MicroRNAs as Potential Biomarkers in the Differential Diagnosis of Lipomatous Tumors and Their Mimics
Abstract
:1. Introduction
2. Results
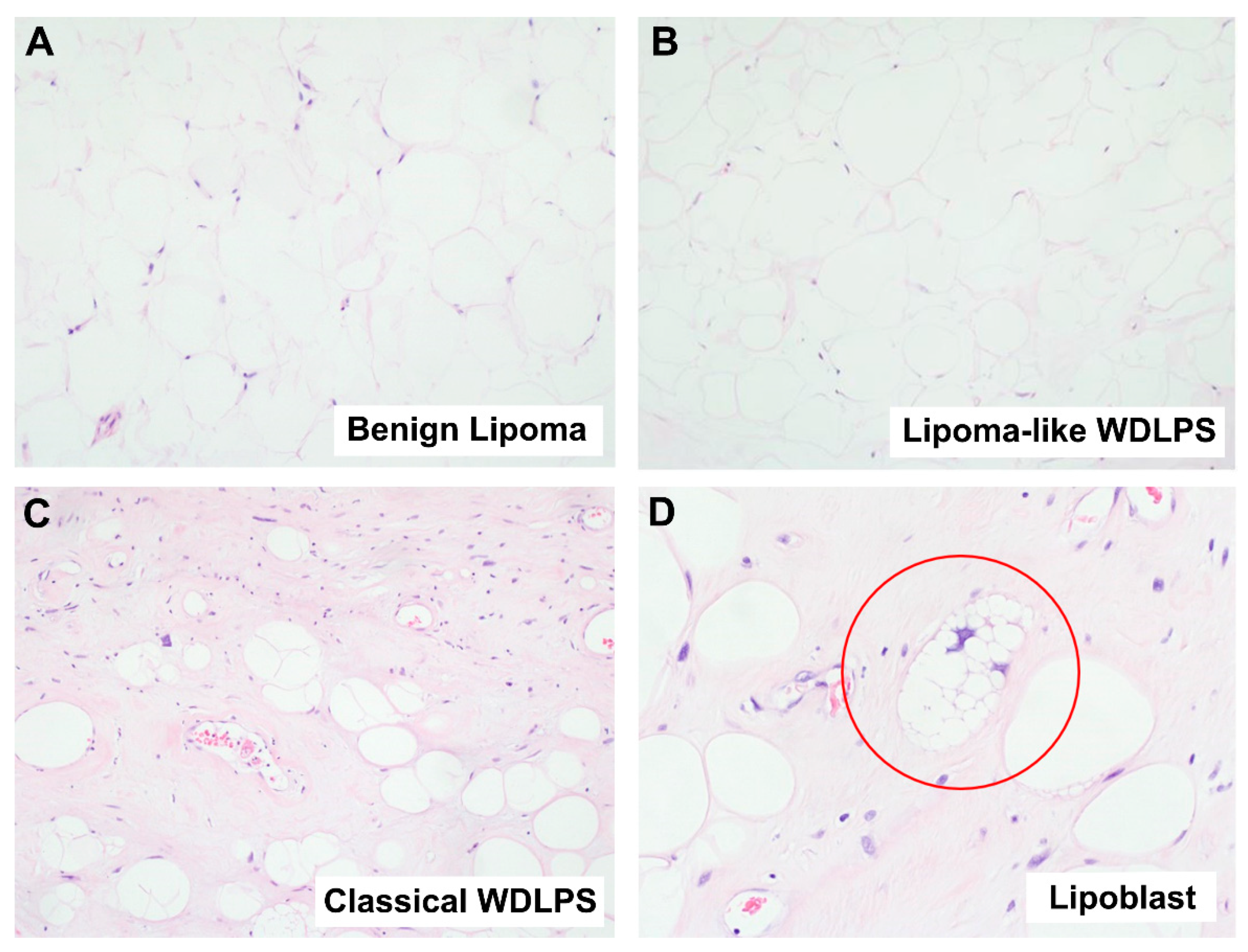
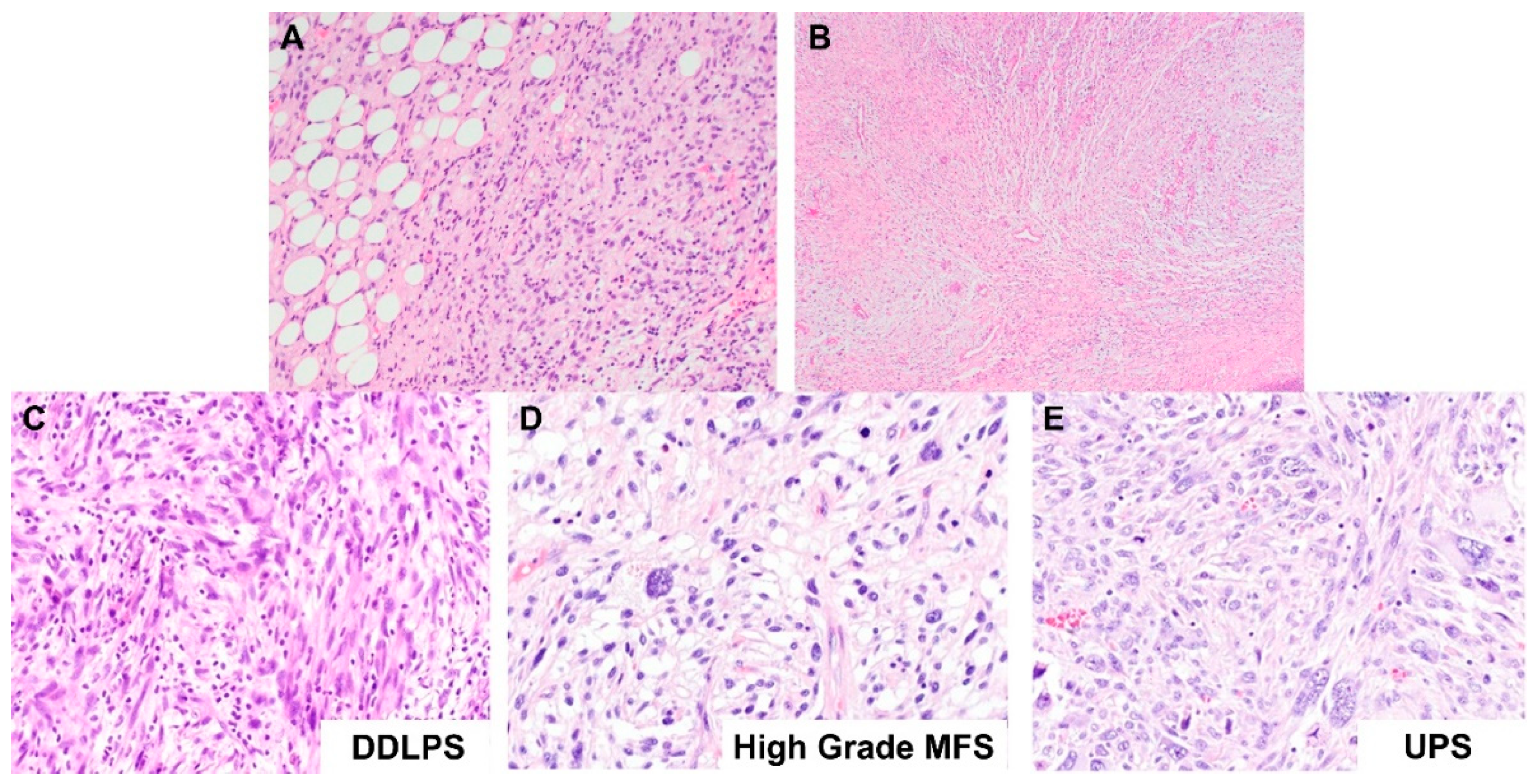
2.1. Diagnostic Challenges of Lipomatous Tumors
2.1.1. Well-Differentiated Lipomatous Tumor
2.1.2. Dedifferentiated Lipomatous Tumor
2.2. miRNAs Expression Profiles Discriminate Benign Lipoma from Liposarcoma (WDLPS and DDLPS) and Other Sarcoma Subtypes (MFS and UPS)
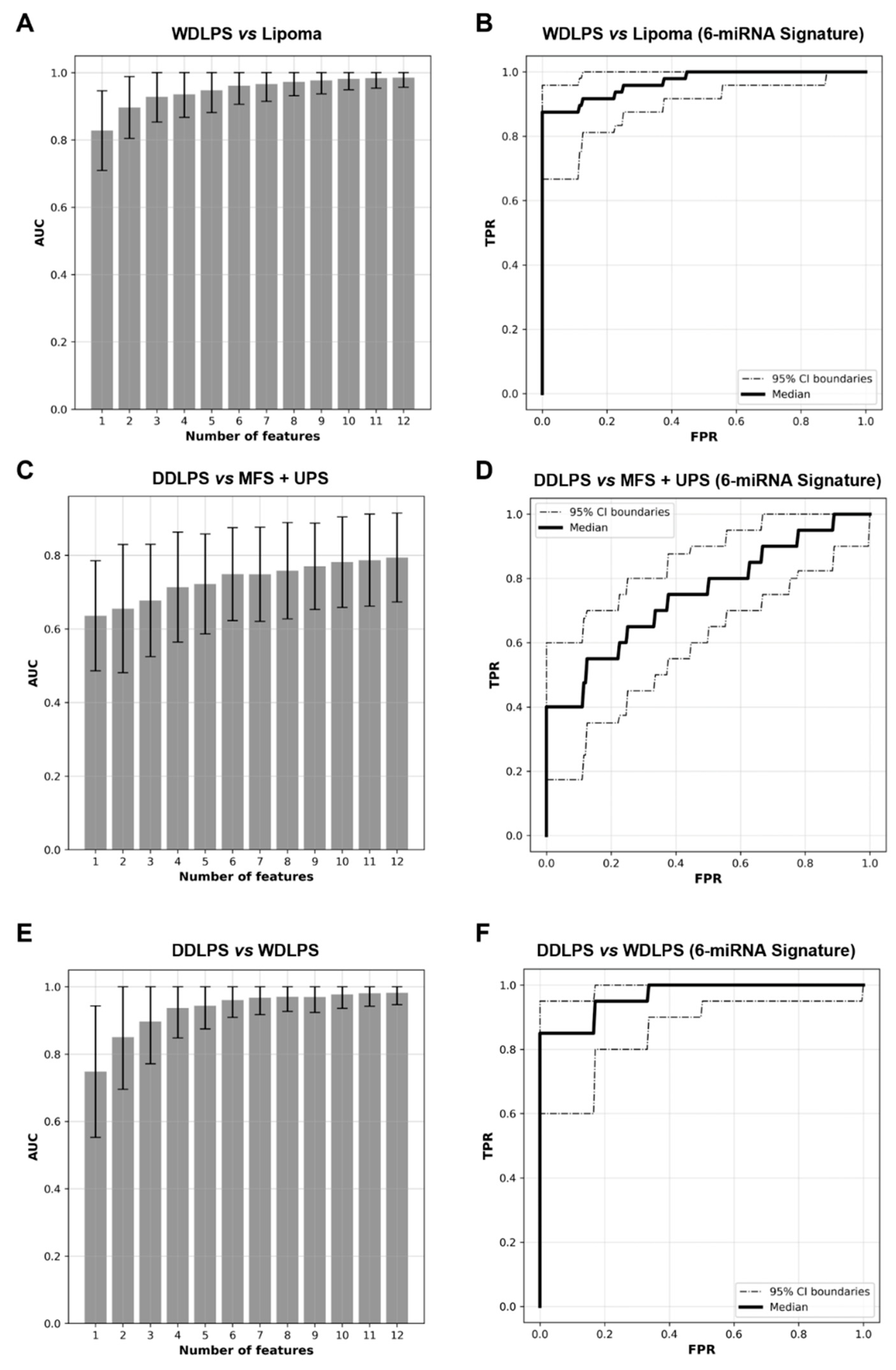
2.3. Expression Levels of Six miRNAs Distinguish Lipoma from WDLPS with High Accuracy
2.4. miRNA Signature Can Distinguish DDLPS from Its Histologic Mimics
2.5. WDLPS and DDLPS Are Associated with Distinct miRNA Expression Profiles
2.6. miRNAs Differentially Expressed between Lipoma and Liposarcoma, and between WDLPS and DDLPS
2.7. Dysregulated Pathways in Liposarcomagenesis—Involvement of PI3K/AKT and MAPK Pathways
3. Discussion
3.1. Ancillary Tests Currently Used in the Diagnosis of Adipocytic Tumors
3.2. miRNAs as Potential Diagnostic Biomarkers
3.3. Biological Relevance of Differentially Regulated miRNAs
3.3.1. miRNAs Differentially Expressed between Lipoma and Liposarcoma
3.3.2. miRNAs Differentially Expressed between WDLPS and DDLPS
3.4. Potential Pathways Involved in Liposarcomagenesis
4. Materials and Methods
4.1. Sample Selection
4.2. Fluorescence In Situ Hybridization (FISH)
4.3. RNA Extraction and miRNA Profiling
4.4. Data Processing and Statistical Analysis
4.5. Feature Selection for Tumor Classification
4.6. Receiver Operating Characteristic (ROC) Curve
4.7. Silico Prediction of the miRNA Regulatory Network
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clark, M.A.; Fisher, C.; Judson, I.; Thomas, J.M. Soft-Tissue Sarcomas in Adults. N. Engl. J. Med. 2005, 353, 701–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO Classification of Tumours Editorial Board. Soft Tissue and Bone Tumours, 5th ed.; World Health Organization: Geneva, Switzerland, 2020; Volume 3, ISBN 978-92-832-4502-5. [Google Scholar]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of Novel Genes Coding for Small Expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis Elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of Hundreds of Conserved and Nonconserved Human MicroRNAs. Nat. Genet. 2005, 37, 766–770. [Google Scholar] [CrossRef]
