramura, T.; Takehara, T.; Anzai, M.; Mitani, T.; Matsumoto, K.; Saeki, K.; Sagawa, N.; Fukuda, K.; Hosoi, Y. Isolation and culture of rabbit primordial germ cells. J. Reprod. Dev. 2008, 54, 352–357. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, C.X.; Wang, W.L.; Zhao, R.Y.; Wang, H.T.; Liu, Y.Y.; Wang, S.Y.; Zhou, H.M. Isolation, culture, and characterization of primordial germ cells in Mongolian sheep. Vitr. Cell. Dev. Biol.-Anim. 2014, 50, 207–213. [Google Scholar] [CrossRef]
- Tilgner, K.; Atkinson, S.P.; Golebiewska, A.; Stojkovic, M.; Lako, M.; Armstrong, L. Isolation of primordial germ cells from differentiating human embryonic stem cells. Stem Cells 2008, 26, 3075–3085. [Google Scholar] [CrossRef]
- Costa, J.; Souza, G.B.; Soares, M.; Ribeiro, R.P.; van den Hurk, R.; Silva, J. In vitro differentiation of primordial germ cells and oocyte-like cells from stem cells. Histol. Histopathol. 2018, 33, 121–132. [Google Scholar] [CrossRef]
- Murase, Y.; Yabuta, Y.; Ohta, H.; Yamashiro, C.; Nakamura, T.; Yamamoto, T.; Saitou, M. Long-term expansion with germline potential of human primordial germ cell-like cells in vitro. EMBO J. 2020, 39, e104929. [Google Scholar] [CrossRef]
- Morohaku, K.; Tanimoto, R.; Sasaki, K.; Kawahara-Miki, R.; Kono, T.; Hayashi, K.; Hirao, Y.; Obata, Y. Complete in vitro generation of fertile oocytes from mouse primordial germ cells. Proc. Natl. Acad. Sci. USA 2016, 113, 9021–9026. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Ogushi, S.; Kurimoto, K.; Shimamoto, S.; Ohta, H.; Saitou, M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science 2012, 338, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Liu, W.; Zimmerman, J.; Pastor, W.A.; Kim, R.; Hosohama, L.; Ho, J.; Aslanyan, M.; Gell, J.J.; Jacobsen, S.E.; et al. The TFAP2C-Regulated OCT4 Naive Enhancer Is Involved in Human Germline Formation. Cell Rep. 2018, 25, 3591–3602. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Sun, N.; Hou, L.; Kim, R.; Faith, J.; Aslanyan, M.; Tao, Y.; Zheng, Y.; Fu, J.; Liu, W.; et al. Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell Rep. 2019, 29, 4568–4582. [Google Scholar] [CrossRef]
- Ghasemi, H.H.; Sobhani, A.; Nazm, B.M. Multipotent SSEA1 Positive Cells Population Differentiation into Primordial Germ Cells and Subsequently Progress into Oocyte-like Cells. Arch. Iran. Med. 2015, 18, 404–410. [Google Scholar]
- Mall, E.M.; Lecanda, A.; Drexler, H.; Raz, E.; Scholer, H.R.; Schlatt, S. Heading towards a dead end: The role of DND1 in germ line differentiation of human iPSCs. PLoS ONE 2021, 16, e258427. [Google Scholar] [CrossRef]
- Pierson, S.M.; Sybirna, A.; Wong, F.; Surani, M.A. Testing the role of SOX15 in human primordial germ cell fate. Wellcome Open Res. 2019, 4, 122. [Google Scholar] [CrossRef]
- Abdyyev, V.K.; Sant, D.W.; Kiseleva, E.V.; Spangenberg, V.E.; Kolomiets, O.L.; Andrade, N.S.; Dashinimaev, E.B.; Vorotelyak, E.A.; Vasiliev, A.V. In vitro derived female hPGCLCs are unable to complete meiosis in embryoid bodies. Exp. Cell Res. 2020, 397, 112358. [Google Scholar] [CrossRef]
