Abstract
The protozoan pathogen Cryptosporidium parvum infects intestinal epithelial cells and causes diarrhea in humans and young animals. Among the more than 20 genes encoding insulinase-like metalloproteinases (INS), two are paralogs with high sequence identity. In this study, one of them, INS-16 encoded by the cgd3_4270 gene, was expressed and characterized in a comparative study of its sibling, INS-15 encoded by the cgd3_4260 gene. A full-length INS-16 protein and its active domain I were expressed in Escherichia coli, and antibodies against the domain I and an INS-16-specific peptide were produced in rabbits. In the analysis of the crude extract of oocysts, a ~60 kDa fragment of INS-16 rather than the full protein was recognized by polyclonal antibodies against the specific peptide, indicating that INS-16 undergoes proteolytic cleavage before maturation. The expression of the ins-16 gene peaked at the invasion phase of in vitro C. parvum culture, with the documented expression of the protein in both sporozoites and merozoites. Localization studies with antibodies showed significant differences in the distribution of the native INS-15 and INS-16 proteins in sporozoites and merozoites. INS-16 was identified as a dense granule protein in sporozoites and macrogamonts but was mostly expressed at the apical end of merozoites. We screened 48 candidate INS-16 inhibitors from the molecular docking of INS-16. Among them, two inhibited the growth of C. parvum in vitro (EC50 = 1.058 µM and 2.089 µM). The results of this study suggest that INS-16 may have important roles in the development of C. parvum and could be a valid target for the development of effective treatments.
1. Introduction
Cryptosporidium spp. are gastrointestinal pathogens that can cause severe diarrhea in humans and various animals [1]. Young children in developing countries infected with Cryptosporidium spp. can develop malnutrition and cognitive impairments in addition to clinical illness [2]. In industrialized countries, waterborne outbreaks of cryptosporidiosis are common [3]. Among the more than 40 named Cryptosporidium species, Cryptosporidium parvum is the main species for cryptosporidiosis in farm animals and one of the two dominant species in humans [4]. Currently, there is a lack of effective drugs against Cryptosporidium spp. Although significant progress has been made in the development of novel drugs against cryptosporidiosis in recent years, we still have poor understanding of the biology of the pathogens [5].
The invasion of host cells by apicomplexan parasites is a complex process mediated by receptors and ligands that involve many proteins on both sides [6]. Secreted proteases and protein kinases in secretory organelles of apicomplexans can modify invasion-related proteins or host cell activities, thus playing important roles in invasion [7]. Comparative genomics analysis of multiple Cryptosporidium species has revealed the presence of numerous genes encoding secreted proteases in the compact genome. Among them, insulinase-like metalloproteinases (INS) are one of the largest protease families with 22 members in C. parvum [8,9]. As the Cryptosporidium genome is only 9 Mb in size and most of the ~4000 genes are single copied, INS probably play important roles in the invasion and development of Cryptosporidium spp.
INS are zinc metalloproteinases of the M16 family, which are widely distributed in nature and divided into the M16A, M16B, and M16C subfamilies. The M16 metalloproteinases are characterized by the presence of a functional domain containing an inversion of the thermolysin zinc-binding motif, HXXEH [10]. They have a wide range of substrates and cleave many proteins and small peptides including insulin, β-amyloid, and glucagon [11,12]. In apicomplexans, falcilysin is an M16C insulinase involved in hemoglobin catabolism and may function as two different proteases in two subcellular organelles of Plasmodium falciparum after proteolytic processing [13,14]. An M16A protease, toxolysin-1 (TLN1) of Toxoplasma gondii, is a rhoptry protein that is released during the invasion of host cells. Both the C-terminus and N-terminus of TLN1 undergo cleavage before the maturation of the protease [15]. Another toxolysin, TLN4, is located in the micronemes and involved in parasite fitness [16].
