Moderate Soil Drying-Induced Alternative Splicing Provides a Potential Novel Approach for the Regulation of Grain Filling in Rice Inferior Spikelets
Abstract
:1. Introduction
2. Results
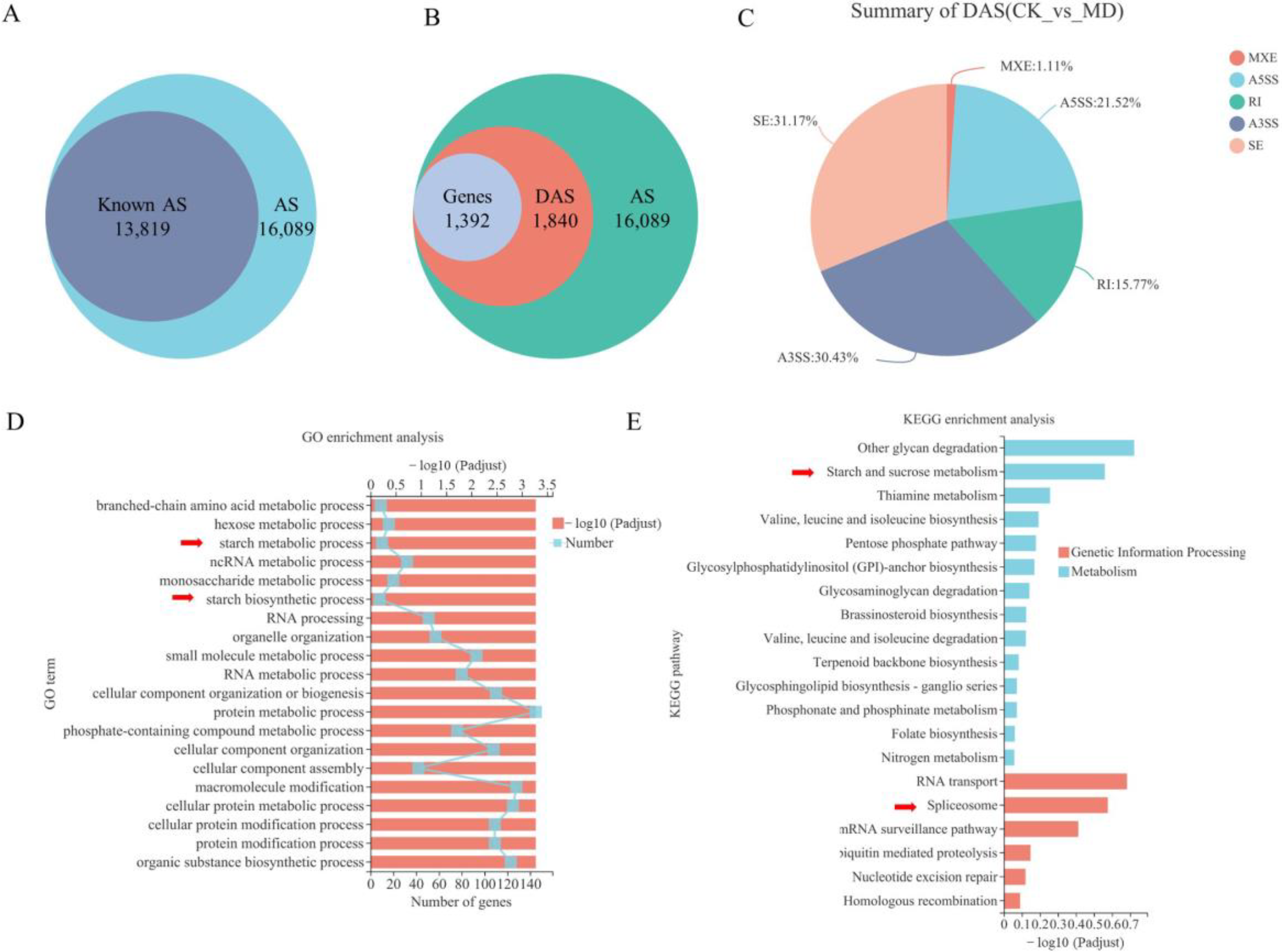
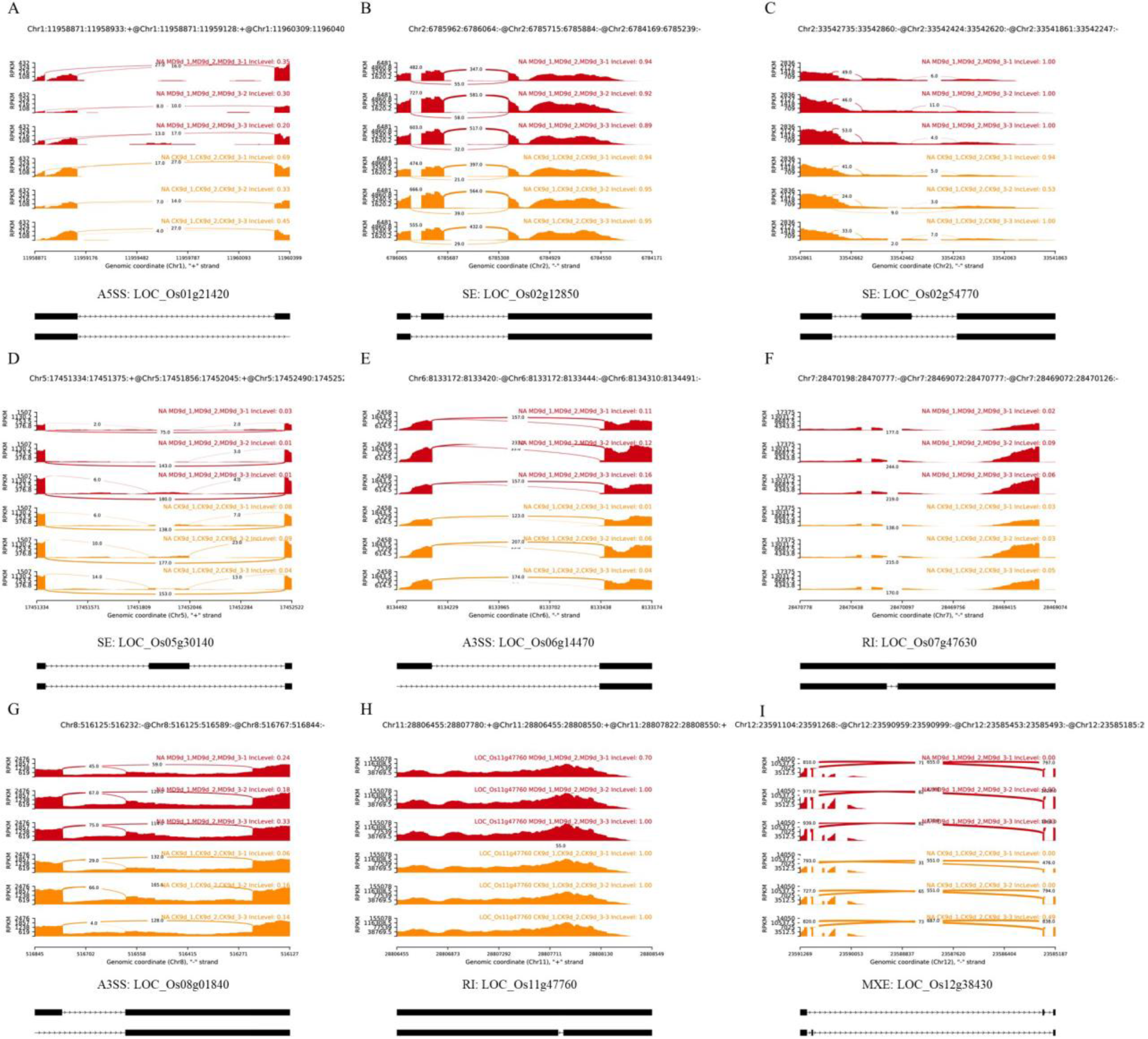
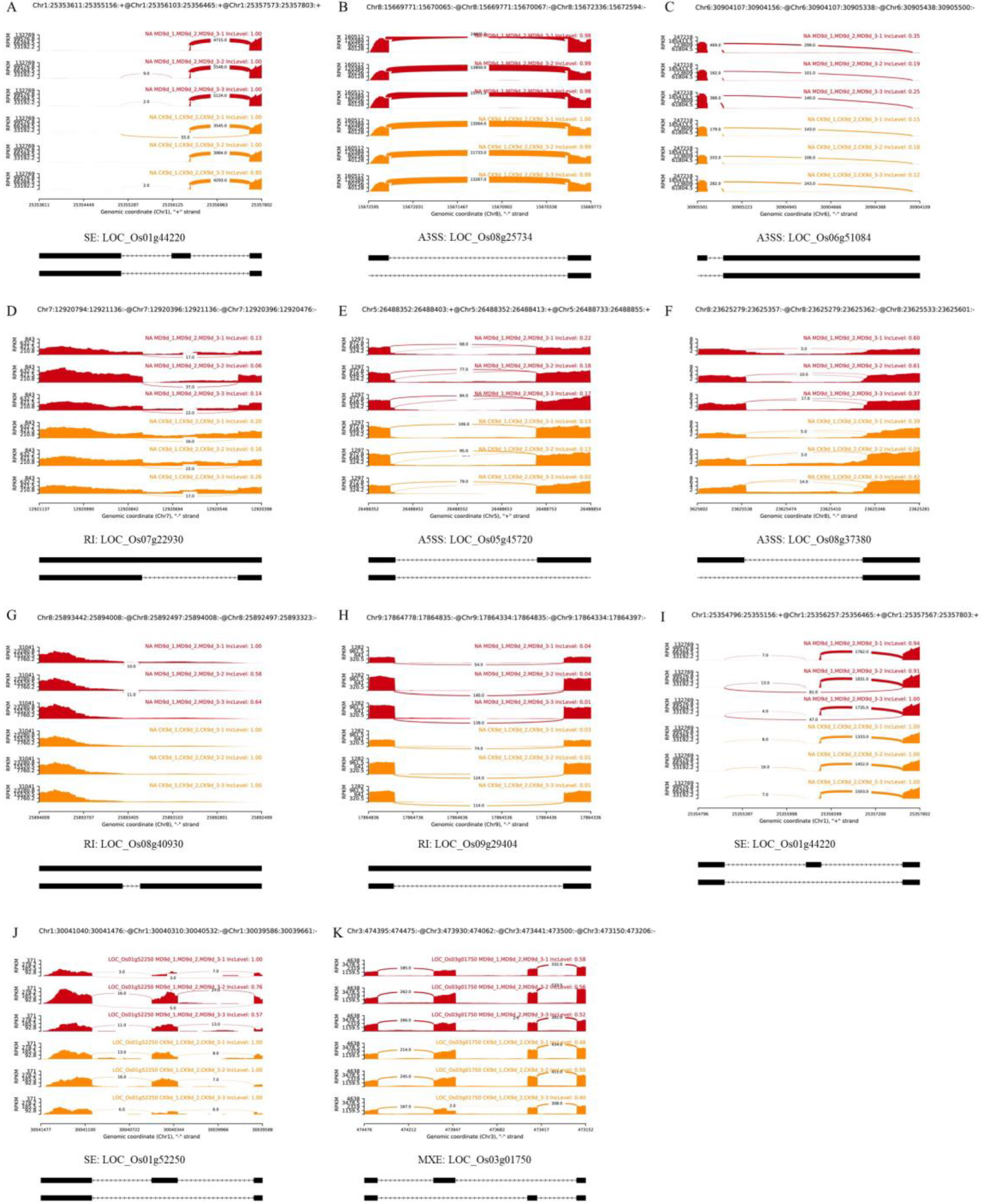
2.1. AS Events of Rice Inferior Spikelets in Response to Moderate Soil Drying during Grain Filling
2.2. MD-Induced AS Might Be Involved in Regulating Grain Filling of Rice Inferior Spikelets
2.3. MD-Induced AS Event Influenced Primary miRNA Expression
2.4. Identification of miRNA Binding Sites Disturbed by AS of Rice Inferior Spikelets under Moderate Soil Drying Post-Anthesis
3. Discussion
3.1. DAS Events in Response to MD Treatment Participate in Starch Biosynthesis and Contribute to Increased Grain Filling in Inferior Spikelets
3.2. AS of Pre-mRNAs of Splicing Factors Increases the Complexity of Inferior Grain Filling Response to MD Treatment
3.3. MD-Induced AS Provides a Mechanism for the Regulation of miRNAProcessing, Leading to Increased Grain Filling in Inferior Spikelets
3.4. AS of Target Genes Increases the Complexity of miRNAs Regulation of Starch Biosynthesis
4. Materials and Methods
4.1. Plant Material and Experimental Design
4.2. RNA-seq Analysis
4.3. Alternative Splicing Analysis
4.4. GO and KEGG Pathway Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Q. Strategies for developing green super rice. Proc. Natl. Acad. Sci. USA 2007, 104, 16402–16409. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Denning, G.; Mew, T. Hybrid rice breeding for super high yield. Denning GL Mew TW Ed. 1998, 3, 10–12. [Google Scholar]
- Yang, J.; Peng, S.; Zhang, Z.; Wang, Z.; Visperas, R.M.; Zhu, Q. Grain and dry matter yields and partitioning of assimilates in japonica/indica hybrid rice. Crop Sci. 2002, 42, 766–772. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J. Grain-filling problem in ‘super’rice. J. Exp. Bot. 2010, 61, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, J. Grain filling of cereals under soil drying. New Phytol. 2006, 169, 223–236. [Google Scholar] [CrossRef]
- Wang, G.-Q.; Li, H.-X.; Feng, L.; Chen, M.-X.; Meng, S.; Ye, N.-H.; Zhang, J. Transcriptomic analysis of grain filling in rice inferior grains under moderate soil drying. J. Exp. Bot. 2019, 70, 1597–1611. [Google Scholar] [CrossRef] [Green Version]
