Inhibition of eIF6 Activity Reduces Hepatocellular Carcinoma Growth: An In Vivo and In Vitro Study
Abstract
:1. Introduction
2. Results
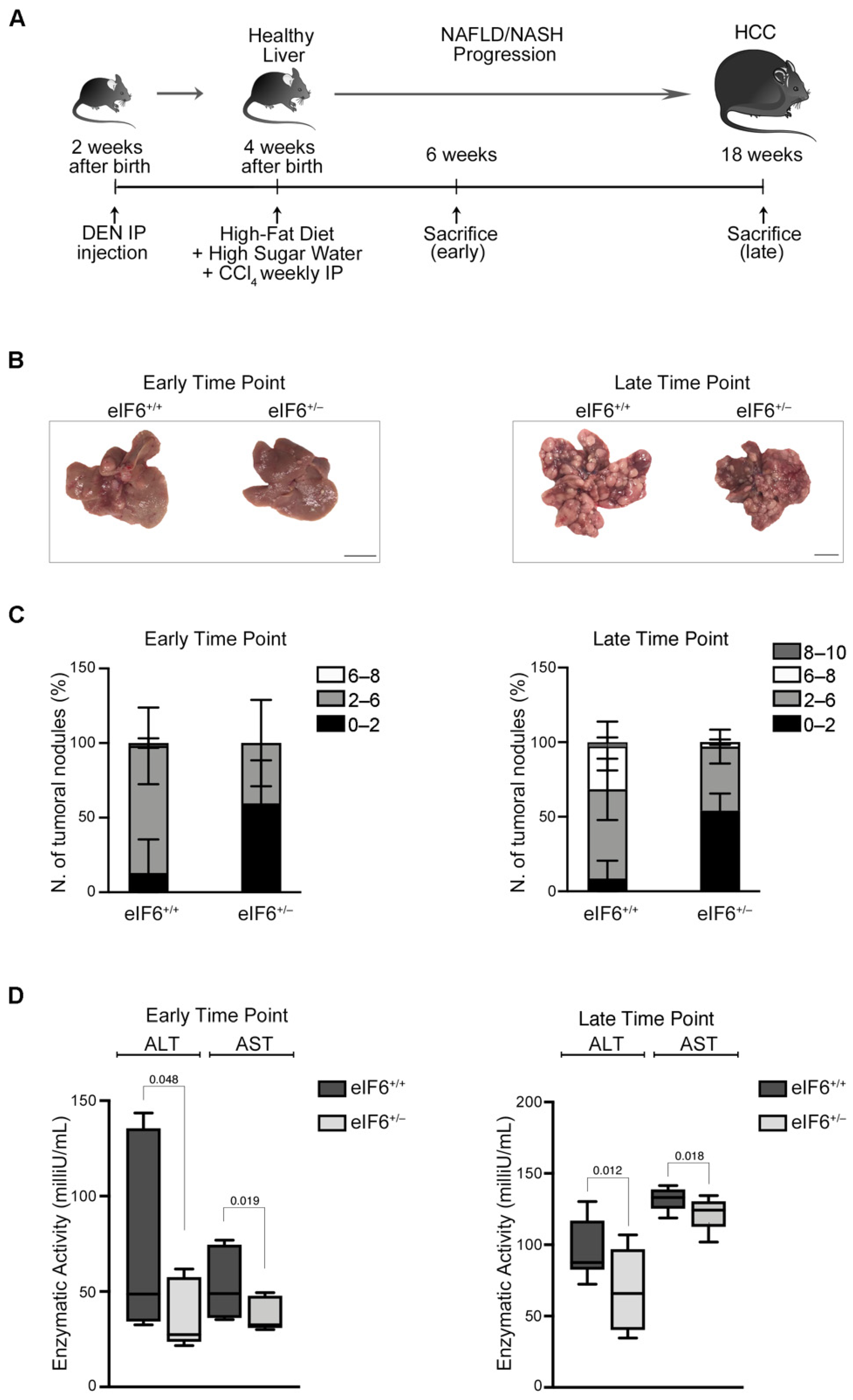
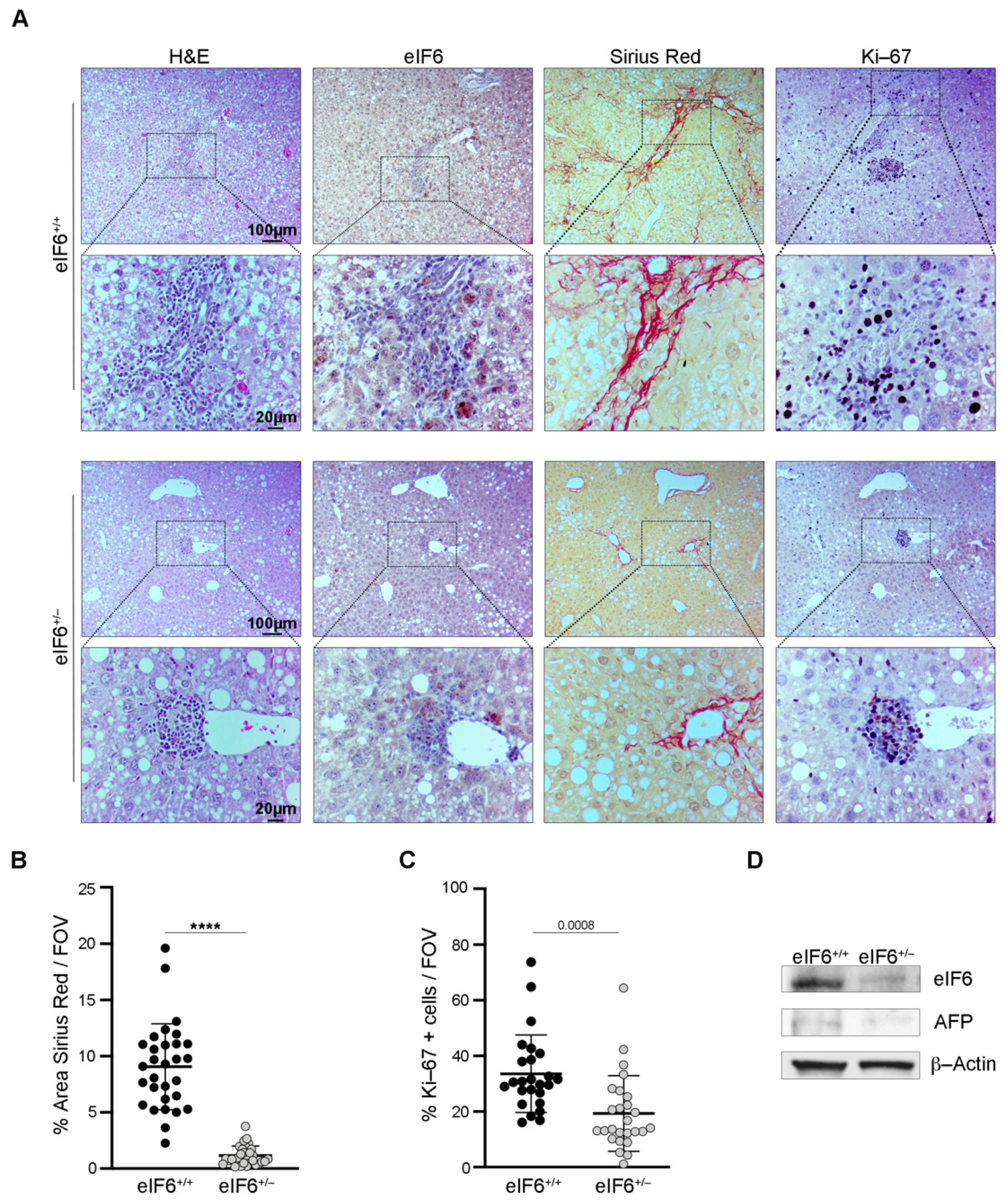
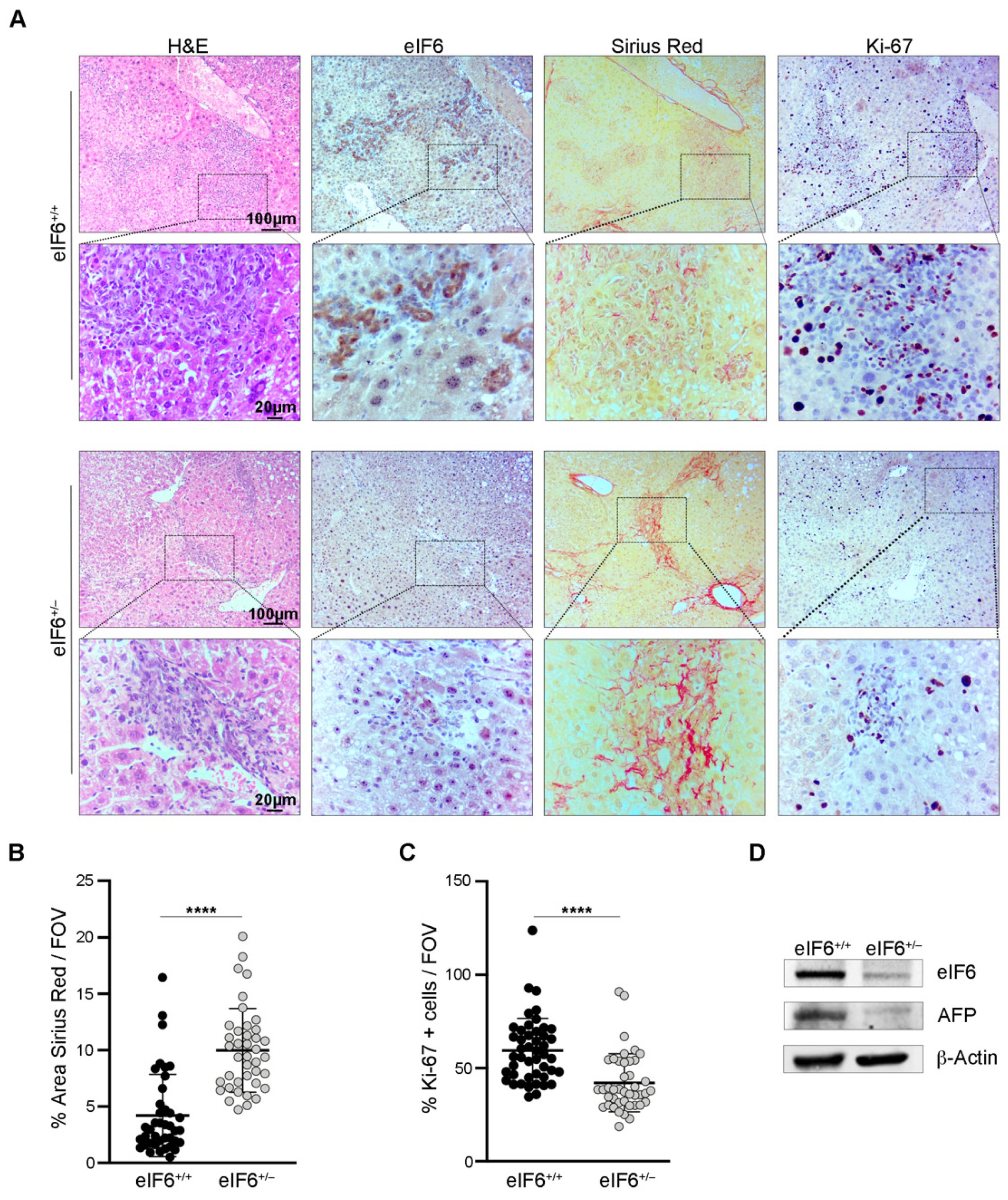
2.1. eIF6 Depletion In Vivo Delays HCC Nodules Formation and Growth without Overt Negative Side Effects
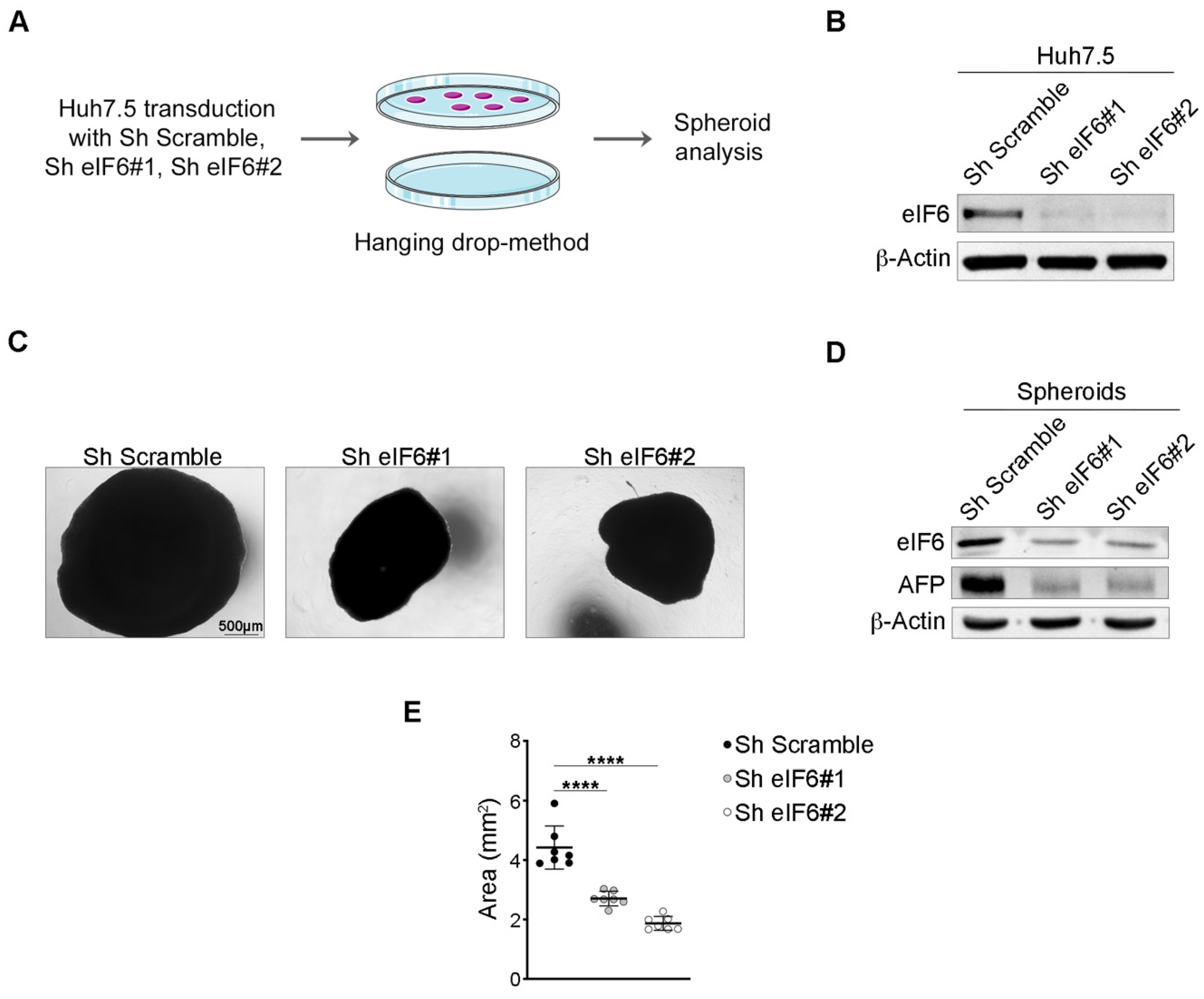
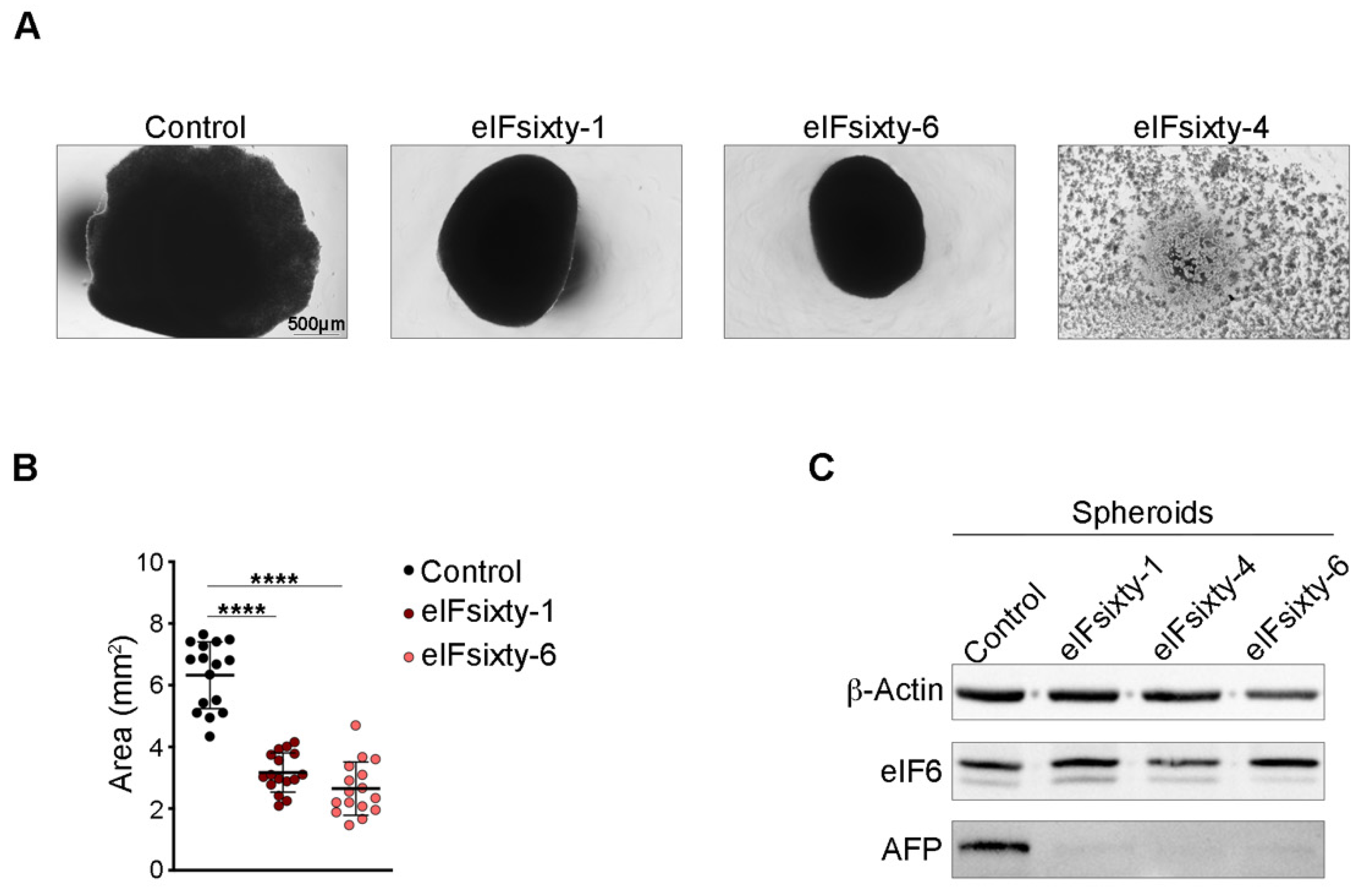
2.2. eIF6 Depletion In Vitro Consistently Reduces HCC Growth in an Established Human 3D Tumor Model
2.3. eIF6 Pharmacological Inhibition Reduces the Growth of HCC Spheroids
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Biochemical Analysis
4.3. Histological Staining and Immunohistochemistry
4.4. iRIA
4.5. eIFsixty-i Compounds
4.6. Cell Cultures
4.7. Polysomal Profiles
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marquardt, J.U.; Andersen, J.B.; Thorgeirsson, S.S. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat. Rev. Cancer 2015, 15, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Pinyol, R.; Torrecilla, S.; Wang, H.; Montironi, C.; Pique-Gili, M.; Torres-Martin, M.; Wei-Qiang, L.; Willoughby, C.E.; Ramadori, P.; Andreu-Oller, C.; et al. Molecular characterisation of hepatocellular carcinoma in patients with non-alcoholic steatohepatitis. J. Hepatol. 2021, 75, 865–878. [Google Scholar] [CrossRef]