Transcriptome data from C. parvum indicate that some INS genes are highly expressed in the invasion stages of the pathogen [17]. In a recent study, one INS protein of C. parvum, INS20-19, was shown to be potentially involved in invasion or early developmental processes [18]. Another C. parvum INS protein, INS-15, appeared to be post-translationally processed as several fragments and have biological activities similar to toxolysins [19]. Another INS protein, INS-1, is expressed in secretory vesicles within the pathogen and contributes to the formation of macrogamonts [20]. Nevertheless, the role, processing, and trafficking of other INS remain unclear.
In this study, we characterized INS-16 encoded by the cgd3_4270 gene and examined its expression patterns in developmental stages of C. parvum. Two inhibitors of the metalloproteinase were identified through virtual screening, which reduced the growth of the pathogen in vitro. In addition, we compared the expression of INS-16 and INS-15, two metalloproteinases with high sequence identity and encoded by neighboring genes. The results suggest that the two INS are located in different organelles and have different biological functions.
2. Results
2.1. Characteristics of INS-16 and INS-16 Domain I
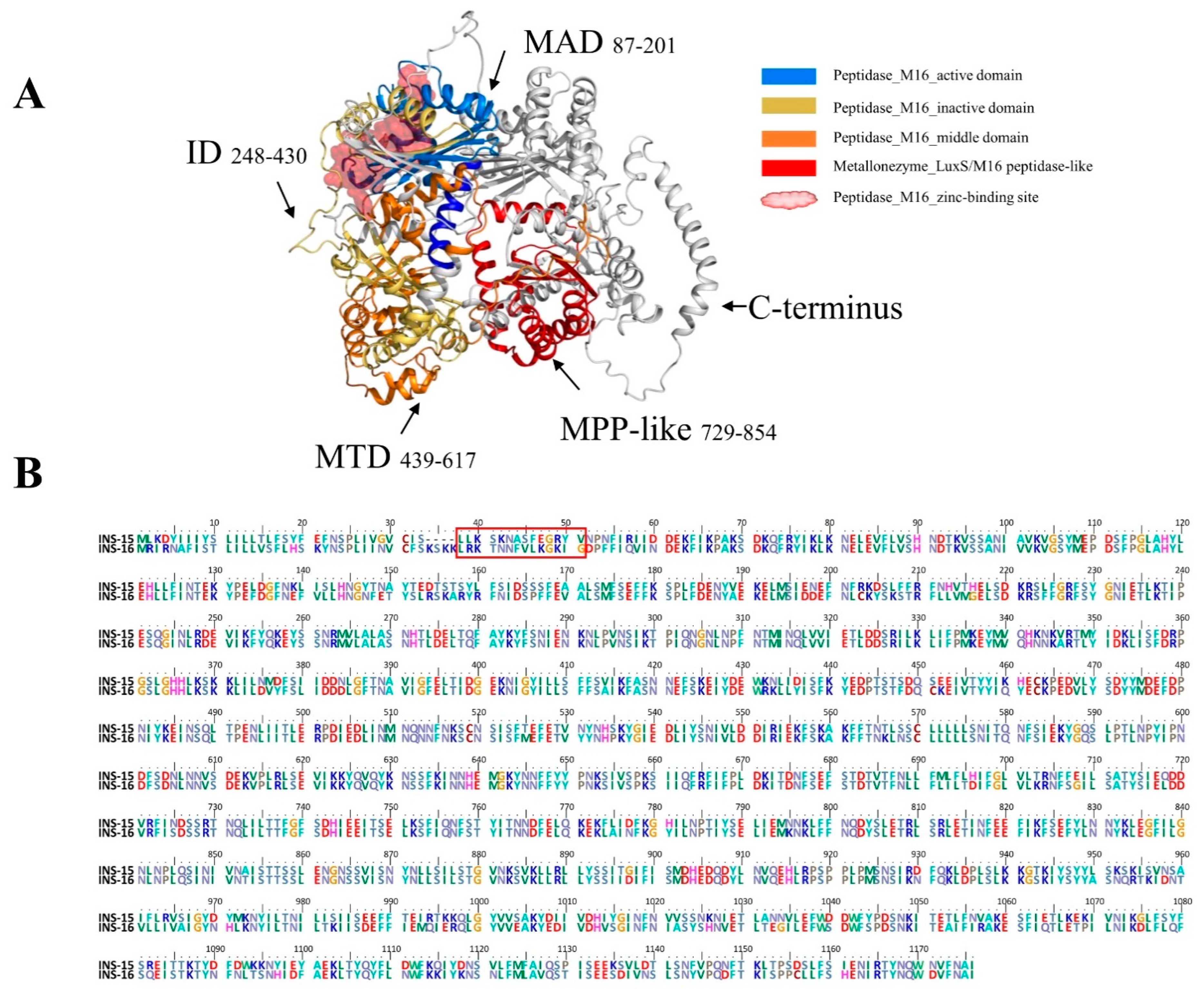
INS-16 is an M16A secretory metalloproteinase of 1176 amino acids and consists of four classic domains of INS with the key functional motif of HXXEH in the M16 active domain (Figure 1A). It differs from INS-15 in the number of domains, and it has one more M16 peptidase-like domain at the C-terminus than INS-15. Most of the sequence differences between the two INS are in the N- terminus and C-terminus, especially amino acids 1 to 60 (Figure 1B). Using DNA extracted from C. parvum oocysts as the template, we successfully cloned the full-length cgd3_4270 gene and its domain I fragment (Figure S1A,D), producing recombinant proteins in E. coli (Figure S1B,E). The recombinant proteins were purified using Ni-NTA affinity chromatography, with the purity being confirmed using SDS-PAGE analysis (Figure S1C,F).
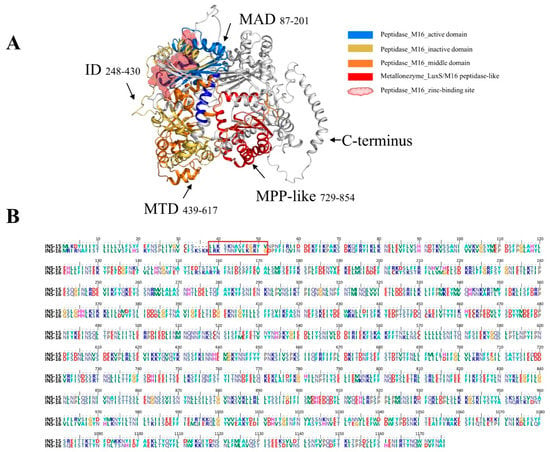
Figure 1.
Domain structure and specific amino acid sequence of INS-16 of Cryptosporidium parvum. (A) Predicted tertiary structures of INS-16. INS-16 have four classic M16 domains; the ribbon model is colored based on the M16 active domain (MAD), inactive domain (ID), the middle domain (MTD), and M16 peptidase-like domain (MPP-like). Black arrows indicate the region of these domains in the model and the amino acid sequence. The red spheroid located between the active domain and the inactive domain indicates the zinc-binding site of INS-16. (B) Alignment of amino acid sequences of INS-15 and INS-16. The red box shows the amino acid sequence of INS-16-specific peptide (LRKTNNFVLKGKIG) used in the study.
2.2. Cross-Reactivity between INS-15 and INS-16
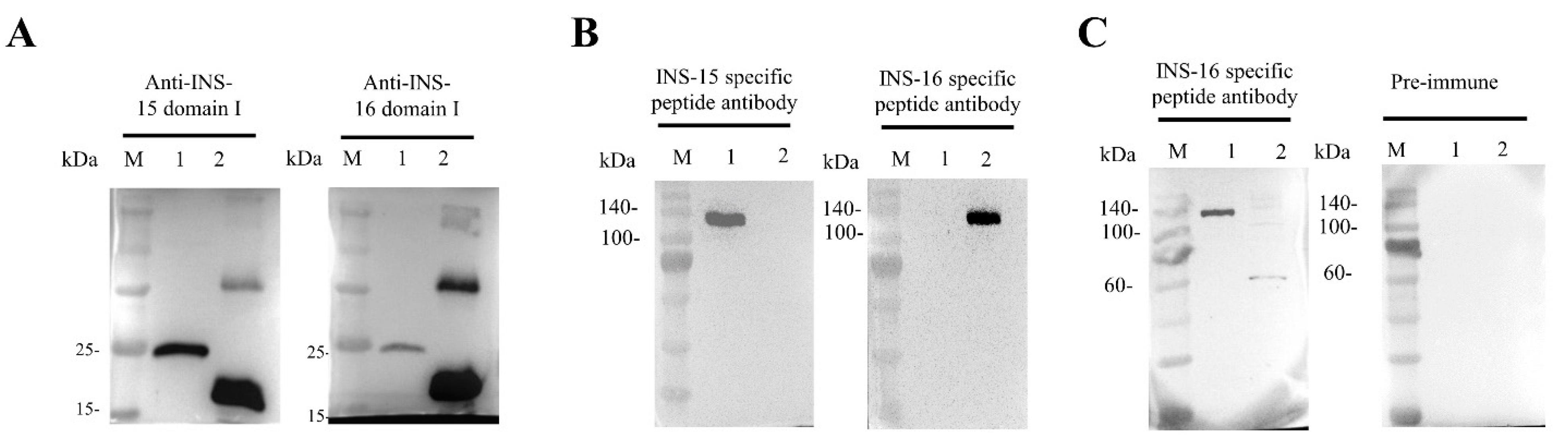
Because INS-16 has high identity to the previously characterized INS-15, we assessed the specificity of the antibodies generated with recombinant INS-16 domain I. In Western blot analysis, the antibodies reacted with the recombinant domain I of both INS-15 and INS-16. Similarly, antibodies against the INS-15 domain I also reacted with the recombinant domain I of both INS-15 (~23 kDa) and INS-16 (~19 kDa) (Figure 2A). However, both antibodies reacted more strongly with their respective proteins. To localize the expression of INS-16, we generated antibodies against the INS-16-specific peptide based on comparisons of the deduced amino acid sequences of INS-15 and INS-16 (Figure 1B). The result of Western blot analysis showed the antibodies against the INS-16-specific peptide only react with the full-length INS-16 protein (~134 kDa). Similarly, the antibodies against the INS-15-specific peptide also only reacted with the full-length INS-15 protein (~137 kDa) (Figure 2B). The antibodies against the two specific peptides did not have any cross-reactivity in Western blot analysis with either INS-15 or INS-16.