- West, F.D.; Machacek, D.W.; Boyd, N.L.; Pandiyan, K.; Robbins, K.R.; Stice, S.L. Enrichment and differentiation of human germ-like cells mediated by feeder cells and basic fibroblast growth factor signaling. Stem Cells 2008, 26, 2768–2776. [Google Scholar] [CrossRef]
- Park, T.S.; Galic, Z.; Conway, A.E.; Lindgren, A.; van Handel, B.J.; Magnusson, M.; Richter, L.; Teitell, M.A.; Mikkola, H.K.; Lowry, W.E.; et al. Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells. Stem Cells 2009, 27, 783–795. [Google Scholar] [CrossRef]
- Geijsen, N.; Horoschak, M.; Kim, K.; Gribnau, J.; Eggan, K.; Daley, G.Q. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 2004, 427, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Wang, J.J.; Cheng, S.F.; Ge, W.; Sun, Y.C.; Sun, X.F.; Sun, R.; Li, L.; Li, B.; Shen, W. Retinoic acid promotes the proliferation of primordial germ cell-like cells differentiated from mouse skin-derived stem cells in vitro. Theriogenology 2016, 85, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Zhai, K.; Chang, Y.; Yao, G.; He, J.; Wang, F.; Kong, H.; Xin, H.; Wang, H.; Jin, M.; et al. CHIR99021 combined with retinoic acid promotes the differentiation of primordial germ cells from human embryonic stem cells. Oncotarget 2017, 8, 7814–7826. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, M.Y.; Tian, Y.; Zhang, S.E.; Yan, H.C.; Ge, W.; Han, B.Q.; Yan, Z.H.; Cheng, S.F.; Shen, W. The proliferation role of LH on porcine primordial germ cell-like cells (pPGCLCs) through ceRNA network construction. Clin. Transl. Med. 2021, 11, e560. [Google Scholar] [CrossRef] [PubMed]
- Mahboudi, S.; Parivar, K.; Mazaheri, Z.; Irani, S.H. Mir-106b Cluster Regulates Primordial Germ Cells Differentiation from Human Mesenchymal Stem Cells. Cell J. 2021, 23, 294–302. [Google Scholar] [CrossRef]
- Xing, M.; Wang, N.; Zeng, H.; Zhang, J. alpha-ketoglutarate promotes the specialization of primordial germ cell-like cells through regulating epigenetic reprogramming. J. Biomed. Res. 2020, 35, 36–46. [Google Scholar] [CrossRef]
- Ando, Y.; Okeyo, K.O.; Adachi, T. Modulation of adhesion microenvironment using mesh substrates triggers self-organization and primordial germ cell-like differentiation in mouse ES cells. APL Bioeng. 2019, 3, 16102. [Google Scholar] [CrossRef]
- Ooi, S.; Jiang, H.; Kang, Y.; Allard, P. Examining the Developmental Trajectory of an in Vitro Model of Mouse Primordial Germ Cells following Exposure to Environmentally Relevant Bisphenol A Levels. Environ. Health Perspect. 2021, 129, 97013. [Google Scholar] [CrossRef]
- Nagamatsu, G.; Saito, S.; Takubo, K.; Suda, T. Integrative Analysis of the Acquisition of Pluripotency in PGCs Reveals the Mutually Exclusive Roles of Blimp-1 and AKT Signaling. Stem Cell Rep. 2015, 5, 111–124. [Google Scholar] [CrossRef]
- Takehara, A.; Matsui, Y. Shortened G1 phase of cell cycle and decreased histone H3K27 methylation are associated with AKT-induced enhancement of primordial germ cell reprogramming. Dev. Growth Differ. 2019, 61, 357–364. [Google Scholar] [CrossRef]