- Loreni, F.; Mancino, M.; Biffo, S. Translation factors and ribosomal proteins control tumor onset and progression: How? Oncogene 2014, 33, 2145–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Hershey, J.W.B.; Sonenberg, N.; Mathews, M.B. Principles of Translational Control. Cold Spring Harb. Perspect. Biol. 2019, 11, a032607. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, J.; Sonenberg, N. The Organizing Principles of Eukaryotic Ribosome Recruitment. Annu. Rev. Biochem. 2019, 88, 307–335. [Google Scholar] [CrossRef]
- Gandin, V.; Miluzio, A.; Barbieri, A.M.; Beugnet, A.; Kiyokawa, H.; Marchisio, P.C.; Biffo, S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 2008, 455, 684–688. [Google Scholar] [CrossRef] [Green Version]
- Truitt, M.L.; Conn, C.S.; Shi, Z.; Pang, X.; Tokuyasu, T.; Coady, A.M.; Seo, Y.; Barna, M.; Ruggero, D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 2015, 162, 59–71. [Google Scholar] [CrossRef] [Green Version]
- Miluzio, A.; Beugnet, A.; Grosso, S.; Brina, D.; Mancino, M.; Campaner, S.; Amati, B.; de Marco, A.; Biffo, S. Impairment of cytoplasmic eIF6 activity restricts lymphomagenesis and tumor progression without affecting normal growth. Cancer Cell 2011, 19, 765–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truitt, M.L.; Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 2016, 16, 288–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Shahbazian, F.M.; Jacobs, M.; Lajtha, A. Rates of protein synthesis in brain and other organs. Int. J. Dev. Neurosci. 1987, 5, 39–42. [Google Scholar] [CrossRef]