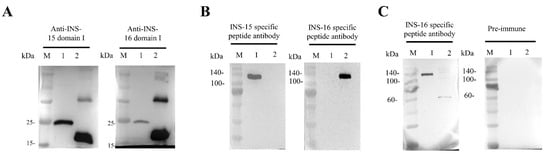
Figure 2.
Western blot analysis of cross-reactivity and native protein expression of INS-15 and INS-16. (A) Cross-reactivity of antibodies between INS-15 and INS-16. Lane M: protein marker; Lane 1: purified recombinant INS-15 domain I; Lane 2: purified recombinant INS-16 domain I. The image on the left shows the result by using anti-INS-15 domain I as the primary antibodies. The image on the right shows the result by using anti-INS-16 domain I as the primary antibodies. (B) Western blot analysis of the specificity of antibodies against INS-16 peptide. Lane M: protein marker; Lane 1: purified recombinant full-length INS-15. Lane 2: purified recombinant full-length INS-16. The picture on the left shows the full-length protein of INS-15 and INS-16 reacting with antibodies against the INS-15 peptide. The picture on the right shows the full-length protein of INS-15 and INS-16 reacting with antibodies against the INS-16 peptide. (C) Expression of native INS-16 protein in C. parvum sporozoites. Lane M: protein marker; Lane 1: purified recombinant full-length INS-16. Lane 2: C. parvum sporozoite lysate. The picture on the right shows the result of proteins reacting with pre-immune serum.
2.3. Proteolytical Processing of Native INS-16
To assess the expression of the native INS-16 protein in C. parvum, we used antibodies generated against the INS-16-specific peptide in the Western blot analysis of sporozoite lysates. The antibodies reacted with a ~60 kDa fragment of the protein instead of the expected 134 kDa full length protein (Figure 2C), indicating that native INS-16 is likely proteolytically processed after its translation.
2.4. Differential Expression of INS-15 and INS-16 in Life Cycle Stages of C. parvum
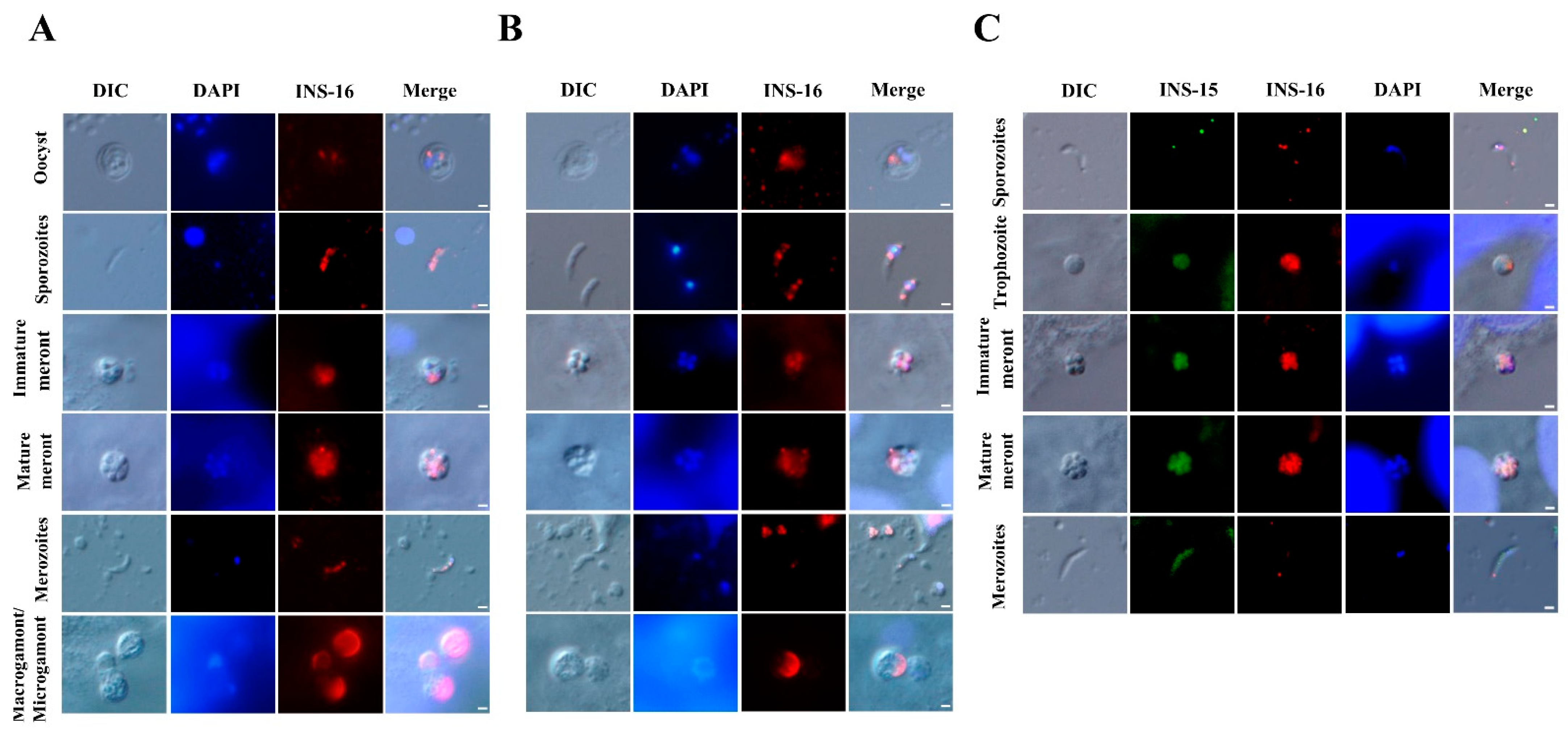
The antibodies against the INS-16 domain I and specific peptide were used to characterize the expression of INS-16 in oocysts, excysted sporozoites, and intracellular stages using immunofluorescence microscopy. The results showed that both antibodies reacted with sporozoites within oocysts, with no significant difference in the staining pattern. However, in excysted sporozoites, anti-INS-16 domain I antibodies reacted with the entire parasites, while antibodies against the INS-16-specific peptide showed a dotty pattern mostly in the middle of the sporozoites in immunofluorescence microscopy. In the immunofluorescence analysis of intracellular developmental stages, both antibodies reacted with meronts with similar staining patterns. The fluorescence signal of antibodies against INS-16 domain I covered almost the entire merozoites, while the fluorescence signal of antibodies against the INS-16-specific peptide was a small dot at the apical end of merozoites. In sexual stage, the antibodies against INS-16 domain I reacted with both the macrogamont and microgamont, while the antibodies against the INS-16-specific peptide reacted mostly with one side of the macrogamont with no reactivity to microgamonts (Figure 3A,B).
