GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc)
Abstract
:1. Introduction
2. Results
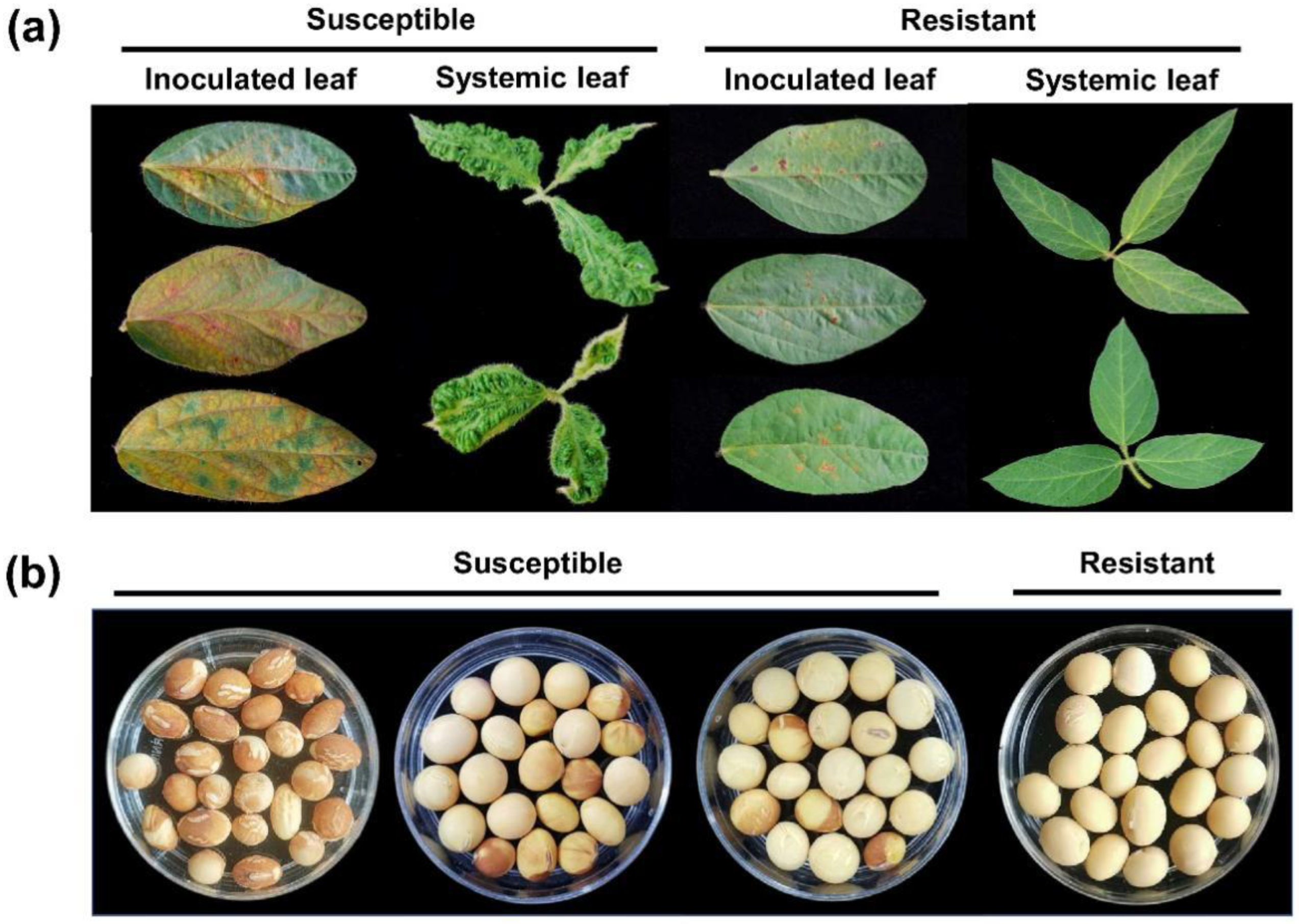
2.1. Response of CSSL Population to SMV
2.2. Identification of Genomic Regions Associated with Resistance to Seed Coat Mottling
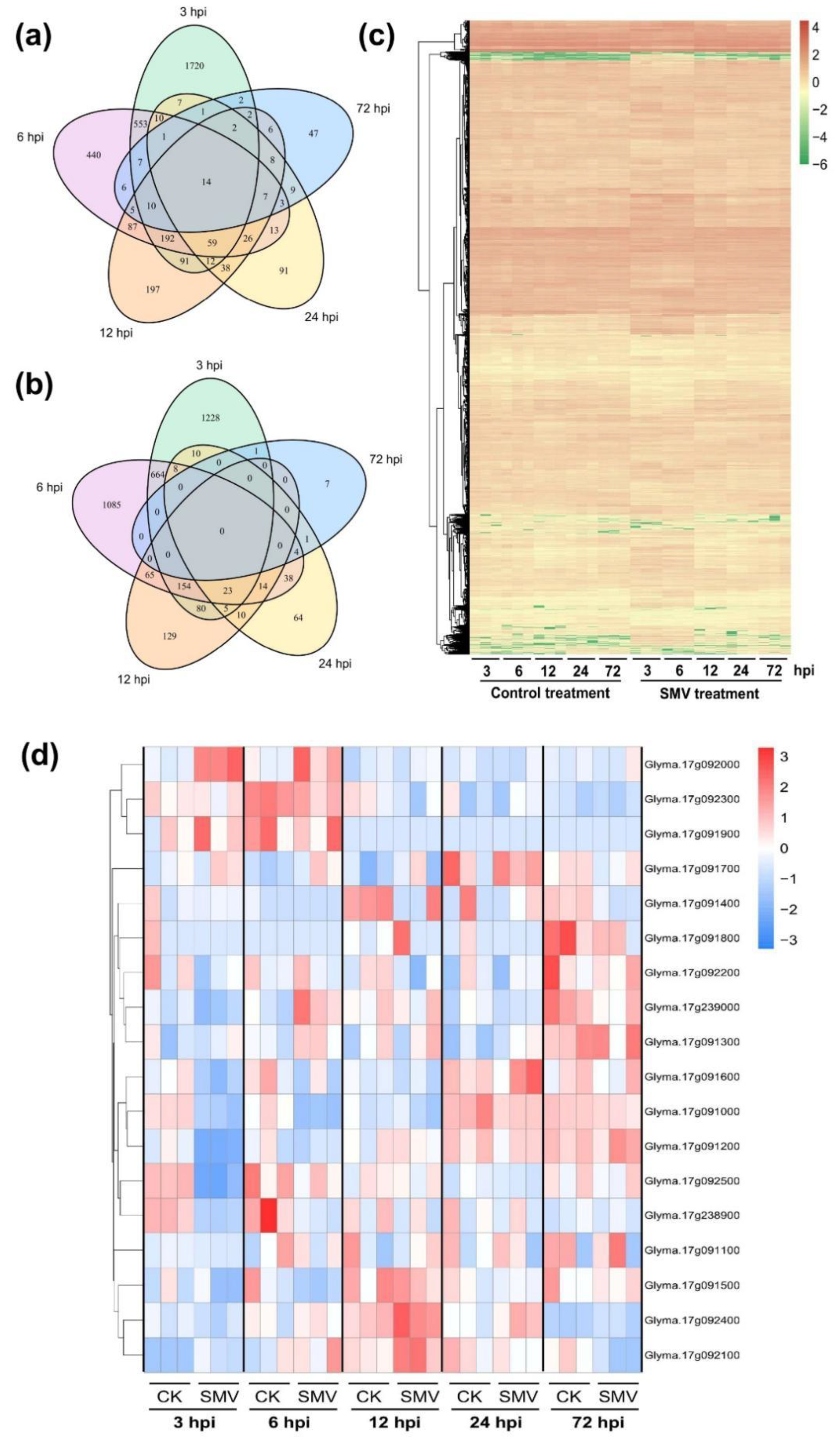
2.3. RNA-Seq Analysis of SN14 Response to SMV Infection
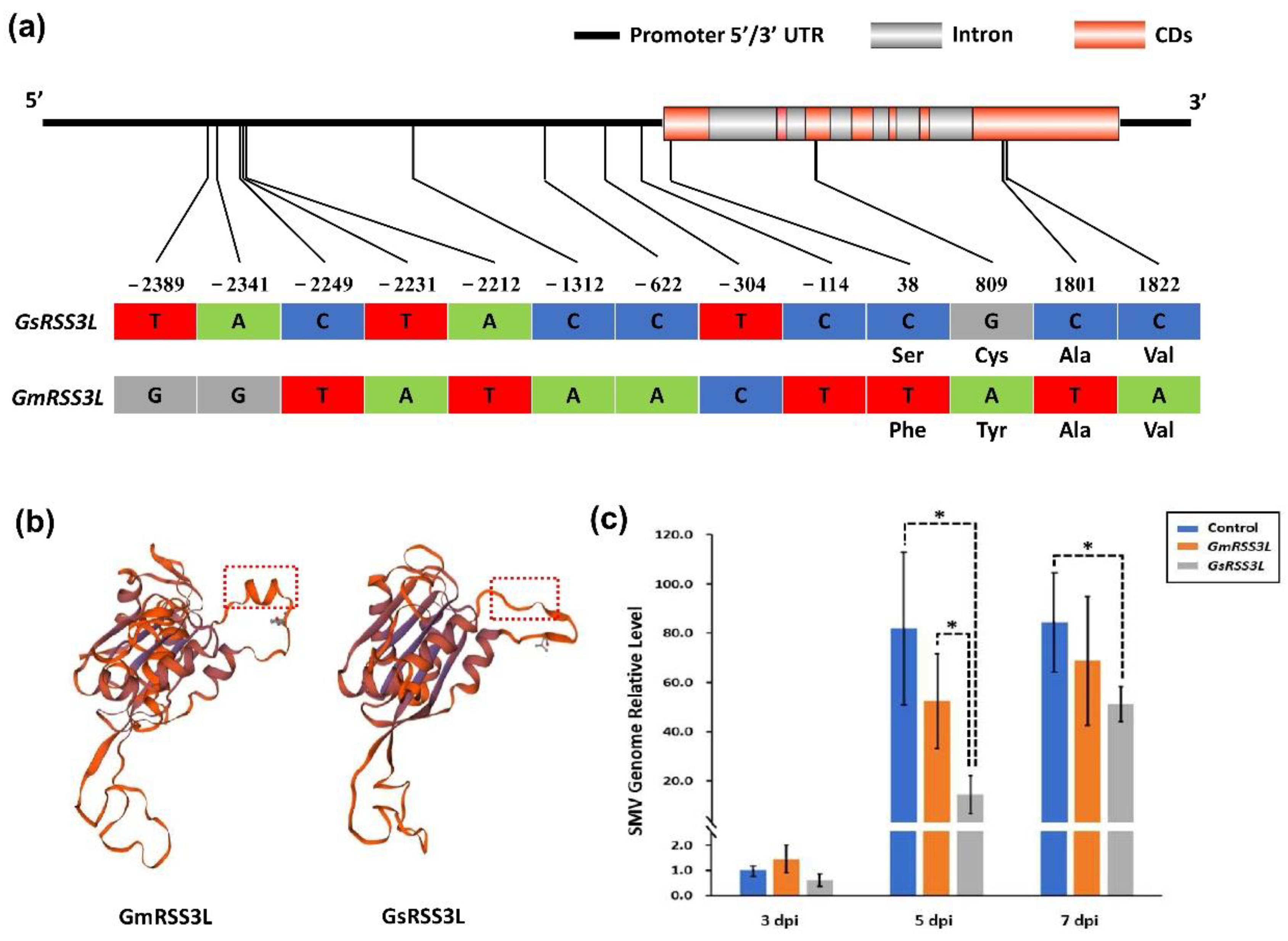
2.4. Prediction and qRT-PCR Validation of Candidate Genes
2.5. Agrobacterium-Mediated Transient Expression of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Genetic Materials
4.2. Virus Culture, Inoculation, and RT-PCR Detection
4.3. QTL Detection
4.4. Transcriptome Analysis
4.5. Prediction and qRT-PCR Validation of Candidate Genes
4.6. Agrobacterium-Mediated Transient Expression of Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoeck, J.A.; Fehr, W.R.; Shoemaker, R.C.; Welke, G.A.; Johnson, S.L.; Cianzio, S.R. Molecular marker analysis of seed size in soybean. Crop Sci. 2003, 43, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Widyasari, K.; Alazem, M.; Kim, K. Soybean resistance to soybean mosaic virus. Plants 2020, 9, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, J.H.; Whitham, S.A. Control of virus disease in soybean. Adv. Virus Res. 2014, 90, 355–390. [Google Scholar] [PubMed]
- Ross, J.P. Effect of aphid-transmitted soybean mosaic virus on yields of closely related resistant and susceptible soybean lines. Crop Sci. 1997, 17, 869–872. [Google Scholar] [CrossRef]