- Samarin, J.; Laketa, V.; Malz, M.; Roessler, S.; Stein, I.; Horwitz, E.; Singer, S.; Dimou, E.; Cigliano, A.; Bissinger, M.; et al. PI3K/AKT/mTOR-dependent stabilization of oncogenic far-upstream element binding proteins in hepatocellular carcinoma cells. Hepatology 2016, 63, 813–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Cigliano, A.; Jiang, L.; Li, X.; Fan, B.; Pilo, M.G.; Liu, Y.; Gui, B.; Sini, M.; Smith, J.W.; et al. 4EBP1/eIF4E and p70S6K/RPS6 axes play critical and distinct roles in hepatocarcinogenesis driven by AKT and N-Ras proto-oncogenes in mice. Hepatology 2015, 61, 200–213. [Google Scholar] [CrossRef]
- Cao, J.; Zhao, M.; Liu, J.; Zhang, X.; Pei, Y.; Wang, J.; Yang, X.; Shen, B.; Zhang, J. RACK1 Promotes Self-Renewal and Chemoresistance of Cancer Stem Cells in Human Hepatocellular Carcinoma through Stabilizing Nanog. Theranostics 2019, 9, 811–828. [Google Scholar] [CrossRef]
- Duan, F.; Wu, H.; Jia, D.; Wu, W.; Ren, S.; Wang, L.; Song, S.; Guo, X.; Liu, F.; Ruan, Y.; et al. O-GlcNAcylation of RACK1 promotes hepatocellular carcinogenesis. J. Hepatol. 2018, 68, 1191–1202. [Google Scholar] [CrossRef]
- Grosso, S.; Volta, V.; Sala, L.A.; Vietri, M.; Marchisio, P.C.; Ron, D.; Biffo, S. PKCbetaII modulates translation independently from mTOR and through RACK1. Biochem. J. 2008, 415, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Wang, W.; Wang, J.; Feng, J.; Wang, Q.; Jin, J.; Lv, M.; Li, X.; Li, Y.; Ma, Y.; et al. Receptor for activated C kinase 1 promotes hepatocellular carcinoma growth by enhancing mitogen-activated protein kinase kinase 7 activity. Hepatology 2013, 57, 140–151. [Google Scholar] [CrossRef]