- Teng, Z.; Yu, H.; Wang, G.; Meng, S.; Liu, B.; Yi, Y.; Chen, Y.; Zheng, Q.; Liu, L.; Yang, J. Synergistic interaction between ABA and IAA due to moderate soil drying promotes grain filling of inferior spikelets in rice. Plant J. 2021, 109, 1457–1472. [Google Scholar] [CrossRef]
- Mohapatra, P.K.; Naik, P.K.; Patel, R. Ethylene inhibitors improve dry matter partitioning and development of late flowering spikelets on rice panicles. Funct. Plant Biol. 2000, 27, 311–323. [Google Scholar] [CrossRef]
- Zhang, H.; Tan, G.; Yang, L.; Yang, J.; Zhang, J.; Zhao, B. Hormones in the grains and roots in relation to post-anthesis development of inferior and superior spikelets in japonica/indica hybrid rice. Plant Physiol. Biochem. 2009, 47, 195–204. [Google Scholar] [CrossRef]
- Chen, Y.; Teng, Z.; Yuan, Y.; Yi, Z.; Zheng, Q.; Yu, H.; Lv, J.; Wang, Y.; Duan, M.; Zhang, J. Excessive nitrogen in field-grown rice suppresses grain filling of inferior spikelets by reducing the accumulation of cytokinin and auxin. Field Crops Res. 2022, 283, 108542. [Google Scholar] [CrossRef]
- Nakamura, Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol. 2002, 43, 718–725. [Google Scholar] [CrossRef]
- Hannah, L.C.; James, M. The complexities of starch biosynthesis in cereal endosperms. Curr. Opin. Biotechnol. 2008, 19, 160–165. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, Y.; Yuan, Y.; Liu, B.; Yu, P.; Song, S.; Yi, Y.; Teng, Z.; Yi, Z.; Zhang, J. Post-anthesis saline-alkali stress inhibits grain filling by promoting ethylene production and signal transduction. Food Energy Secur. 2022, e384. [Google Scholar] [CrossRef]
- Pang, Y.; Hu, Y.; Bao, J. Comparative phosphoproteomic analysis reveals the response of starch metabolism to high-temperature stress in rice endosperm. Int. J. Mol. Sci. 2021, 22, 10546. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, Y.; Chen, T.; Zhang, H.; Yang, J.; Zhang, J. Abscisic acid and the key enzymes and genes in sucrose-to-starch conversion in rice spikelets in response to soil drying during grain filling. Planta 2015, 241, 1091–1107. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.; Yuan, L.; Wang, Z.; Yang, J.; Zhang, J. Post-anthesis alternate wetting and moderate soil drying enhances activities of key enzymes in sucrose-to-starch conversion in inferior spikelets of rice. J. Exp. Bot. 2012, 63, 215–227. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-X.; Zhu, F.-Y.; Wang, F.-Z.; Ye, N.-H.; Gao, B.; Chen, X.; Zhao, S.-S.; Fan, T.; Cao, Y.-Y.; Liu, T.-Y. Alternative splicing and translation play important roles in hypoxic germination in rice. J. Exp. Bot. 2019, 70, 817–833. [Google Scholar] [CrossRef]
- Muñoz, M.J.; Santangelo, M.S.P.; Paronetto, M.P.; de la Mata, M.; Pelisch, F.; Boireau, S.; Glover-Cutter, K.; Ben-Dov, C.; Blaustein, M.; Lozano, J.J. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 2009, 137, 708–720. [Google Scholar] [CrossRef] [Green Version]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef]
- Yan, K.; Liu, P.; Wu, C.-A.; Yang, G.-D.; Xu, R.; Guo, Q.-H.; Huang, J.-G.; Zheng, C.-C. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 2012, 48, 521–531. [Google Scholar] [CrossRef] [Green Version]
- Marrs, K.A.; Walbot, V. Expression and RNA splicing of the maize glutathione S-transferase Bronze2 gene is regulated by cadmium and other stresses. Plant Physiol. 1997, 113, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-X.; Zhang, Y.; Fernie, A.R.; Liu, Y.-G.; Zhu, F.-Y. SWATH-MS-based proteomics: Strategies and applications in plants. Trends Biotechnol. 2021, 39, 433–437. [Google Scholar] [CrossRef]
- Quesada, V.; Macknight, R.; Dean, C.; Simpson, G.G. Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 2003, 22, 3142–3152. [Google Scholar] [CrossRef] [Green Version]
- Isshiki, M.; Morino, K.; Nakajima, M.; Okagaki, R.J.; Wessler, S.R.; Izawa, T.; Shimamoto, K. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 1998, 15, 133–138. [Google Scholar] [CrossRef] [Green Version]
- Isshiki, M.; Nakajima, M.; Satoh, H.; Shimamoto, K. dull: Rice mutants with tissue-specific effects on the splicing of the waxy pre-mRNA. Plant J. 2000, 23, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Tian, Y.; Chi, W.; Zhang, H.; Yu, J.; Chen, G.; Wu, W.; Jiang, X.; Wang, S.; Lin, Z. Alternative splicing of OsGS1; 1 affects nitrogen-use efficiency, grain development, and amylose content in rice. Plant J. 2022, 110, 1751–1762. [Google Scholar] [CrossRef]