- Rui, R.; Liu, S.; Karthikeyan, A.; Wang, T.; Niu, H.; Yin, J.; Yang, Y.; Wang, L.; Yang, Q.; Zhi, H.; et al. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theor. Appl. Genet. 2017, 130, 2395–2410. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Domier, L.L.; Tolin, S.A.; Whitham, S.A.; Saghai Maroof, M.A. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Yang, Q.H.; Zhi, H.J.; Gai, J.Y. Identification and distribution of Soybean mosaic virus strains in Southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Wu, Y.; An, J.; Huang, W.; Liu, X.; Xue, Y.; Luan, X.; Lin, F.; Sun, L. Pathogenicity and genome-wide sequence analysis reveals relationships between soybean mosaic virus strains. Arch. Virol. 2022, 167, 517–529. [Google Scholar] [CrossRef]
- Koning, G.; TeKrony, D.M.; Ghabrial, S.A. Soybean seedcoat mottling: Association with Soybean mosaic virus and Phomopsis spp. seed infection. Plant Dis. 2003, 87, 413–417. [Google Scholar] [CrossRef] [Green Version]
- Senda, M. Patterning of virus-infected Glycine max seedcoat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 2004, 16, 807–818. [Google Scholar] [CrossRef] [Green Version]
- Karthikeyan, A.; Li, K.; Jiang, H.; Ren, R.; Li, C.; Zhi, H.; Chen, S.; Gai, J. Inheritance, fine-mapping, and candidate gene analyses of resistance to soybean mosaic virus strain SC5 in soybean. Mol. Genet. Genom. 2017, 292, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, W.; Zhang, L.; Liu, C.Y.; Zhu, X.S.; Chen, Q.S.; Hu, G. Genetic analysis and molecular mapping of two SMV-resistance traits in soybean: Adult-plant resistance and resistance to seedcoat mottling. J. Integr. Agric. 2010, 9, 11–18. [Google Scholar] [CrossRef]
- Cooper, R.L. A major gene for resistance to seedcoat mottling in soybean. Crop Sci. 1966, 6, 290–292. [Google Scholar] [CrossRef]