- Ruan, Y.; Sun, L.; Hao, Y.; Wang, L.; Xu, J.; Zhang, W.; Xie, J.; Guo, L.; Zhou, L.; Yun, X.; et al. Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J. Clin. Investig. 2012, 122, 2554–2566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceci, M.; Gaviraghi, C.; Gorrini, C.; Sala, L.A.; Offenhauser, N.; Marchisio, P.C.; Biffo, S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 2003, 426, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Golob-Schwarzl, N.; Krassnig, S.; Toeglhofer, A.M.; Park, Y.N.; Gogg-Kamerer, M.; Vierlinger, K.; Schroder, F.; Rhee, H.; Schicho, R.; Fickert, P.; et al. New liver cancer biomarkers: PI3K/AKT/mTOR pathway members and eukaryotic translation initiation factors. Eur. J. Cancer 2017, 83, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liu, S.; Wang, X.; Zheng, X.; Chen, Y.; Shen, H. eIF6 promotes the malignant progression of human hepatocellular carcinoma via the mTOR signaling pathway. J. Transl. Med. 2021, 19, 216. [Google Scholar] [CrossRef] [PubMed]
- Calvisi, D.F.; Wang, C.; Ho, C.; Ladu, S.; Lee, S.A.; Mattu, S.; Destefanis, G.; Delogu, S.; Zimmermann, A.; Ericsson, J.; et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 2011, 140, 1071–1083. [Google Scholar] [CrossRef] [Green Version]
- Biffo, S.; Manfrini, N.; Ricciardi, S. Crosstalks between translation and metabolism in cancer. Curr. Opin. Genet. Dev. 2018, 48, 75–81. [Google Scholar] [CrossRef]