- Gu, S.; Kay, M.A. How Do MiRNAs Mediate Translational Repression? Silence 2010, 1, 11. [Google Scholar] [CrossRef] [Green Version]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and Cancer—A Brief Overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Tsongalis, G.J.; Calin, G.; Cordelier, P.; Croce, C.; Monzon, F.; Szafranska-Schwarzbach, A.E. MicroRNA Analysis: Is It Ready for Prime Time? Clin. Chem. 2013, 59, 343–347. [Google Scholar] [CrossRef] [Green Version]
- Thway, K. Well-Differentiated Liposarcoma and Dedifferentiated Liposarcoma: An Updated Review. Semin. Diagn. Pathol. 2019, 36, 112–121. [Google Scholar] [CrossRef]
- Thway, K.; Jones, R.L.; Noujaim, J.; Zaidi, S.; Miah, A.B.; Fisher, C. Dedifferentiated Liposarcoma: Updates on Morphology, Genetics, and Therapeutic Strategies. Adv. Anat. Pathol. 2016, 23, 30. [Google Scholar] [CrossRef]
- Demicco, E.G. Molecular Updates in Adipocytic Neoplasms. Semin. Diagn. Pathol. 2019, 36, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Thway, K.; Wang, J.; Swansbury, J.; Min, T.; Fisher, C. Fluorescence In Situ Hybridization for MDM2 Amplification as a Routine Ancillary Diagnostic Tool for Suspected Well-Differentiated and Dedifferentiated Liposarcomas: Experience at a Tertiary Center. Sarcoma 2015, 2015, 812089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, J.; Downs-Kelly, E.; Goldblum, J.R.; Turner, S.; Kulkarni, S.; Tubbs, R.R.; Rubin, B.P.; Skacel, M. Fluorescence in Situ Hybridization for MDM2 Gene Amplification as a Diagnostic Tool in Lipomatous Neoplasms. Mod. Pathol. 2008, 21, 943–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sciot, R. MDM2 Amplified Sarcomas: A Literature Review. Diagnostics 2021, 11, 496. [Google Scholar] [CrossRef]
- Clay, M.R.; Martinez, A.P.; Weiss, S.W.; Edgar, M.A. MDM2 and CDK4 Immunohistochemistry: Should It Be Used in Problematic Differentiated Lipomatous Tumors? A New Perspective. Am. J. Surg. Pathol. 2016, 40, 1647–1652. [Google Scholar] [CrossRef]
- Thway, K.; Flora, R.; Shah, C.; Olmos, D.; Fisher, C. Diagnostic Utility of P16, CDK4, and MDM2 as an Immunohistochemical Panel in Distinguishing Well-Differentiated and Dedifferentiated Liposarcomas From Other Adipocytic Tumors. Am. J. Surg. Pathol. 2012, 36, 462–469. [Google Scholar] [CrossRef]