- Berezikov, E.; Chung, W.-J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian mirtron genes. Molecular cell 2007, 28, 328–336. [Google Scholar] [CrossRef] [Green Version]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008, 15, 902–909. [Google Scholar] [CrossRef]
- Hirsch, J.; Lefort, V.; Vankersschaver, M.; Boualem, A.; Lucas, A.; Thermes, C.; d’Aubenton-Carafa, Y.; Crespi, M. Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 2006, 140, 1192–1204. [Google Scholar] [CrossRef] [Green Version]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zhang, B. MicroRNAs in control of plant development. J. Cell. Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, H.; Li, L. Alternative mRNA processing increases the complexity of microRNA-based gene regulation in Arabidopsis. Plant J. 2012, 70, 421–431. [Google Scholar] [CrossRef]
- Teng, Z.; Chen, Y.; Yuan, Y.; Peng, Y.; Yi, Y.; Yu, H.; Yi, Z.; Yang, J.; Peng, Y.; Duan, M.; et al. Identification of microRNAs regulating grain filling of rice inferior spikelets in response to moderate soil drying post-anthesis. Crop J. 2021. [Google Scholar] [CrossRef]
- Zhao, Y.F.; Peng, T.; Sun, H.Z.; Teotia, S.; Wen, H.L.; Du, Y.X.; Zhang, J.; Li, J.Z.; Tang, G.L.; Xue, H.W. miR1432-Os ACOT (Acyl-CoA thioesterase) module determines grain yield via enhancing grain filling rate in rice. Plant Biotechnol. J. 2019, 17, 712–723. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-C.; Yu, Y.; Wang, C.-Y.; Li, Z.-Y.; Liu, Q.; Xu, J.; Liao, J.-Y.; Wang, X.-J.; Qu, L.-H.; Chen, F. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848–852. [Google Scholar] [CrossRef]
- Peng, T.; Lv, Q.; Zhang, J.; Li, J.; Du, Y.; Zhao, Q. Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa). J. Exp. Bot. 2011, 62, 4943–4954. [Google Scholar] [CrossRef] [Green Version]
- Isshiki, M.; Tsumoto, A.; Shimamoto, K. The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 2006, 18, 146–158. [Google Scholar] [CrossRef] [Green Version]
- Barta, A.; Kalyna, M.; Reddy, A.S. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 2010, 22, 2926–2929. [Google Scholar] [CrossRef]