- Brina, D.; Miluzio, A.; Ricciardi, S.; Biffo, S. eIF6 anti-association activity is required for ribosome biogenesis, translational control and tumor progression. Biochim. Biophys. Acta 2015, 1849, 830–835. [Google Scholar] [CrossRef]
- Scagliola, A.; Miluzio, A.; Ventura, G.; Oliveto, S.; Cordiglieri, C.; Manfrini, N.; Cirino, D.; Ricciardi, S.; Valenti, L.; Baselli, G.; et al. Targeting of eIF6-driven translation induces a metabolic rewiring that reduces NAFLD and the consequent evolution to hepatocellular carcinoma. Nat. Commun. 2021, 12, 4878. [Google Scholar] [CrossRef]
- Brina, D.; Miluzio, A.; Ricciardi, S.; Clarke, K.; Davidsen, P.K.; Viero, G.; Tebaldi, T.; Offenhauser, N.; Rozman, J.; Rathkolb, B.; et al. eIF6 coordinates insulin sensitivity and lipid metabolism by coupling translation to transcription. Nat. Commun. 2015, 6, 8261. [Google Scholar] [CrossRef] [Green Version]
- Pesce, E.; Miluzio, A.; Turcano, L.; Minici, C.; Cirino, D.; Calamita, P.; Manfrini, N.; Oliveto, S.; Ricciardi, S.; Grifantini, R.; et al. Discovery and Preliminary Characterization of Translational Modulators that Impair the Binding of eIF6 to 60S Ribosomal Subunits. Cells 2020, 9, 172. [Google Scholar] [CrossRef] [Green Version]
- Ayvaz, I.; Sunay, D.; Sariyar, E.; Erdal, E.; Karagonlar, Z.F. Three-Dimensional Cell Culture Models of Hepatocellular Carcinoma—A Review. J. Gastrointest. Cancer 2021, 52, 1294–1308. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Lee, Y.A.; Fujiwara, N.; Ybanez, M.; Allen, B.; Martins, S.; Fiel, M.I.; Goossens, N.; Chou, H.I.; Hoshida, Y.; et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J. Hepatol. 2018, 69, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Lamerz, R. AFP isoforms and their clinical significance (overview). Anticancer Res. 1997, 17, 2927–2930. [Google Scholar] [PubMed]