- Tam, W. The Emergent Role of MicroRNAs in Molecular Diagnostics of Cancer. J. Mol. Diagn. 2008, 10, 411–414. [Google Scholar] [CrossRef] [Green Version]
- Croce, C.M. Causes and Consequences of MicroRNA Dysregulation in Cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA Expression Profiles Classify Human Cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A MicroRNA Expression Signature of Human Solid Tumors Defines Cancer Gene Targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Rosenfeld, N.; Aharonov, R.; Meiri, E.; Rosenwald, S.; Spector, Y.; Zepeniuk, M.; Benjamin, H.; Shabes, N.; Tabak, S.; Levy, A.; et al. MicroRNAs Accurately Identify Cancer Tissue Origin. Nat. Biotechnol. 2008, 26, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Bentwich, I. A Postulated Role for MicroRNA in Cellular Differentiation. FASEB J. 2005, 19, 875–879. [Google Scholar] [CrossRef] [Green Version]
- Meiri, E.; Mueller, W.C.; Rosenwald, S.; Zepeniuk, M.; Klinke, E.; Edmonston, T.B.; Werner, M.; Lass, U.; Barshack, I.; Feinmesser, M.; et al. A Second-Generation MicroRNA-Based Assay for Diagnosing Tumor Tissue Origin. Oncologist 2012, 17, 801–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barshack, I.; Lithwick-Yanai, G.; Afek, A.; Rosenblatt, K.; Tabibian-Keissar, H.; Zepeniuk, M.; Cohen, L.; Dan, H.; Zion, O.; Strenov, Y.; et al. MicroRNA Expression Differentiates between Primary Lung Tumors and Metastases to the Lung. Pathol. Res. Pract. 2010, 206, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Gilad, S.; Lithwick-Yanai, G.; Barshack, I.; Benjamin, S.; Krivitsky, I.; Edmonston, T.B.; Bibbo, M.; Thurm, C.; Horowitz, L.; Huang, Y.; et al. Classification of the Four Main Types of Lung Cancer Using a MicroRNA-Based Diagnostic Assay. J. Mol. Diagn. 2012, 14, 510–517. [Google Scholar] [CrossRef]
- Lebanony, D.; Benjamin, H.; Gilad, S.; Ezagouri, M.; Dov, A.; Ashkenazi, K.; Gefen, N.; Izraeli, S.; Rechavi, G.; Pass, H.; et al. Diagnostic Assay Based on Hsa-MiR-205 Expression Distinguishes Squamous From Nonsquamous Non–Small-Cell Lung Carcinoma. J. Clin. Oncol. 2009, 27, 2030–2037. [Google Scholar] [CrossRef]
- Fridman, E.; Dotan, Z.; Barshack, I.; David, M.B.; Dov, A.; Tabak, S.; Zion, O.; Benjamin, S.; Benjamin, H.; Kuker, H.; et al. Accurate Molecular Classification of Renal Tumors Using MicroRNA Expression. J. Mol. Diagn. 2010, 12, 687–696. [Google Scholar] [CrossRef]