- Hu, G.; Gao, F.; Wu, Z. Studies for linkage of resistance to seedcoat mottling on soybean. Acta Agron. Sin. 1996, 22, 555–559. [Google Scholar]
- Capistrano-Gossmann, G.G.; Ries, D.; Holtgrawe, D.; Minoche, A.; Kraft, T.; Frerichmann, S.L.M.; Soerensen, T.R.; Dohm, J.C.; Gonzalez, I.; Schilhabel, M.; et al. Crop wild relative populations of Beta vulgaris allow direct mapping of agronomically important genes. Nat. Commun. 2017, 8, 15708. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [Green Version]
- Xin, D.; Qi, Z.; Jiang, H.; Hu, Z.; Zhu, R.; Hu, J.; Han, H.; Hu, G.; Liu, C.; Chen, Q. QTL location and epistatic effect analysis of 100-seed weight using wild soybean (Glycine soja Sieb. & Zucc.) chromosome segment substitution lines. PLoS ONE 2016, 11, e0149380. [Google Scholar] [CrossRef] [Green Version]
- Toda, Y.; Yoshida, M.; Hattori, T.; Takeda, S. RICE SALT SENSITIVE3 binding to bHLH and JAZ factors mediates control of cell wall plasticity in the root apex. Plant Signal. Behav. 2013, 8, e26256. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.-F.; Wu, X.-Y.; Chen, Y.-X.; Liu, Y.-N.; Shao, Z.-Q.; Wu, P.; Wu, M.; Liu, C.-C.; Wu, W.-P.; Yang, J.-Y.; et al. Fine mapping of the Rsv1-h gene in the soybean cultivar Suweon 97 that confers resistance to two Chinese strains of the soybean mosaic virus. Theor. Appl. Genet. 2016, 129, 2227–2236. [Google Scholar] [CrossRef]
- Tran, P.T.; Widyasari, K.; Seo, J.K.; Kim, K.H. Isolation and validation of a candidate Rsv3 gene from a soybean genotype that confers strain-specific resistance to soybean mosaic virus. Virology 2018, 53, 153–159. [Google Scholar] [CrossRef]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 