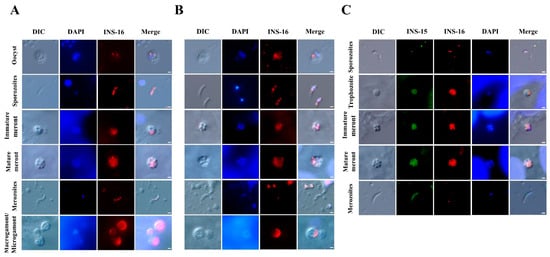
Figure 3.
Patterns of INS-16 expression in oocysts, sporozoites, and intracellular stages of Cryptosporidium parvum in HCT-8 cells as indicated with immunofluorescence microscopy. (A) The localization of INS-16 with antibodies against INS-16 domain I. (B) The localization of INS-16 with antibodies against INS-16-specific peptide. The reactivity of the antibodies with oocysts, free sporozoites, and merozoites in infected HCT-8 cells is shown (red). Nuclei were counter-stained with DAPI (blue). Scale bars = 1 μm. (C) Co-localization of INS-15 and INS-16 in sporozoite, trophozoite, meront and free merozoite. The reaction of antibodies against INS-15-specific peptide is shown in green fluorescence, while the reaction of antibodies against INS-16-specific peptide is shown in red fluorescence. Nuclei were counter-stained blue with DAPI. Scale bars = 1 μm.
To compare the expression between INS-15 and INS-16 in different C. parvum life cycle stages, antibodies against INS-15 and INS-16-specific peptides were used in co-localization analysis. The results showed that the signals of INS-15 and INS-16 antibodies in free sporozoites did not overlap completely, with INS-15 being localized in the nucleus of the parasite and INS-16 having more spotty expression over the entire sporozoites, as indicated above. While the staining patterns of INS-15 and INS-16 antibodies were largely similar in trophozoites and meronts, the fluorescence signal of INS-15 antibodies covered almost the entire free merozoites compared with only a small dot near the apical end of free merozoites in the immunofluorescence analysis with INS-16-specific antibodies (Figure 3C).
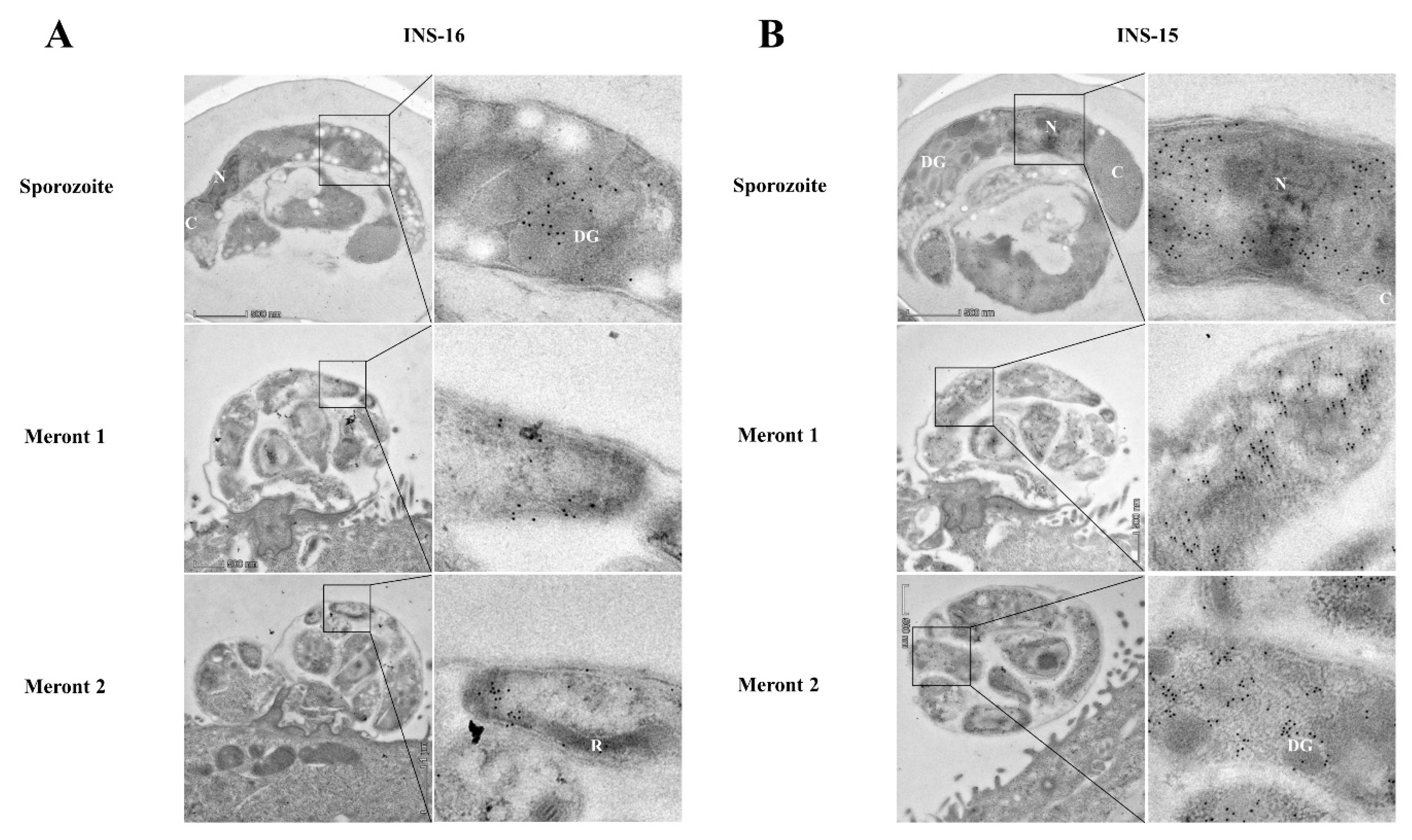
The expression of INS-16 in sporozoites and merozoites was further examined via immunoelectron microscopy using antibodies against the INS-16-specific peptide. In oocysts, INS-16 expression was mainly in the anterior end and middle of the sporozoites, mostly in dense granules, with some gold particles in the oocyst matrix also (Figure 4A). In contrast, the expression of INS-15 was mainly concentrated around the nucleus of sporozoites, with some gold particles in dense granules (Figure 4B). In meronts of infected HCT-8 cells, INS-16 expression was mainly in the apical end of merozoites, while the expression of INS-15 covered the entire merozoites (Figure 4A,B). This result is consistent with immunofluorescence analysis of INS-15 and INS-16, as indicated above.
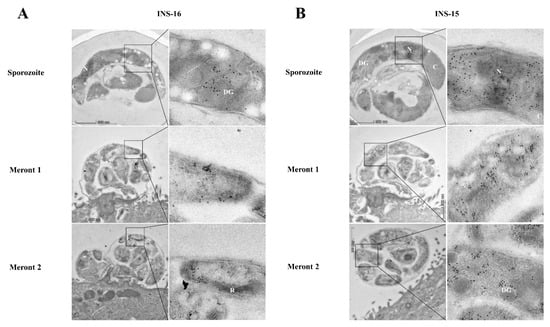
Figure 4.