- Spector, Y.; Fridman, E.; Rosenwald, S.; Zilber, S.; Huang, Y.; Barshack, I.; Zion, O.; Mitchell, H.; Sanden, M.; Meiri, E. Development and Validation of a MicroRNA-Based Diagnostic Assay for Classification of Renal Cell Carcinomas. Mol. Oncol. 2013, 7, 732–738. [Google Scholar] [CrossRef]
- Kowalik, C.G.; Palmer, D.A.; Sullivan, T.B.; Teebagy, P.A.; Dugan, J.M.; Libertino, J.A.; Burks, E.J.; Canes, D.; Rieger-Christ, K.M. Profiling MicroRNA from Nephrectomy and Biopsy Specimens: Predictors of Progression and Survival in Clear Cell Renal Cell Carcinoma. BJU Int. 2017, 120, 428–440. [Google Scholar] [CrossRef] [Green Version]
- Benjamin, H.; Lebanony, D.; Rosenwald, S.; Cohen, L.; Gibori, H.; Barabash, N.; Ashkenazi, K.; Goren, E.; Meiri, E.; Morgenstern, S.; et al. A Diagnostic Assay Based on MicroRNA Expression Accurately Identifies Malignant Pleural Mesothelioma. J. Mol. Diagn. 2010, 12, 771–779. [Google Scholar] [CrossRef]
- Doleshal, M.; Magotra, A.A.; Choudhury, B.; Cannon, B.D.; Labourier, E.; Szafranska, A.E. Evaluation and Validation of Total RNA Extraction Methods for MicroRNA Expression Analyses in Formalin-Fixed, Paraffin-Embedded Tissues. J. Mol. Diagn. 2008, 10, 203–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bovell, L.; Shanmugam, C.; Katkoori, V.R.; Zhang, B.; Vogtmann, E.; Grizzle, W.E.; Manne, U. MiRNAs Are Stable in Colorectal Cancer Archival Tissue Blocks. Front. Biosci. 2012, 4, 1937–1940. [Google Scholar] [CrossRef]
- Smolle, M.A.; Leithner, A.; Posch, F.; Szkandera, J.; Liegl-Atzwanger, B.; Pichler, M. MicroRNAs in Different Histologies of Soft Tissue Sarcoma: A Comprehensive Review. Int. J. Mol. Sci. 2017, 18, 1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, T.; Kunisada, T.; Takeda, K.; Uotani, K.; Yoshida, A.; Ochiya, T.; Ozaki, T. MicroRNAs in Soft Tissue Sarcomas: Overview of the Accumulating Evidence and Importance as Novel Biomarkers. BioMed Res. Int. 2014, 2014, 592868. [Google Scholar] [CrossRef] [PubMed]
- Drury, R.; Verghese, E.T.; Hughes, T.A. The Roles of MicroRNAs in Sarcomas. J. Pathol. 2012, 227, 385–391. [Google Scholar] [CrossRef]