- Pesce, E.; Minici, C.; Babetaler, J.; Hurt, E.; Degano, M.; Calamita, P.; Biffo, S. Direct and high throughput (HT) interactions on the ribosomal surface by iRIA. Sci. Rep. 2015, 5, 15401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Koval, A.; Katanaev, V.L. Beyond TNBC: Repositioning of Clofazimine Against a Broad Range of Wnt-Dependent Cancers. Front. Oncol. 2020, 10, 602817. [Google Scholar] [CrossRef]
- Ji, Y.; Shah, S.; Soanes, K.; Islam, M.N.; Hoxter, B.; Biffo, S.; Heslip, T.; Byers, S. Eukaryotic initiation factor 6 selectively regulates Wnt signaling and beta-catenin protein synthesis. Oncogene 2008, 27, 755–762. [Google Scholar] [CrossRef] [Green Version]
- Ye, F.; Zhang, W.; Lu, W.; Xie, Y.; Jiang, H.; Jin, J.; Luo, C. Identification of Novel Inhibitors against Coactivator Associated Arginine Methyltransferase 1 Based on Virtual Screening and Biological Assays. Biomed. Res. Int. 2016, 2016, 7086390. [Google Scholar] [CrossRef] [Green Version]
- Klinge, S.; Voigts-Hoffmann, F.; Leibundgut, M.; Arpagaus, S.; Ban, N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 2011, 334, 941–948. [Google Scholar] [CrossRef] [Green Version]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef]