4033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, C.; Chen, P.; Gergerich, R. Characterization of resistance to Soybean mosaic virus in diverse soybean germplasm. Crop Sci. 2005, 45, 2503–2509. [Google Scholar] [CrossRef]
- Li, C.; Adhimoolam, K.; Yuan, Y.; Yin, J.; Ren, R.; Yang, Y.; Zhi, H. Identification of candidate genes for resistance to Soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses. Crop Pasture Sci. 2017, 68, 156–166. [Google Scholar] [CrossRef]
- Fu, S.; Zhan, Y.; Zhi, H.; Gai, J.; Yu, D. Mapping of SMV resistance gene Rsc-7 by SSR markers in soybean. Genetica 2006, 128, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ma, Y.; Yang, Y.; Liu, N.; Li, C.; Song, Y.; Zhi, H. Fine mapping and analyses of RSC8 resistance candidate genes to soybean mosaic virus in soybean. Theor. Appl. Genet. 2011, 122, 555–565. [Google Scholar] [CrossRef]
- Ma, Y.; Li, H.; Wang, D.; Liu, N.; Zhi, H. Molecular mapping and marker assisted selection of soybean mosaic virus resistance gene RSC12 in soybean. Legume Genom. Genet. 2010, 1, 41–46. [Google Scholar]
- Li, K.; Ren, R.; Adhimoolam, K.; Gao, L.; Yuan, Y.; Liu, Z.; Zhong, Y.; Zhi, H. Genetic analysis and identification of two soybean mosaic virus resistance genes in soybean [Glycine max (L.) Merr]. Plant Breed. 2015, 134, 684–695. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Li, C.; Yin, J.; Li, N.; Yang, Y.; Song, Y.; Ren, R.; Zhi, H.; Gai, J.; et al. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor. Appl. Genet. 2018, 131, 461–476. [Google Scholar] [CrossRef]
- Tuyen, D.D.; Lal, S.K.; Xu, D.H. Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor. Appl. Genet. 2010, 121, 229–236. [Google Scholar] [CrossRef]