- Oliveros-Etter, M.; Li, Z.; Nee, K.; Hosohama, L.; Hargan-Calvopina, J.; Lee, S.A.; Joti, P.; Yu, J.; Clark, A.T. PGC Reversion to Pluripotency Involves Erasure of DNA Methylation from Imprinting Control Centers followed by Locus-Specific Re-methylation. Stem Cell Rep. 2015, 5, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Bazley, F.A.; Liu, C.F.; Yuan, X.; Hao, H.; All, A.H.; De Los, A.A.; Zambidis, E.T.; Gearhart, J.D.; Kerr, C.L. Direct Reprogramming of Human Primordial Germ Cells into Induced Pluripotent Stem Cells: Efficient Generation of Genetically Engineered Germ Cells. Stem Cells Dev. 2015, 24, 2634–2648. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, V.; Salehi, M.; Nourozian, M.; Fadaei, F.; Farahani, R.M.; Piryaei, A.; Delbari, A. The ability of mouse nuclear transfer embryonic stem cells to differentiate into primordial germ cells. Genet. Mol. Biol. 2015, 38, 220–226. [Google Scholar] [CrossRef][Green Version]
- Makiyan, Z. Endometriosis origin from primordial germ cells. Organogenesis 2017, 13, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Molyneaux, K.; Wylie, C. Primordial germ cell migration. Int. J. Dev. Biol. 2004, 48, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Richardson, B.E.; Lehmann, R. Mechanisms guiding primordial germ cell migration: Strategies from different organisms. Nat. Rev. Mol. Cell Biol. 2010, 11, 37–49. [Google Scholar] [CrossRef]
- De Felici, M. The Formation and Migration of Primordial Germ Cells in Mouse and Man. In Molecular Mechanisms of Cell Differentiation in Gonad Development; Springer: Cham, Switzerland, 2016; Volume 58, pp. 23–46. [Google Scholar] [CrossRef]
- Lange, U.C.; Adams, D.J.; Lee, C.; Barton, S.; Schneider, R.; Bradley, A.; Surani, M.A. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol. Cell. Biol. 2008, 28, 4688–4696. [Google Scholar] [CrossRef]
- Sun, J.; Ting, M.C.; Ishii, M.; Maxson, R. Msx1 and Msx2 function together in the regulation of primordial germ cell migration in the mouse. Dev. Biol. 2016, 417, 11–24. [Google Scholar] [CrossRef]
- Laird, D.J.; Altshuler-Keylin, S.; Kissner, M.D.; Zhou, X.; Anderson, K.V. Ror2 enhances polarity and directional migration of primordial germ cells. PLoS Genet. 2011, 7, e1002428. [Google Scholar] [CrossRef]
- Park, E.; Lee, B.; Clurman, B.E.; Lee, K. NUP50 is necessary for the survival of primordial germ cells in mouse embryos. Reproduction 2016, 151, 51–58. [Google Scholar] [CrossRef]
- Senft, A.D.; Bikoff, E.K.; Robertson, E.J.; Costello, I. Genetic dissection of Nodal and Bmp signalling requirements during primordial germ cell development in mouse. Nat. Commun. 2019, 10, 1089. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, K.; Mukumoto, Y.; Tanigawa, Y.; Komiya, T. Xenopus Vasa Homolog XVLG1 is Essential for Migration and Survival of Primordial Germ Cells. Zool. Sci. 2017, 34, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Lejong, M.; Choa-Duterre, M.; Vanmuylder, N.; Louryan, S. Geldanamycin administration reduces the amount of primordial germ cells in the mouse embryo. Morphologie 2018, 102, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, C.; Hirota, T.; Kurimoto, K.; Nakamura, T.; Yabuta, Y.; Nagaoka, S.I.; Ohta, H.; Yamamoto, T.; Saitou, M. Persistent Requirement and Alteration of the Key Targets of PRDM1 During Primordial Germ Cell Development in Mice. Biol. Reprod. 2016, 94, 7. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, J.; Seppala, M.; Cobourne, M.T.; Kim, S.H. Ptch2/Gas1 and Ptch1/Boc differentially regulate Hedgehog signalling in murine primordial germ cell migration. Nat. Commun. 2020, 11, 1994. [Google Scholar] [CrossRef]