- Gu, L.; Zhu, Y.; Lin, X.; Tan, X.; Lu, B.; Li, Y. Stabilization of FASN by ACAT1-mediated GNPAT acetylation promotes lipid metabolism and hepatocarcinogenesis. Oncogene 2020, 39, 2437–2449. [Google Scholar] [CrossRef]
- Li, L.; Pilo, G.M.; Li, X.; Cigliano, A.; Latte, G.; Che, L.; Joseph, C.; Mela, M.; Wang, C.; Jiang, L.; et al. Inactivation of fatty acid synthase impairs hepatocarcinogenesis driven by AKT in mice and humans. J. Hepatol. 2016, 64, 333–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piscaglia, F.; Svegliati-Baroni, G.; Barchetti, A.; Pecorelli, A.; Marinelli, S.; Tiribelli, C.; Bellentani, S.; HCC-NAFLD Italian Study Group. Clinical patterns of hepatocellular carcinoma in nonalcoholic fatty liver disease: A multicenter prospective study. Hepatology 2016, 63, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Miluzio, A.; Ricciardi, S.; Manfrini, N.; Alfieri, R.; Oliveto, S.; Brina, D.; Biffo, S. Translational control by mTOR-independent routes: How eIF6 organizes metabolism. Biochem. Soc. Trans. 2016, 44, 1667–1673. [Google Scholar] [CrossRef] [PubMed]
- Biffo, S.; Sanvito, F.; Costa, S.; Preve, L.; Pignatelli, R.; Spinardi, L.; Marchisio, P.C. Isolation of a novel beta4 integrin-binding protein (p27(BBP)) highly expressed in epithelial cells. J. Biol. Chem. 1997, 272, 30314–30321. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.R.; Kang, H.M.; Ryu, J.W.; Kim, D.S.; Noh, K.H.; Kim, E.S.; Lee, H.J.; Chung, K.S.; Cho, H.S.; Kim, N.S.; et al. Cell Spheroids with Enhanced Aggressiveness to Mimic Human Liver Cancer In Vitro and In Vivo. Sci. Rep. 2017, 7, 10499. [Google Scholar] [CrossRef] [Green Version]