- Cao, P.; Zhao, Y.; Wu, F.; Xin, D.; Liu, C.; Wu, X.; Lv, J.; Chen, Q.; Qi, Z. Multi-omics techniques for soybean molecular breeding. Int. J. Mol. Sci. 2022, 23, 4994. [Google Scholar] [CrossRef]
- Guo, X.; Jiang, J.; Liu, Y.; Yu, L.; Chang, R.; Guan, R.; Qiu, L. Identification of a novel salt tolerance-related locus in wild soybean (Glycine soja Sieb. & Zucc.). Front. Plant Sci. 2021, 12, 791175. [Google Scholar] [CrossRef] [PubMed]
- Toda, Y.; Tanaka, M.; Ogawa, D.; Kurata, K.; Kurotani, K.; Habu, Y.; Ando, T.; Sugimoto, K.; Mitsuda, N.; Katoh, E.; et al. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and Class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 2013, 25, 1709–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Ye, J. Manipulation of jasmonate signaling by plant viruses and their insect vectors. Viruses 2020, 12, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzo, O.; Solano, R. Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 2005, 8, 532–540. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. Jasmonate signaling: Toward and integrated view. Plant Physiol. 2008, 146, 1459–1468. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Li, Y. Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog. 2021, 17, e1009242. [Google Scholar] [CrossRef]
- Laurie-Berry, N.; Joardar, V.; Street, I.H.; Kunkel, B.N. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defense during infection by Pseudomonas syringae. Mol. Plant Microbe Int. 2006, 19, 789–800. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Zhang, H.; Sun, Z.; Li, J.; Hong, G.; Zhu, Q.; Zhou, X.; MacFarlane, S.; Yan, F.; Chen, J. Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 2017, 214, 388–399. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Huang, Y.; Yang, J.; Yao, S.; Zhao, K.; Wang, D.; Qin, Q.; Bian, Z.; Li, Y.; Lan, Y.; et al. Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 2020, 28, 89–103. [Google Scholar] [CrossRef]
- Yamada, T.; Takagi, K.; Ishimoto, M. Recent advances in soybean transformation and their application to molecular breeding and genomic analysis. Breed. Sci. 2012, 61, 480–494. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhai, R.; Zhong, Y.K.; Karthikeyan, A.; Ren, R.; Zhang, K.; Li, K.; Zhi, H.J. Screening isolates of Soybean mosaic virus for infectivity in a model plant, Nicotiana benthamiana. Plant Dis. 2015, 99, 442–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodin, M.M.; Zaitlin, D.; Naidu, R.A.; Lommel, S.A. Nicotiana benthamiana: Its history and future as a model for plant-pathology interactions. Mol. Plant Microbe Int. 2008, 21, 1015–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarty, R.; Banerjee, R.; Chung, S.M.; Farman, M.; Citovsky, V.; Hogenhout, S.A.; Tzfira, T.; Goodin, M.M. pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: Probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Int. 2007, 20, 740–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.; Xi, D.; Yuan, S.; Xu, F.; Zhang, D.; Lin, H. Salicylic acid and jasmonic acid are essential for systemic resistance against Tobacco mosaic virus in Nicotiana benthamiana. Mol. Plant Microbe Int. 2014, 27, 567–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, W.; Yan, T.; Deng, X.; Wuriyanghan, H. Synthesis of full-length cDNA infectious clones of Soybean mosaic virus and functional identification of a key amino acid in the silencing suppressor Hc-Pro. Viruses 2020, 12, 886. [Google Scholar] [CrossRef] [PubMed]