- Mallol, A.; Guirola, M.; Payer, B. PRDM14 controls X-chromosomal and global epigenetic reprogramming of H3K27me3 in migrating mouse primordial germ cells. Epigenetics Chromatin 2019, 12, 38. [Google Scholar] [CrossRef]
- Hoyer, P.E.; Byskov, A.G.; Mollgard, K. Stem cell factor and c-Kit in human primordial germ cells and fetal ovaries. Mol. Cell. Endocrinol. 2005, 234, 1–10. [Google Scholar] [CrossRef]
- Mollgard, K.; Jespersen, A.; Lutterodt, M.C.; Yding, A.C.; Hoyer, P.E.; Byskov, A.G. Human primordial germ cells migrate along nerve fibers and Schwann cells from the dorsal hind gut mesentery to the gonadal ridge. Mol. Hum. Reprod. 2010, 16, 621–631. [Google Scholar] [CrossRef]
- Wolff, E.; Suplicki, M.M.; Behr, R. Primordial germ cells do not migrate along nerve fibres in marmoset monkey and mouse embryos. Reproduction 2019, 157, 101–109. [Google Scholar] [CrossRef]
- Luo, Y.; Schimenti, J.C. MCM9 deficiency delays primordial germ cell proliferation independent of the ATM pathway. Genesis 2015, 53, 678–684. [Google Scholar] [CrossRef]
- Kim, S.; Gunesdogan, U.; Zylicz, J.J.; Hackett, J.A.; Cougot, D.; Bao, S.; Lee, C.; Dietmann, S.; Allen, G.E.; Sengupta, R.; et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell 2014, 56, 564–579. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.C.; Schimenti, J.C. Sexually dimorphic DNA damage responses and mutation avoidance in the mouse germline. Genes Dev. 2020, 34, 1637–1649. [Google Scholar] [CrossRef] [PubMed]
- Cantu, A.V.; Altshuler-Keylin, S.; Laird, D.J. Discrete somatic niches coordinate proliferation and migration of primordial germ cells via Wnt signaling. J. Cell Biol. 2016, 214, 215–229. [Google Scholar] [CrossRef]
- Risal, S.; Zhang, J.; Adhikari, D.; Liu, X.; Shao, J.; Hu, M.; Busayavalasa, K.; Tu, Z.; Chen, Z.; Kaldis, P.; et al. MASTL is essential for anaphase entry of proliferating primordial germ cells and establishment of female germ cells in mice. Cell Discov. 2017, 3, 16052. [Google Scholar] [CrossRef] [PubMed]
- Teng, H.; Sui, X.; Zhou, C.; Shen, C.; Yang, Y.; Zhang, P.; Guo, X.; Huo, R. Fatty acid degradation plays an essential role in proliferation of mouse female primordial germ cells via the p53-dependent cell cycle regulation. Cell Cycle 2016, 15, 425–431. [Google Scholar] [CrossRef]
- Sorrenti, M.; Klinger, F.G.; Iona, S.; Rossi, V.; Marcozzi, S.; De Felici, M. Expression and possible roles of extracellular signal-related kinases 1–2 (ERK1–2) in mouse primordial germ cell development. J. Reprod. Dev. 2020, 66, 399–409. [Google Scholar] [CrossRef]
- Ulu, F.; Kim, S.M.; Yokoyama, T.; Yamazaki, Y. Dose-dependent functions of fibroblast growth factor 9 regulate the fate of murine XY primordial germ cells. Biol. Reprod. 2017, 96, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Tian-Zhong, M.; Bi, C.; Ying, Z.; Xia, J.; Cai-Ling, P.; Yun-Shan, Z.; Mei-Wen, H.; Yan-Ru, N. Critical role of Emx2 in the pluripotency-differentiation transition in male gonocytes via regulation of FGF9/NODAL pathway. Reproduction 2016, 151, 673–681. [Google Scholar] [CrossRef]