- Gits, C.M.M.; van Kuijk, P.F.; Jonkers, M.B.E.; Boersma, A.W.M.; Smid, M.; van Ijcken, W.F.; Coindre, J.-M.; Chibon, F.; Verhoef, C.; Mathijssen, R.H.J.; et al. MicroRNA Expression Profiles Distinguish Liposarcoma Subtypes and Implicate MiR-145 and MiR-451 as Tumor Suppressors. Int. J. Cancer 2014, 135, 348–361. [Google Scholar] [CrossRef]
- Ugras, S.; Brill, E.; Jacobsen, A.; Hafner, M.; Socci, N.D.; DeCarolis, P.L.; Khanin, R.; O’Connor, R.; Mihailovic, A.; Taylor, B.S.; et al. Small RNA Sequencing and Functional Characterization Reveals MicroRNA-143 Tumor Suppressor Activity in Liposarcoma. Cancer Res. 2011, 71, 5659–5669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravid, Y.; Formanski, M.; Smith, Y.; Reich, R.; Davidson, B. Uterine Leiomyosarcoma and Endometrial Stromal Sarcoma Have Unique MiRNA Signatures. Gynecol. Oncol. 2016, 140, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Pazzaglia, L.; Novello, C.; Conti, A.; Pollino, S.; Picci, P.; Benassi, M.S. MiR-152 down-Regulation Is Associated with MET up-Regulation in Leiomyosarcoma and Undifferentiated Pleomorphic Sarcoma. Cell Oncol. 2017, 40, 77–88. [Google Scholar] [CrossRef]
- Tang, F.; Hajkova, P.; O’Carroll, D.; Lee, C.; Tarakhovsky, A.; Lao, K.; Surani, M.A. MicroRNAs Are Tightly Associated with RNA-Induced Gene Silencing Complexes in Vivo. Biochem. Biophys. Res. Commun. 2008, 372, 24–29. [Google Scholar] [CrossRef]
- Siebolts, U.; Varnholt, H.; Drebber, U.; Dienes, H.-P.; Wickenhauser, C.; Odenthal, M. Tissues from Routine Pathology Archives Are Suitable for MicroRNA Analyses by Quantitative PCR. J. Clin. Pathol. 2009, 62, 84–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Ying, L.; Tian, Y.; Yang, P.; Zhu, Y.; Wang, Z.; Qiu, F.; Lin, J. MiR-144 Downregulation Increases Bladder Cancer Cell Proliferation by Targeting EZH2 and Regulating Wnt Signaling. FEBS J. 2013, 280, 4531–4538. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, P.; Li, J.; Wang, Y.; Du, Y.; Chen, X.; Zang, W.; Wang, H.; Chu, H.; Zhao, G.; et al. MiR-144 Inhibits Proliferation and Induces Apoptosis and Autophagy in Lung Cancer Cells by Targeting TIGAR. Cell. Physiol. Biochem. 2015, 35, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Yu, H.; Hu, M.; Xia, T.; Yue, X. MiR-144-3p Exerts Anti-Tumor Effects in Glioblastoma by Targeting c-Met. J. Neurochem. 2015, 135, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Rossing, M.; Borup, R.; Henao, R.; Winther, O.; Vikesaa, J.; Niazi, O.; Godballe, C.; Krogdahl, A.; Glud, M.; Hjort-Sørensen, C.; et al. Down-Regulation of MicroRNAs Controlling Tumourigenic Factors in Follicular Thyroid Carcinoma. J. Mol. Endocrinol. 