Differences in the distribution of INS-15 and INS-16 expression in organelles of developmental stages of Cryptosporidium parvum. The localization of INS-15 and INS-16 in subcellular structures was analyzed using immuno-transmission electron microscopy. The distribution of INS-16 (A) and INS-15 (B) in sporozoites and meronts. C. parvum oocysts and meronts in infected HCT-8 cells were fixed and stained with antibodies against the INS-16 or INS-15-specific peptide followed by 10 nm colloidal gold-conjugated goat anti-rabbit IgG. N, nucleus; DG, dense granule; C, crystalloid body; R, rhoptry. Scale bars, 500 nm.
2.5. Anti-Cryptosporidial Effects of Candidate Inhibitors of INS-16
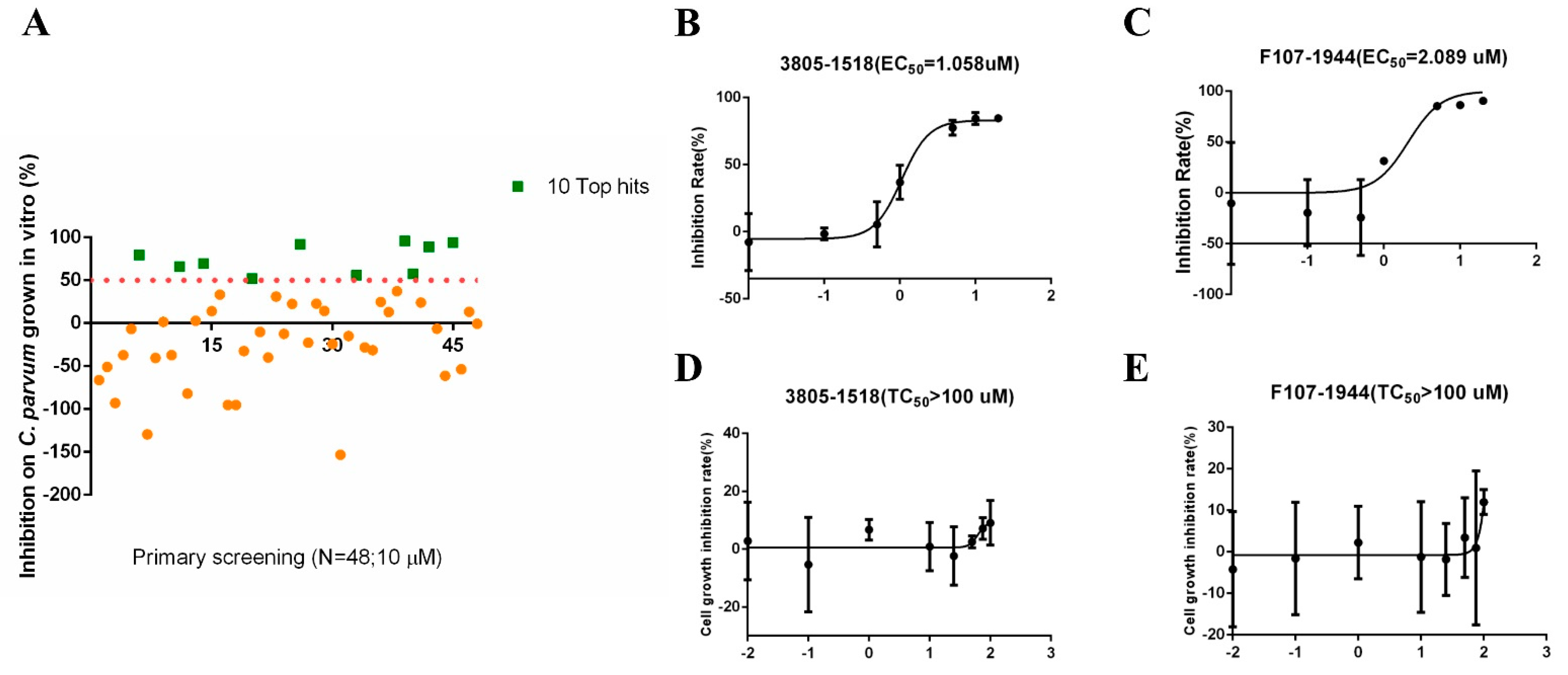
The molecular docking of the simulated structure of INS-16 identified 100 potential inhibitors of the metalloproteinase. Among them, 48 compounds were evaluated for in vitro effects on the invasion and growth of C. parvum in HCT-8 cells using a qRT-PCR assay (Table 1). Ten of the compounds showed >50% growth inhibition at the concentration of 10 µM used in the initial evaluation (Figure 5A). The efficacy of these was further assessed in dose–response experiments. Among them, 3805-1518 and F107-1944 had inhibition rates of over 80% at the concentration of 5 µM, with EC50 values of 1.058 µM and 2.089 µM, respectively (Figure 5B,C). The two compounds displayed low cytotoxicity on HCT-8 cells, with TC50 values > 100 µM (Figure 5D,E).

Table 1.
Candidate inhibitors selected based on molecular docking of INS-16 of Cryptosporidium parvum.
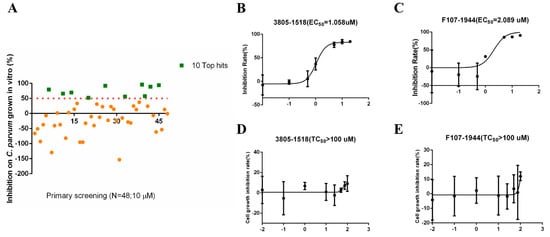
Figure 5.
Inhibitory efficacy of candidate INS-16 inhibitors on the development of Cryptosporidium parvum in HCT-8 cells. (A) Efficacy of all 48 compounds at 10 µM in primary evaluations. Ten compounds with high levels of efficacy (>50%) are marked as green squares. Thirty-eight compounds with low levels of efficacy (<50%) are marked as yellow dots. (B,C) Dose–response curves of compound 3805-1518 and F107-1944 on C. parvum growth. The two compounds can inhibit C. parvum growth by 50% at the concentration of 1.058 µM and 2.089 µM, respectively. The anti-cryptosporidial activities were determined by using qRT-PCR. (D,E) Dose–response curves of compound 3805-1518 and F107-1944 on HCT-8 cell growth. The maximum used on HCT-8 cells without any inhibitory effect above 100 µM for 3805-1518 and F107-1944. The data shown are means ± SD (n ≥ 3) from one representative of at least three independent experiments.
3. Discussion
In this study, a comparative study of INS-16 was conducted together with INS-15 expressed by a paralogous gene. The results obtained indicated that despite the high sequence identity and similar gene expression, the two M16 metalloproteinases appear to be expressed in different organelles of the pathogen, and therefore have different biological functions. Previously, several INS members of C. parvum have been characterized, including INS1, INS4, INS5, INS6, INS20-19, INS-21, and INS-23 [18,20,21,22,23]. In agreement with data generated from the present study, they seemingly have diverse expression patterns and biological functions.