- Chen, M.-X.; Zhang, K.-L.; Zhang, M.; Das, D.; Fang, Y.-M.; Dai, L.; Zhang, J.; Zhu, F.-Y. Alternative splicing and its regulatory role in woody plants. Tree Physiol. 2020, 40, 1475–1486. [Google Scholar] [CrossRef]
- Ohdan, T.; Francisco Jr, P.B.; Sawada, T.; Hirose, T.; Terao, T.; Satoh, H.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef] [Green Version]
- Tuncel, A.; Kawaguchi, J.; Ihara, Y.; Matsusaka, H.; Nishi, A.; Nakamura, T.; Kuhara, S.; Hirakawa, H.; Nakamura, Y.; Cakir, B. The rice endosperm ADP-glucose pyrophosphorylase large subunit is essential for optimal catalysis and allosteric regulation of the heterotetrameric enzyme. Plant Cell Physiol. 2014, 55, 1169–1183. [Google Scholar] [CrossRef] [Green Version]
- Ying, S.-Y.; Lin, S.-L. Intronic microRNAs. Biochem. Biophys. Res. Commun. 2005, 326, 515–520. [Google Scholar] [CrossRef]
- Peng, T.; Sun, H.; Qiao, M.; Zhao, Y.; Du, Y.; Zhang, J.; Li, J.; Tang, G.; Zhao, Q. Differentially expressed microRNA cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice. BMC Plant Biol. 2014, 14, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Li, Y.-F.; Sunkar, R.; Zhang, W. SeqTar: An effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucleic Acids Res. 2012, 40, e28. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. 2009, 25, 21–44. [Google Scholar] [CrossRef] [Green Version]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [Green Version]
- Castiglioni, P.; Warner, D.; Bensen, R.J.; Anstrom, D.C.; Harrison, J.; Stoecker, M.; Abad, M.; Kumar, G.; Salvador, S.; D’Ordine, R. Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 2008, 147, 446–455. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Quilichini, T.D.; Zhai, C.; Qin, L.; Nilsen, K.T.; Li, Q.; Sharpe, A.G.; Kochian, L.V.; Zou, J.; Reddy, A.S. Alternative splicing dynamics and evolutionary divergence during embryogenesis in wheat species. Plant Biotechnol. J. 2021, 19, 1624. [Google Scholar] [CrossRef]
- Yu, J.; Miao, J.; Zhang, Z.; Xiong, H.; Zhu, X.; Sun, X.; Pan, Y.; Liang, Y.; Zhang, Q.; Abdul Rehman, R.M. Alternative splicing of Os LG 3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 2018, 16, 1667–1678. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Zhi, L.; Liu, L.; Meng, D.; Su, Q.; Batool, A.; Ji, J.; Song, L.; Zhang, N.; Guo, L. Alternative Splicing of TaGS3 Differentially Regulates Grain Weight and Size in Bread Wheat. Int. J. Mol. Sci. 2021, 22, 11692. [Google Scholar] [CrossRef]