2012, 48, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Zhou, X.; Wei, M. MicroRNA-144 Suppresses Osteosarcoma Growth and Metastasis by Targeting ROCK1 and ROCK2. Oncotarget 2015, 6, 10297–10308. [Google Scholar] [CrossRef] [Green Version]
- Minna, E.; Romeo, P.; Dugo, M.; Cecco, L.D.; Todoerti, K.; Pilotti, S.; Perrone, F.; Seregni, E.; Agnelli, L.; Neri, A.; et al. MiR-451a Is Underexpressed and Targets AKT/MTOR Pathway in Papillary Thyroid Carcinoma. Oncotarget 2016, 7, 12731–12747. [Google Scholar] [CrossRef]
- Renner, M.; Czwan, E.; Hartmann, W.; Penzel, R.; Brors, B.; Eils, R.; Wardelmann, E.; Büttner, R.; Lichter, P.; Schirmacher, P.; et al. MicroRNA Profiling of Primary High-Grade Soft Tissue Sarcomas. Genes Chromosomes Cancer 2012, 51, 982–996. [Google Scholar] [CrossRef]
- Gonzalez dos Anjos, L.; De Almeida, B.C.; Gomes de Almeida, T.; Mourão Lavorato Rocha, A.; De Nardo Maffazioli, G.; Soares, F.A.; Werneck da Cunha, I.; Baracat, E.C.; Carvalho, K.C. Could MiRNA Signatures Be Useful for Predicting Uterine Sarcoma and Carcinosarcoma Prognosis and Treatment? Cancers 2018, 10, 315. [Google Scholar] [CrossRef] [Green Version]
- Xin, H.; Wang, C.; Liu, Z. MiR-196a-5p Promotes Metastasis of Colorectal Cancer via Targeting IκBα. BMC Cancer 2019, 19, 30. [Google Scholar] [CrossRef]
- Xiong, M.; Wang, P.; Pan, B.; Nie, J.; Wang, S.; He, B. The Diagnostic and Prognostic Values of MicroRNA-196a in Cancer. Biosci. Rep. 2021, 41, BSR20203559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yao, C.; Li, H.; Wang, G.; He, X. Combined Elevation of MicroRNA-196a and MicroRNA-196b in Sera Predicts Unfavorable Prognosis in Patients with Osteosarcomas. Int. J. Mol. Sci. 2014, 15, 6544–6555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhang, L.; Zhang, X.; Xing, C.; Liu, R.; Zhang, F. MiR-196a Promoted Cell Migration, Invasion and the Epithelial-Mesenchymal Transition by Targeting HOXA5 in Osteosarcoma. Cancer Biomark. 2020, 29, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Bill, K.; Liu, J.; Young, E.; Peng, T.; Bolshakov, S.; Hoffman, A.; Song, Y.; Demicco, E.G.; Terrada, D.L.; et al. MiR-155 Is a Liposarcoma Oncogene That Targets Casein Kinase-1α and Enhances β-Catenin Signaling. Cancer Res. 2012, 72, 1751–1762. [Google Scholar] [CrossRef] [Green Version]