INS-16, like insulin-degrading enzyme (IDE) in humans, appears to be a classical zinc metalloproteinase that is proteolytically processed before maturation. Domain analysis shows that INS-16 contains one active domain and three inactive or middle domains. This special structure allows the N-terminal zinc-binding active domain of classical zinc metalloproteinase to be connected to the C-terminal domain, forming a closed proteolytic chamber to exert activity [24,25]. In the Western blot analysis, it was shown that INS-16-specific antibodies recognize a ~60 kDa product in C. parvum sporozoites, which is much smaller than the recombinant protein expressed in E. coli. This proteolytic processing appears to be common in INS of C. parvum. In previous studies, several products of different sizes were observed in the detection of INS4, INS6, INS-15, and INS20-19 in crude extracts of sporozoites despite the fact that some Cryptosporidium INS do not have four domains. This was attributed to the presence of several putative SΦX(E/D) cleavage sites in the sequences [18,19,21]. The proteolytic processing of INS-16 is similar to two toxolysins in Toxoplasma gondii. The Western blot analysis of native TLN4 showed that TLN4 antibodies principally recognized a ~55 kDa product in tachyzoite lysate, which contained the active domain and the first inactive domain of TLN4 [16]. Another toxolysin, TLN1, was also shown to go through cleavage at the C-terminal, generating a product smaller than the predicted size. It was believed that the cleavage of the C-terminal domain provided additional flexibility for substrate binding, allowing the enzyme to cleave larger substrates [15].
Although related, INS-16 and INS-15 could play different biological functions in the invasion or development of C. parvum. These two metalloproteinases have significant sequence homology to each other and are encoded by two neighboring genes. Although both genes had the highest expression at 0–2 h of the in vitro infection, the localizations of the protein expression in sporozoites and merozoites are different between the two metalloproteinases. Members of the INS family are located in different organelles and play different functions, as indicted in comparative studies of INS4 and INS6, and INS-21 and INS-23 [21,23]. In immunoelectron microscopy analysis, the expression of INS-16 was mainly detected in dense granules located in the anterior end and middle of sporozoites and merozoites. In contrast, INS-15 expression in sporozoites was mainly confined to areas around the nucleus, as shown in the present (Figure 4A) and previous studies [19]. INS-15, nevertheless, appears to be a dense granule protein present over the entire merozoites of C. parvum (Figure 4B). The only other known dense granule protein of C. parvum, CpClec, is expressed in both sporozoites and merozoites and mediates the infection of C. parvum through Ca2+-dependent binding with sulfated proteoglycans on host intestinal epithelial cells [26]. In other apicomplexan parasites, dense granule proteins are involved in the formation of parasitophorous vacuoles and the modification of the host cell activities [27]. Further studies are needed to elucidate the precise functions of diverse members of the INS family of C. parvum.
INS-16 is highly expressed in macrogamonts and may play a role in the sexual life stage of C. parvum. Indirect immunofluorescence microscopy showed high INS-16 expression in macrogamonts, and this expression pattern was similar but not identical to the previously reported INS1 expression [20]. INS1 expression is more likely located in small vesicles within macrogamonts, while INS-16 is more likely located in the dense granules within macrogamonts. In addition, the transcriptional activity of the ins-16 gene was high at 72 h of in vitro C. parvum infection (Figure S2). Therefore, INS-16 may interact with INS1 or other wall-forming proteins to participate in the development of the sexual stages and oocysts of C. parvum.
Inhibitors of INS-16 can effectively inhibit the growth of C. parvum in vitro, indicating that INS-16 may be an important drug target. Although we failed to obtain the full INS-16 protein with enzymatic activity, based on the structure of human IDE together with known inhibitors, we were able to simulate the active structure of INS-16, leading to the identification of 48 potential inhibitors from the ChemDiv database through molecular docking. Among them, only 3805-1518 and F107-1944 effectively inhibited the growth of C. parvum in vitro without significant cytotoxicity to the host cells. Prior to this, there have been no studies on the inhibitors of C. parvum INS. In Plasmodium falciparum, it was known that piperazine-based hydroxamic acids kill parasites by blocking falcilysin (FLN). These inhibitors can competitively bind to active and substrate recognition sites in the protease therefore inhibit FLN activity [28,29]. At present, the mechanism of INS-16 in the invasion and development of C. parvum is not clear. Further structural and genetic manipulation studies are needed to identify the action mechanism of INS-16 and its candidate inhibitors.
4. Materials and Methods
4.1. Parasite, Host Cells, and In Vitro Infection
Oocysts of the C. parvum IOWA isolates were purchased from Waterborne, Inc. (New Orleans, LA, USA), stored at 4 °C, and used within three months. For infection experiments, oocysts were treated with 0.5% sodium hypochlorite on ice for 10 min and washed three times with PBS via centrifugation at 13,200× g for 3 min. The treated oocysts were resuspended with RPMI 1640 culture medium for in vitro infection. HCT-8 cells (ATCC, CCL-244, Chinese Academy of Sciences Shanghai Branch) were seeded into 12-well plates, cultured to ~80% confluence, and inoculated with RPMI 1640 culture medium containing C. parvum oocysts. Free sporozoites were obtained from the sodium-hypochlorite-treated oocysts via incubation in PBS (pH 7.4) containing 0.25% trypsin and 0.5% taurodeoxycholic acid at 37 °C for 1 h. Free merozoites were collected from RPMI 1640 culture medium harvested from C. parvum-infected HCT-8 culture at 36 h via centrifugation.
4.2. Expression of Full-Length INS-16 and INS-16 Domain I
Domains in INS-16 were predicted using InterPro 88.0 (https://www.ebi.ac.uk/interpro/ (accessed on 10 March 2022)) and Pfam 31.0 (https://pfam.xfam.org/ (accessed on 10 March 2022)). Cryptosporidium genomic DNA was extracted from oocysts by using the QIAamp DNA MINI kit (Qiagen, Dusseldorf, Germany). The full-length cgd3_4270 gene and its domain I sequence were amplified by using the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA). The primers used in the PCR amplification of the full cgd3_4270 gene were 5′-CATGCCATGGAATACAATTCACCACTAATAA-3′ (the NcoI restriction site underlined) and 5ʹ-AAATCTCGAGGATCGCATTAAAAACATCC-3ʹ (the XhoI restriction site underlined), while those for domain I were 5′-GCGGATCCTATATTAAGTTGAAAAATGAACTTGA-3′ (the BamH I restriction site underlined) and 5′-CTCTCGAGAAAACGTGTTGATTTGGAATA-3′ (the XhoI restriction site underlined). The PCR amplification of the full cgd3_4270 gene was performed under the following conditions: 95 °C for 30 s; 34 cycles of 95 °C for 10 s, 55 °C for 10 s, and 72 °C for 2 min; and 72 °C for 5 min. The PCR amplification of the domain I fragment was performed under the following conditions: 95 °C for 30 s; 34 cycles of 95 °C for 10 s, 56 °C for 10 s, and 72 °C for 1 min; and 72 °C for 5 min. The PCR products generated were purified using the MiniBEST Agarose Gel DNA Extraction Kit (TaKaRa, Tokyo, Japan), digested with corresponding enzymes and inserted into the expression vector pET28a (Novagen, Madison, WI, USA). The recombinant plasmids were transformed into E. coli DH5α cells (Weidi Biotech, Shanghai, China) which were subsequently cultured on Luria–Bertani plates with 50 μg/mL kanamycin for 12 h. The recombinant plasmid with the correct sequence was extracted from positive colonies using the MiniBEST Plasmid Purification Kit (TaKaRa) and transformed into BL21-Codon plus (DE3)-RIPL cells (Weidi Biotech). The protein expression was induced by treating the suspension culture with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 18 °C for 6 h. The recombinant protein was purified using Ni-NTA beads (Thermo Fisher Scientific) and analyzed for purity using SDS-PAGE with Coomassie blue G-250 staining.