- Hyten, D.L.; Choi, I.; Song, Q.; Specht, J.E.; Carter, T.E.; Shoemaker, R.C.; Hwang, E.; Matukumalli, L.K.; Cregan, P.B. A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci. 2010, 50, 960–968. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–570. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
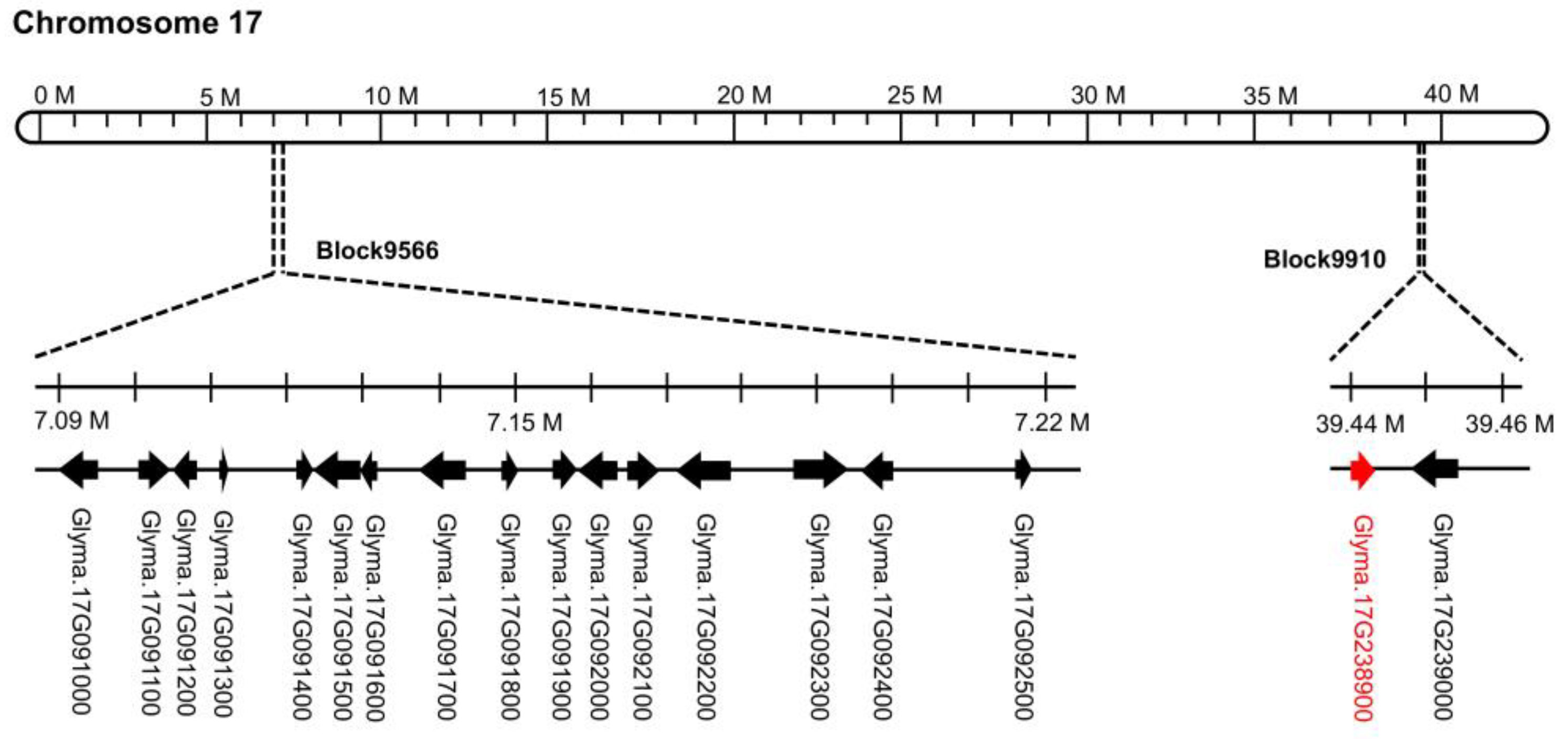

| Block | Chromosome | Physical location 1 | LOD 2 | PVE (%) 3 | Add 4 | |
|---|---|---|---|---|---|---|
| Start | End | |||||
| block9566 | Chr.17 | 7,086,191 | 7,227,595 | 2.50 | 4.57 | −0.02 |
| block9910 | Chr.17 | 39,435,988 | 39,459,788 | 7.90 | 14.65 | −0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; Wang, J.; Yang, X.; Zhang, X.; Xin, X.; Liu, C.; Zou, J.; Cheng, X.; Zhang, N.; Hu, Y.; et al. GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc). Int. J. Mol. Sci. 2022, 23, 7577. https://doi.org/10.3390/ijms23147577
Song S, Wang J, Yang X, Zhang X, Xin X, Liu C, Zou J, Cheng X, Zhang N, Hu Y, et al. GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc). International Journal of Molecular Sciences. 2022; 23(14):7577. https://doi.org/10.3390/ijms23147577
Chicago/Turabian StyleSong, Shuang, Jing Wang, Xingqi Yang, Xuan Zhang, Xiuli Xin, Chunyan Liu, Jianan Zou, Xiaofei Cheng, Ning Zhang, Yuxi Hu, and et al. 2022. "GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc)" International Journal of Molecular Sciences 23, no. 14: 7577. https://doi.org/10.3390/ijms23147577