- Hu, Y.C.; Nicholls, P.K.; Soh, Y.Q.; Daniele, J.R.; Junker, J.P.; van Oudenaarden, A.; Page, D.C. Licensing of primordial germ cells for gametogenesis depends on genital ridge signaling. PLoS Genet. 2015, 11, e1005019. [Google Scholar] [CrossRef]
- Overeem, A.W.; Chang, Y.W.; Spruit, J.; Roelse, C.M.; Chuva, D.S.L.S. Ligand-Receptor Interactions Elucidate Sex-Specific Pathways in the Trajectory From Primordial Germ Cells to Gonia During Human Development. Front. Cell Dev. Biol. 2021, 9, 661243. [Google Scholar] [CrossRef]
- Chassot, A.A.; Le Rolle, M.; Jourden, M.; Taketo, M.M.; Ghyselinck, N.B.; Chaboissier, M.C. Constitutive WNT/CTNNB1 activation triggers spermatogonial stem cell proliferation and germ cell depletion. Dev. Biol. 2017, 426, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Le Rolle, M.; Massa, F.; Siggers, P.; Turchi, L.; Loubat, A.; Koo, B.K.; Clevers, H.; Greenfield, A.; Schedl, A.; Chaboissier, M.C.; et al. Arrest of WNT/beta-catenin signaling enables the transition from pluripotent to differentiated germ cells in mouse ovaries. Proc. Natl. Acad. Sci. USA 2021, 118, e2023376118. [Google Scholar] [CrossRef] [PubMed]
- Yadu, N.; Kumar, P.G. Retinoic acid signaling in regulation of meiosis during embryonic development in mice. Genesis 2019, 57, e23327. [Google Scholar] [CrossRef]
- Barrios, F.; Filipponi, D.; Pellegrini, M.; Paronetto, M.P.; Di Siena, S.; Geremia, R.; Rossi, P.; De Felici, M.; Jannini, E.A.; Dolci, S. Opposing effects of retinoic acid and FGF9 on Nanos2 expression and meiotic entry of mouse germ cells. J. Cell Sci. 2010, 123, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Saga, Y. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev. 2008, 22, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Fukuda, K.; Weinstein, M.; Graff, J.M.; Saga, Y. SMAD2 and p38 signaling pathways act in concert to determine XY primordial germ cell fate in mice. Development 2015, 142, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Miao, Y.; Li, X.H.; Zhang, N.; Wang, Q.; Yue, W.; Sun, S.C.; Xiong, B.; Qiao, J.; Li, M. Proteome landscape and spatial map of mouse primordial germ cells. Sci. China Life Sci. 2021, 64, 966–981. [Google Scholar] [CrossRef]
- Eguizabal, C.; Herrera, L.; De Onate, L.; Montserrat, N.; Hajkova, P.; Izpisua, B.J. Characterization of the Epigenetic Changes During Human Gonadal Primordial Germ Cells Reprogramming. Stem Cells 2016, 34, 2418–2428. [Google Scholar] [CrossRef]
- Tang, W.W.; Dietmann, S.; Irie, N.; Leitch, H.G.; Floros, V.I.; Bradshaw, C.R.; Hackett, J.A.; Chinnery, P.F.; Surani, M.A. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell 2015, 161, 1453–1467. [Google Scholar] [CrossRef]
- Seki, Y. PRDM14 Is a Unique Epigenetic Regulator Stabilizing Transcriptional Networks for Pluripotency. Front. Cell Dev. Biol. 2018, 6, 12. [Google Scholar] [CrossRef]