4.3. Preparation of INS-16 Antibodies
Polyclonal antibodies were prepared through immunizations of rabbits with the recombinant INS-16 domain I and the INS-16-specific peptide (amino acid sequence 38LRKTNNFVLKGKIG51) by GenScript Ltd. (Nanjing, China). They were purified from the immune sera using protein A Sepharose affinity chromatography (Beyotime, Shanghai, China).
4.4. Assessment of the Expression of ins-16 Gene
The relative expression levels of the cgd3_4270 gene during the intracellular development of C. parvum in HCT-8 cells were determined using qRT-PCR analysis as described previously [17]. Total RNA was extracted from C. parvum cultures using an RNeasy Kit (Qiagen), and cDNA was synthesized from it using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The primers of the cgd3_4270 gene used in qPCR were 5′-CGCCAATTCAAAACGGTAAT-3′ and 5′-ATTTCAAATGATGGCCCAAG-3′, while those of Cp18S were 5′-CTCCACCAACTAAGAACGGCC-3′ and 5′-TAGAGATTGGAGGTTGTTCCT-3′. The data presented are the results of three independent experiments performed in duplicate.
4.5. Western Blot Analysis of Cross-Reactivity of Antibodies
Western blot analysis was used to assess the cross-reactivity of INS-16 domain I antibodies, using the recombinant INS-15 domain I and antibodies against the INS-15 domain I from the previous study as the control [19]. The recombinant domain I and its native proteins extracted from sporozoites via boiling were separated with 10% SDS-PAGE and transferred onto a polyvinylidene fluoride (PVDF) membrane (Merck Millipore, Billerica, MA, USA) at 300 mA for 80 min. The membrane was blocked at room temperature with 5% nonfat milk-TBST for 1 h and incubated with primary antibodies (against the domain I or specific peptide of INS-15 and INS-16) diluted 1:1000 in blocking buffer (5% nonfat milk in TBST) at 4 °C overnight. After being washed three times with TBST, the membrane was incubated at room temperature with horseradish peroxidase (HRP)-conjugated goat-anti-rabbit antibodies (Yeasen, Shanghai, China) diluted 1:2500 in TBST for 1 h. The membrane was washed three times, treated with High-sig ECL Western Blotting Substrate (Tanon, Shanghai, China), and analyzed using Tanon 5200 (Tanon).
4.6. Immunofluorescence Assay
Oocysts or sporozoites resuspended in PBS were dried onto slides and fixed with methanol at room temperature for 15 min. For the intracellular stages of C. parvum, infected HCT-8 cells grown on coverslips for 24, 30, and 48 h were washed with PBS and fixed. These slides or coverslips were treated with 0.5% Triton-X for 15 min and blocked for nonspecific binding with 5% BSA for 1 h. After being washed with PBS, the slides or coverslips were incubated sequentially with antibodies against the INS-16 domain I or INS-16-specific peptide and Alexa Fluor 594-conjugated Goat Anti-rabbit IgG (Cell Signaling Technology, Danvers, MA, USA), with the nuclei being counter-stained with 4′6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO, USA). For co-localization, antibodies against the INS-15 domain I and specific peptide were used in some of the immunofluorescence microscopy analyses.
4.7. Immunoelectron Microscopy
Oocysts and free sporozoites were fixed at 4 °C in freshly prepared 4% paraformaldehyde (Leagene, Beijing, China) and 0.1% glutaraldehyde (Leagene) for 60 min. They were processed for immunoelectron microscopy (IEM) as described [23].
4.8. Inhibition of C. parvum Invasion and Growth by Candidate INS-16 Inhibitors
A total of 48 compounds were selected from the ChemDiv database through the molecular docking of INS-16 as described previously [30] (Table 1). The anti-cryptosporidial effect of these compounds at the concentration of 10 µM were assessed by using qRT-PCR in 44 h infection assays as described [31]. At least two technical replicates were used in the qRT-PCR analysis of each RNA preparation. In a secondary analysis of selective compounds, various concentrations (from 10 nM to 20 µM) of the compounds were used to treat C. parvum cultures. The cytotoxicity of 3805-1518 and F107-1944 on HCT-8 cells were assessed using a 24 h MTS assay (Abcam, Cambridge, MA, USA), with OD490 being determined using a microplate analyzer (BioTek, Winooski, VT, USA). These tests were performed in triplicate.
5. Conclusions
In conclusion, the results of the present studies revealed clear differences in the expression and subcellular localizations of two INS members, and while INS-16 is highly expressed in dense granules of sporozoites and in macrogamonts, its expression in merozoites is mostly confined to the apical end, indicating that INS-16 likely exerts multiple functions in the invasion and growth of C. parvum. These observations need validation through characterizations of the protein and its function using genetic tools developed recently for C. parvum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23147617/s1.