- Xu, H.; Li, X.; Zhang, H.; Wang, L.; Zhu, Z.; Gao, J.; Li, C.; Zhu, Y. High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell Environ. 2020, 43, 1879–1896. [Google Scholar] [CrossRef]
- Lee, S.-K.; Hwang, S.-K.; Han, M.; Eom, J.-S.; Kang, H.-G.; Han, Y.; Choi, S.-B.; Cho, M.-H.; Bhoo, S.H.; An, G. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 531–546. [Google Scholar] [CrossRef]
- Szydlowski, N.; Ragel, P.; Raynaud, S.; Lucas, M.M.; Roldán, I.; Montero, M.; Muñoz, F.J.; Ovecka, M.; Bahaji, A.; Planchot, V. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 2009, 21, 2443–2457. [Google Scholar] [CrossRef] [Green Version]
- Utsumi, Y.; Utsumi, C.; Sawada, T.; Fujita, N.; Nakamura, Y. Functional diversity of isoamylase oligomers: The ISA1 homo-oligomer is essential for amylopectin biosynthesis in rice endosperm. Plant Physiol. 2011, 156, 61–77. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.X.; Zhang, K.L.; Gao, B.; Yang, J.F.; Tian, Y.; Das, D.; Fan, T.; Dai, L.; Hao, G.F.; Yang, G.F. Phylogenetic comparison of 5′ splice site determination in central spliceosomal proteins of the U1-70K gene family, in response to developmental cues and stress conditions. Plant J. 2020, 103, 357–378. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, M.-X.; Zhu, F.-Y.; Zhang, J.; Liu, Y.-G. Emerging functions of plant Serine/Arginine-Rich (SR) proteins: Lessons from animals. Crit. Rev. Plant Sci. 2020, 39, 173–194. [Google Scholar] [CrossRef]
- Brown, J.W.; Marshall, D.F.; Echeverria, M. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008, 13, 335–342. [Google Scholar] [CrossRef]
- Yang, G.; Yan, K.; Wu, B.; Wang, Y.; Gao, Y.; Zheng, C. Genomewide analysis of intronic microRNAs in rice and Arabidopsis. J. Genet. 2012, 91, 313–324. [Google Scholar] [CrossRef]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [Green Version]