- Hill, P.; Leitch, H.G.; Requena, C.E.; Sun, Z.; Amouroux, R.; Roman-Trufero, M.; Borkowska, M.; Terragni, J.; Vaisvila, R.; Linnett, S.; et al. Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 2018, 555, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Osada, A.; Ohta, M.; Yokota, K.; Nishiyama, A.; Niikura, Y.; Tamura, T.; Sekita, Y.; Kimura, T. SWI/SNF chromatin remodeling complex is required for initiation of sex-dependent differentiation in mouse germline. Sci. Rep. 2021, 11, 24074. [Google Scholar] [CrossRef] [PubMed]
- Ruthig, V.A.; Friedersdorf, M.B.; Garness, J.A.; Munger, S.C.; Bunce, C.; Keene, J.D.; Capel, B. The RNA-binding protein DND1 acts sequentially as a negative regulator of pluripotency and a positive regulator of epigenetic modifiers required for germ cell reprogramming. Development 2019, 146, dev175950. [Google Scholar] [CrossRef] [PubMed]
- Moratilla, A.; Sainz, D.L.M.D.; Cadenas, M.M.; Lopez-Iglesias, P.; Gonzalez-Peramato, P.; De Miguel, M.P. Inhibition of PKCepsilon induces primordial germ cell reprogramming into pluripotency by HIF1&2 upregulation and histone acetylation. Am. J. Stem Cells 2021, 10, 1–17. [Google Scholar]
- Fernandez-Perez, D.; Brieno-Enriquez, M.A.; Isoler-Alcaraz, J.; Larriba, E.; Del, M.J. MicroRNA dynamics at the onset of primordial germ and somatic cell sex differentiation during mouse embryonic gonad development. RNA 2018, 24, 287–303. [Google Scholar] [CrossRef]
- Boyer, A.; Yeh, J.R.; Zhang, X.; Paquet, M.; Gaudin, A.; Nagano, M.C.; Boerboom, D. CTNNB1 signaling in sertoli cells downregulates spermatogonial stem cell activity via WNT4. PLoS ONE 2012, 7, e29764. [Google Scholar] [CrossRef]
- Liu, C.F.; Parker, K.; Yao, H.H. WNT4/beta-catenin pathway maintains female germ cell survival by inhibiting activin betaB in the mouse fetal ovary. PLoS ONE 2010, 5, e10382. [Google Scholar] [CrossRef]
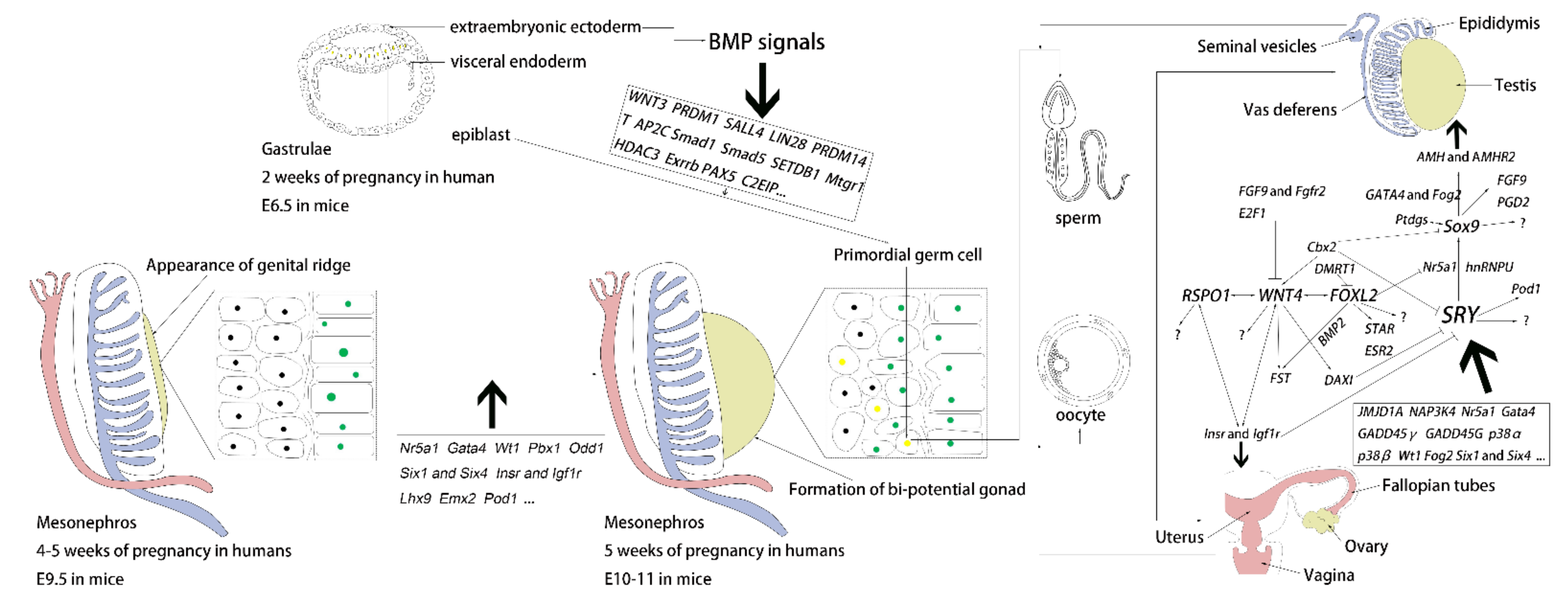
| Genes | Function in Organogenesis and DSD | References |
|---|---|---|
| NR5A1 | NR5A1 is involved in the development of the gonad, adrenal gland, and pituitary. | [25,26,27] |
| NR5A1 variants are associated with male infertility and DSD in humans males and females. | [28,29,30,31,32,33,34] | |
| GATA4 | GATA4 is required for the proliferation of coelomic epithelial cells and is involved in gonadal development by interacting with FOG2. | [35,36] |
| The GATA4 mutant protein failed to bind with FOG2, resulting in DSD. | [37] | |
| WT1 | WT1 plays a distinct role in gonadal formation and development by maintaining somatic cell survival. | [38] |
| WT1 mutations are responsible for Frasier syndrome with streak gonads. | [39,40] | |
| LHX9 | LHX9 participates in gonad formation by regulating cell proliferation. | [41] |
| EMX2 | EMX2 is required for the migration and survival of cells in the mesenchymal compartment and involves GR formation by regulating NR5A1 expression. | [42] |
| SIX1 and SIX4 | SIX1 and SIX4 have a functional redundancy and mainly function in the proliferation of supporting cell precursors and steroidogenic cell precursors. | [43] |
| POD1 | POD1 is essential for gonadal development by regulating Nr5a1 expression. | [44] |
| CBX2 | CBX2 is required for splenic vascular, adrenal gland, and gonad formation. | [45,46] |