Author Contributions
Conceptualization, Y.F. and N.L.; investigation, H.C., R.X. and Y.G.; analysis, Y.L. and Z.Z.; writing—original draft preparation, H.C., R.X. and Y.L.; writing—review and editing, L.X., Y.F. and N.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32150710530 and 32030110), the Laboratory of Lingnan Modern Agriculture Project (NT2021007), the 111 Project (D20008), and the Innovation Team Project of Guangdong University (2019KCXTD001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, and further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Checkley, W.; White, A.C., Jr.; Jaganath, D.; Arrowood, M.J.; Chalmers, R.M.; Chen, X.M.; Fayer, R.; Griffiths, J.K.; Guerrant, R.L.; Hedstrom, L.; et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015, 15, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Efstratiou, A.; Ongerth, J.; Karanis, P. Evolution of monitoring for Giardia and Cryptosporidium in water. Water Res. 2017, 123, 96–112. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Feng, Y.; Xiao, L. An update on zoonotic Cryptosporidium species and genotypes in humans. Animals 2021, 11, 3307. [Google Scholar] [CrossRef]
- Chavez, M.A.; White, A.C., Jr. Novel treatment strategies and drugs in development for cryptosporidiosis. Expert Rev. Anti. Infect. Ther. 2018, 16, 655–661. [Google Scholar] [CrossRef]
- Guerin, A.; Striepen, B. The biology of the intestinal intracellular parasite Cryptosporidium. Cell Host Microbe 2020, 28, 509–515. [Google Scholar] [CrossRef]
- Hunter, C.A.; Sibley, L.D. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat. Rev. Microbiol. 2012, 10, 766–778. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Tang, K.; Rowe, L.A.; Li, N.; Roellig, D.M.; Knipe, K.; Frace, M.; Yang, C.; Feng, Y.; Xiao, L. Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. BMC Genom. 2015, 16, 320. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Roellig, D.M.; Guo, Y.; Li, N.; Frace, M.A.; Tang, K.; Zhang, L.; Feng, Y.; Xiao, L. Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genom. 2016, 17, 1006. [Google Scholar] [CrossRef] [Green Version]
- Becker, A.B.; Roth, R.A. An unusual active site identified in a family of zinc metalloendopeptidases. Proc. Natl. Acad. Sci. USA 1992, 89, 3835–3839. [Google Scholar] [CrossRef] [Green Version]
- Kurochkin, I.V.; Guarnera, E.; Berezovsky, I.N. Insulin-degrading enzyme in the fight against Alzheimer's disease. Trends Pharmacol. Sci. 2018, 39, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Maianti, J.P.; Tan, G.A.; Vetere, A.; Welsh, A.J.; Wagner, B.K.; Seeliger, M.A.; Liu, D.R. Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme. Nat. Chem. Biol. 2019, 15, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Murata, C.E.; Goldberg, D.E. Plasmodium falciparum falcilysin: A metalloprotease with dual specificity. J. Biol. Chem. 2003, 278, 38022–38028. [Google Scholar] [CrossRef] [Green Version]
- Murata, C.E.; Goldberg, D.E. Plasmodium falciparum falcilysin: An unprocessed food vacuole enzyme. Mol. Biochem. Parasitol. 2003, 129, 123–126. [Google Scholar] [CrossRef] [Green Version]
- Hajagos, B.E.; Turetzky, J.M.; Peng, E.D.; Cheng, S.J.; Ryan, C.M.; Souda, P.; Whitelegge, J.P.; Lebrun, M.; Dubremetz, J.F.; Bradley, P.J. Molecular dissection of novel trafficking and processing of the Toxoplasma gondii rhoptry metalloprotease toxolysin-1. Traffic 2012, 13, 292–304. [Google Scholar] [CrossRef] [Green Version]
- Laliberte, J.; Carruthers, V.B. Toxoplasma gondii toxolysin 4 is an extensively processed putative metalloproteinase secreted from micronemes. Mol. Biochem. Parasitol. 2011, 177, 49–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauzy, M.J.; Enomoto, S.; Lancto, C.A.; Abrahamsen, M.S.; Rutherford, M.S. The Cryptosporidium parvum transcriptome during in vitro development. PLoS ONE 2012, 7, e31715. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wang, Y.; Wu, H.; Li, N.; Jiang, J.; Guo, Y.; Feng, Y.; Xiao, L. Characterization of a species-specific insulinase-like protease in Cryptosporidium parvum. Front. Microbiol. 2019, 10, 354. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Guo, Y.; Li, N.; Zhang, Q.; Wu, H.; Ryan, U.; Feng, Y.; Xiao, L. Characterization of INS-15, a metalloprotease potentially involved in the invasion of Cryptosporidium parvum. Microorganisms 2019, 7, 452. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Feng, Y.; Xiao, L.; Sibley, L.D. Insulinase-like protease 1 contributes to macrogamont formation in Cryptosporidium parvum. mBio 2021, 12, e03405-20. [Google Scholar] [CrossRef]
- He, W.; Lai, C.; Yang, F.; Li, Y.; Li, N.; Guo, Y.; Zhang, Z.; Xiao, L.; Feng, Y. Comparative study of two insulinlike proteases in Cryptosporidium parvum. Microorganisms 2021, 9, 861. [Google Scholar] [CrossRef] [PubMed]
- Ni, N.; Jia, R.; Guo, Y.; Li, N.; Wu, H.; Feng, Y.; Xiao, L. Expression and functional studies of INS-5, an insulinase-like protein in Cryptosporidium parvum. Front. Microbiol. 2020, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Lai, C.; Yang, F.; Zhang, Q.; Li, N.; Guo, Y.; Xiao, L.; Feng, Y. Preliminary characterization of two small insulinase-like proteases in Cryptosporidium parvum. Front. Microbiol. 2021, 12, 651512. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Kuo, W.L.; Yousef, M.; Rosner, M.R.; Tang, W.J. The C-terminal domain of human insulin degrading enzyme is required for dimerization and substrate recognition. Biochem. Biophys. Res. Commun. 2006, 343, 1032–1037. [Google Scholar] [CrossRef]
- Shen, Y.; Joachimiak, A.; Rosner, M.R.; Tang, W.J. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature 2006, 443, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Ocana, J.; Maravilla, P.; Olivo-Diaz, A. Interaction between human mucins and parasite glycoproteins: The role of lectins and glycosidases in colonization by intestinal protozoa. Rev. Inst. Med. Trop. Sao Paulo 2020, 62, e64. [Google Scholar] [CrossRef]
- Panas, M.W.; Boothroyd, J.C. Seizing control: How dense granule effector proteins enable Toxoplasma to take charge. Mol. Microbiol. 2021, 115, 466–477. [Google Scholar] [CrossRef]
- Chance, J.P.; Fejzic, H.; Hernandez, O.; Istvan, E.S.; Andaya, A.; Maslov, N.; Aispuro, R.; Crisanto, T.; Nguyen, H.; Vidal, B.; et al. Development of piperazine-based hydroxamic acid inhibitors against falcilysin, an essential malarial protease. Bioorg. Med. Chem. Lett. 2018, 28, 1846–1848. [Google Scholar] [CrossRef]
- Kahlon, G.; Lira, R.; Masvlov, N.; Pompa, E.; Brar, N.; Eagon, S.; Anderson, M.O.; Andaya, A.; Chance, J.P.; Fejzic, H.; et al. Structure guided development of potent piperazine-derived hydroxamic acid inhibitors targeting falcilysin. Bioorg. Med. Chem. Lett. 2021, 32, 127683. [Google Scholar] [CrossRef]
- Shu, F.; Li, Y.; Chu, W.; Chen, X.; Zhang, Z.; Guo, Y.; Feng, Y.; Xiao, L.; Li, N. Characterization of calcium-dependent protein kinase 2A, a potential drug target against cryptosporidiosis. Front. Microbiol. 2022, 13, 883674. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, F.; Zhu, G. Involvement of host cell integrin alpha2 in Cryptosporidium parvum infection. Infect. Immun. 2012, 80, 1753–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).