- Chandra, T.; Mishra, S.; Panda, B.B.; Sahu, G.; Dash, S.K.; Shaw, B.P. Study of expressions of miRNAs in the spikelets based on their spatial location on panicle in rice cultivars provided insight into their influence on grain development. Plant Physiol. Biochem. 2021, 159, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Campo, S.; Peris-Peris, C.; Siré, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; San Segundo, B. Identification of a novel micro RNA (mi RNA) from rice that targets an alternatively spliced transcript of the N ramp6 (N atural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227. [Google Scholar] [CrossRef]
- Bian, H.; Xie, Y.; Guo, F.; Han, N.; Ma, S.; Zeng, Z.; Wang, J.; Yang, Y.; Zhu, M. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 2012, 196, 149–161. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.-x.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.X.; Mei, L.C.; Wang, F.; Dewayalage, I.K.W.B.; Yang, J.F.; Dai, L.; Yang, G.F.; Gao, B.; Cheng, C.L.; Liu, Y.G. PlantSPEAD: A web resource towards comparatively analysing stress-responsive expression of splicing-related proteins in plant. Plant Biotechnol. J. 2021, 19, 227. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, Z.; Zheng, Q.; Liu, B.; Meng, S.; Zhang, J.; Ye, N. Moderate Soil Drying-Induced Alternative Splicing Provides a Potential Novel Approach for the Regulation of Grain Filling in Rice Inferior Spikelets. Int. J. Mol. Sci. 2022, 23, 7770. https://doi.org/10.3390/ijms23147770
Teng Z, Zheng Q, Liu B, Meng S, Zhang J, Ye N. Moderate Soil Drying-Induced Alternative Splicing Provides a Potential Novel Approach for the Regulation of Grain Filling in Rice Inferior Spikelets. International Journal of Molecular Sciences. 2022; 23(14):7770. https://doi.org/10.3390/ijms23147770
Chicago/Turabian StyleTeng, Zhenning, Qin Zheng, Bohan Liu, Shuan Meng, Jianhua Zhang, and Nenghui Ye. 2022. "Moderate Soil Drying-Induced Alternative Splicing Provides a Potential Novel Approach for the Regulation of Grain Filling in Rice Inferior Spikelets" International Journal of Molecular Sciences 23, no. 14: 7770. https://doi.org/10.3390/ijms23147770
APA StyleTeng, Z., Zheng, Q., Liu, B., Meng, S., Zhang, J., & Ye, N. (2022). Moderate Soil Drying-Induced Alternative Splicing Provides a Potential Novel Approach for the Regulation of Grain Filling in Rice Inferior Spikelets. International Journal of Molecular Sciences, 23(14), 7770. https://doi.org/10.3390/ijms23147770