- Bianchini, L.; Saâda, E.; Gjernes, E.; Marty, M.; Haudebourg, J.; Birtwisle-Peyrottes, I.; Keslair, F.; Chignon-Sicard, B.; Chamorey, E.; Pedeutour, F. Let-7 MicroRNA and HMGA2 Levels of Expression Are Not Inversely Linked in Adipocytic Tumors: Analysis of 56 Lipomas and Liposarcomas with Molecular Cytogenetic Data. Genes Chromosomes Cancer 2011, 50, 442–455. [Google Scholar] [CrossRef]
- Tap, W.D.; Eilber, F.C.; Ginther, C.; Dry, S.M.; Reese, N.; Barzan-Smith, K.; Chen, H.-W.; Wu, H.; Eilber, F.R.; Slamon, D.J.; et al. Evaluation of Well-Differentiated/de-Differentiated Liposarcomas by High-Resolution Oligonucleotide Array-Based Comparative Genomic Hybridization. Genes Chromosomes Cancer 2011, 50, 95–112. [Google Scholar] [CrossRef]
- Hisaoka, M.; Matsuyama, A.; Nakamoto, M. Aberrant Calreticulin Expression Is Involved in the Dedifferentiation of Dedifferentiated Liposarcoma. Am. J. Pathol. 2012, 180, 2076–2083. [Google Scholar] [CrossRef]
- Taylor, B.S.; DeCarolis, P.L.; Angeles, C.V.; Brenet, F.; Schultz, N.; Antonescu, C.R.; Scandura, J.M.; Sander, C.; Viale, A.J.; Socci, N.D.; et al. Frequent Alterations and Epigenetic Silencing of Differentiation Pathway Genes in Structurally Rearranged Liposarcomas. Cancer Discov. 2011, 1, 587–597. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Xiao, Z.; Luo, J.; Zhang, Y.; Lin, L. MiR-139-5p, MiR-940 and MiR-193a-5p Inhibit the Growth of Hepatocellular Carcinoma by Targeting SPOCK1. J. Cell. Mol. Med. 2019, 23, 2475–2488. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Hosany, S.; Zhong, S.; Jiang, Y.; Zhang, F.; Lin, L.; Wang, X.; Gao, S.; Hu, X. MicroRNA-193a Inhibits Breast Cancer Proliferation and Metastasis by Downregulating WT1. PLoS ONE 2017, 12, e0185565. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Li, J.; Yan, M.; Liu, L.; Lin, H.; Zhao, F.; Sun, L.; Zhang, Y.; Cui, Y.; Zhang, F.; et al. MicroRNA-193a-3p and -5p Suppress the Metastasis of Human Non-Small-Cell Lung Cancer by Downregulating the ERBB4/PIK3R3/MTOR/S6K2 Signaling Pathway. Oncogene 2015, 34, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Zhao, F.; Cai, W.; Meng, X.; Li, Y.; Cai, S. MiR-193a-3p and MiR-193a-5p Suppress the Metastasis of Human Osteosarcoma Cells by