- Gallo, S.; Ricciardi, S.; Manfrini, N.; Pesce, E.; Oliveto, S.; Calamita, P.; Mancino, M.; Maffioli, E.; Moro, M.; Crosti, M.; et al. RACK1 Specifically Regulates Translation through Its Binding to Ribosomes. Mol. Cell. Biol. 2018, 38, e00230-18. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scagliola, A.; Miluzio, A.; Mori, G.; Ricciardi, S.; Oliveto, S.; Manfrini, N.; Biffo, S. Inhibition of eIF6 Activity Reduces Hepatocellular Carcinoma Growth: An In Vivo and In Vitro Study. Int. J. Mol. Sci. 2022, 23, 7720. https://doi.org/10.3390/ijms23147720
Scagliola A, Miluzio A, Mori G, Ricciardi S, Oliveto S, Manfrini N, Biffo S. Inhibition of eIF6 Activity Reduces Hepatocellular Carcinoma Growth: An In Vivo and In Vitro Study. International Journal of Molecular Sciences. 2022; 23(14):7720. https://doi.org/10.3390/ijms23147720
Chicago/Turabian StyleScagliola, Alessandra, Annarita Miluzio, Giada Mori, Sara Ricciardi, Stefania Oliveto, Nicola Manfrini, and Stefano Biffo. 2022. "Inhibition of eIF6 Activity Reduces Hepatocellular Carcinoma Growth: An In Vivo and In Vitro Study" International Journal of Molecular Sciences 23, no. 14: 7720. https://doi.org/10.3390/ijms23147720
APA StyleScagliola, A., Miluzio, A., Mori, G., Ricciardi, S., Oliveto, S., Manfrini, N., & Biffo, S. (2022). Inhibition of eIF6 Activity Reduces Hepatocellular Carcinoma Growth: An In Vivo and In Vitro Study. International Journal of Molecular Sciences, 23(14), 7720. https://doi.org/10.3390/ijms23147720