| INSR and IGF1R | INSR and IGFIR regulate somatic progenitor cell proliferation by mediating Insulin and its growth factors (IGF1 and IGF2) during GR formation. | [47] |
| PBX1 | PBX1 is involved in progenitor cell proliferation in GR by regulating the expression of NR5A1. | [48] |
| PBX1 mutation abolishes its interaction with CBX2 and EMX2, causing gonadal dysgenesis and radiocubital synostosis in humans. | [49] | |
| ODD1 | ODD1 regulates the expression of LHX1, PAX2, and WT1, inhibiting cell apoptosis in nephrogenic mesenchyme and participating in gonadal development. | [50] |
| Genes | Functions | References |
|---|---|---|
| JMJD1A | JMJD1A is involved in the H3K9 demethylation in SRY, and JMJD1A deficiency presents a decrease in SRY expression. | [135] |
| MAP3K4 | MAP3K4 is involved in SRY expression, and loss of MAP3K4 will lead to a male sex reversal. | [136] |
| GADD45γ | GADD45γ is upstream of MAP3K4, without which will cause male sexual reversal. | [72] |
| p38α and p38β | p38α and p38β are members of p38 MAPK family, and loss of p38α and p38β will causes disruption to SRY expression and XY embryonic gonadal sex reversal. | |
| GADD45G | GADD45G is necessary for SRY expression. | [137] |
| HNRNPU | HNRNPU enhances the expression of SOX8 and SOX9 by interacting with WT1 and SOX9. | [138] |
| AMH and AMHR2 | AMH and its receptor AMHR2 are involved in the normal development of the accessory gland. | [139] |
| NR5A1 | NR5A1 is involved in testis formation by cooperating with other regulators such as WT1, DAX1, SRY, and SOX9. | [140] |
| GATA4 | The interaction of GATA4 and FOG2 is important in sex differentiation because it regulates the expression of SRY and AMH. | [36,141] |
| FOG2 | ||
| PTGDS | PTGDS is one of the downstream targets of SOX9, which involves the production of prostaglandin D2, maintaining the sustained expression of SOX9 in testis. | [142,143] |
| WT1 | WT1 is a potential upstream of SRY and controls somatic cells’ fate through regulating NR5A1 expression. | [38,144] |
| WT1 variants lead to 46,XX testicular DSD in humans. | [145,146] | |
| SIX1 and SIX4 | SIX1 and SIX4 regulate SRY expression by activating FOG2 expression, regulating male sex determination. | [43] |
| POD1 | The downstream target of SRY is POD1, which is involved in the formation of testicular cords and testis-specific coelomic vessels. | [44,147] |
| CBX2 | CBX2 regulates SRY expression by interfering with upstream steps. | [46,148] |
| INSR and IGF1R | INSR and IGF1R have potential feedback interactions between WNT4 and RSPO1 signaling pathways. | [84] |
| INSR and IGF1R are involved in the adrenal specification, testicular differentiation, and ovarian development by regulating the expression of sex-related genes, including WT1, LHX9, and NR5A1. | [149] | |
| FGF9 and PGD2 | FGF9 and PGD2 are downstream of SOX9 and are involved in supporting-to-Sertoli cell differentiation by activating testis-related genes and repressing anti-testis genes. | [79] |
| E2F1 | E2F1 regulates testicular descent and controls spermatogenesis by repressing WNT4 expression | [150] |
| FGF9 and FGFR2 | FGF9 and FGFR2 are required in testis development by repressing the expression of WNT4 and FOXL2. | [151,152] |
| FST | FST prevents testis-specific vasculature formation by antagonizing Activin B action through WNT4 activation. | [153] |
| DAX1 | DAX1 is downstream of WNT4 and is involved in gonadal sexual differentiation by antagonizing SRY expression. | [154] |
| BMP2 | BMP2 acts cooperatively with FOXL2 to regulate FST gene expression during ovarian development. | [155] |
| DMRT1 | DMRT1 is required for sexual differentiation of somatic and germ cells by silencing FOXL2 expression. | [156] |
| Genes | Functions | References |
|---|---|---|
| WNT3 | WNT3 enables PE cells to receive a BMP4 signal. | [168] |
| PRDM1 | PRDM1 is Likely downstream of BMP4. | [169] |
| PRDM1 is involved in PGCs formation by repressing somatic cell program genes through selective recruitment of HDAC3. | [170] | |
| SALL4 | SALL4 participates in the specification of PGCs by suppressing the expression of somatic cell program genes. | [171] |
| LIN28 | LIN28 is involved in PGCs development by regulating PRDM1 transcript translation. | [172] |
| PRDM14 | PRDM14 establishes germ cell lineage by inducing SOX2 expression and cooperating with TFAP2C and PRDM1, which upregulates pluripotency genes and represses somatic markers. | [173,174] |
| T | T specifies germ cell fate by activating the expression of PRDM1 and PRDM14. | [175] |
| AP2C | AP2C is most likely downstream of PRDM1 and is involved in maintaining PGCs. | [176] |
| SMAD1 | SMAD1 is a downstream signal mediator for BMPs and is essential for PGCs formation. | [177] |
| SMAD5 | SMAD5 is a downstream signal mediator for BMPs and is required for PGCs development. | [178] |
| SETDB1 | SETDB1 is involved in PGCs fate determination by ensuring BMP signaling through repressing the expression of Dppa2, Otx2, and Utf1. | [179] |
| MTGR1 | MTGR1 is involved in stem cell maintenance and PGCs formation by mediating PRDM14 functions. | [180] |
| ESRRB | ESRRB functions as an upstream factor of BMP4 and regulates PGCs development. | [181] |
| PAX5 | PAX5 participates in PGCs specification by activating germline and repressing somatic program genes through a PAX5-OCT4-PRDM1 core transcriptional network. | [182] |
| C2EIP | C2EIP promotes the generation of PGCs by activating the Hedgehog (Hh) signaling pathway via PTCH2 ubiquitination. | [183] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Wu, C.; Li, Z.; Wu, Z.; Hong, L. Early Gonadal Development and Sex Determination in Mammal. Int. J. Mol. Sci. 2022, 23, 7500. https://doi.org/10.3390/ijms23147500
Xie Y, Wu C, Li Z, Wu Z, Hong L. Early Gonadal Development and Sex Determination in Mammal. International Journal of Molecular Sciences. 2022; 23(14):7500. https://doi.org/10.3390/ijms23147500
Chicago/Turabian StyleXie, Yanshe, Changhua Wu, Zicong Li, Zhenfang Wu, and Linjun Hong. 2022. "Early Gonadal Development and Sex Determination in Mammal" International Journal of Molecular Sciences 23, no. 14: 7500. https://doi.org/10.3390/ijms23147500
APA StyleXie, Y., Wu, C., Li, Z., Wu, Z., & Hong, L. (2022). Early Gonadal Development and Sex Determination in Mammal. International Journal of Molecular Sciences, 23(14), 7500. https://doi.org/10.3390/ijms23147500