down-Regulating Rab27B and SRR, Respectively. Clin. Exp. Metastasis 2016, 33, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Zeng, X.; Huang, Y.; Chen, S.; Lin, F.; Yang, G.; Yang, N. MiR-423-5p Serves as a Diagnostic Indicator and Inhibits the Proliferation and Invasion of Ovarian Cancer. Exp. Ther. Med. 2018, 15, 4723–4730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, W.; Yu, T.; An, Q.; Cao, X.; Pan, H. MicroRNA-423-5p Inhibits Colon Cancer Growth by Promoting Caspase-Dependent Apoptosis. Exp. Ther. Med. 2018, 16, 1225–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, Y.; Xiong, F.; Yang, L.; Bo, H.; Gong, Z.; Wang, Y.; Wei, F.; Tang, Y.; Li, X.; Liao, Q.; et al. Long Noncoding RNA AFAP1-AS1 Acts as a Competing Endogenous RNA of MiR-423-5p to Facilitate Nasopharyngeal Carcinoma Metastasis through Regulating the Rho/Rac Pathway. J. Exp. Clin. Cancer Res. 2018, 37, 253. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Peng, L.; Gong, X.; Zhang, X.; Sun, R.; Du, J. MiR-423-5p Inhibits Osteosarcoma Proliferation and Invasion Through Directly Targeting STMN1. Cell. Physiol. Biochem. 2018, 50, 2249–2259. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, H.; Jin, W.; Ding, Z.; Zheng, S.; Yu, Y. Tissue MicroRNA-21 Expression Predicted Recurrence and Poor Survival in Patients with Colorectal Cancer—A Meta-Analysis. OncoTargets Ther. 2016, 9, 2615–2624. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Cai, Q.; Huang, Y.; Li, S.; Yang, H.; Sun, L.; Chen, K.; Wang, Y. MicroRNA-21 as a Potential Diagnostic Biomarker for Breast Cancer Patients: A Pooled Analysis of Individual Studies. Oncotarget 2016, 7, 34498–34506. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Nie, Y.; Qu, S.; Liao, J.-Y.; Cui, X.; Yao, H.; Zeng, Y.; Su, F.; Song, E.; Liu, Q. MiR-21 Induces Myofibroblast Differentiation and Promotes the Malignant Progression of Breast Phyllodes Tumors. Cancer Res. 2014, 74, 4341–4352. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Tao, F.; Wang, W.; Ji, K. Prognostic Value of MicroRNA-21 in Pancreatic Ductal Adenocarcinoma: A Meta-Analysis. World J. Surg. Oncol. 2016, 14, 82. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.-J.; Li, Y.; Zheng, W.; Zhao, J.; Guo, M.; Zhou, Y.; Qin, N.; Zheng, J.; Xu, L. Antisense Oligonucleotides against MicroRNA-21 Reduced the Proliferation and Migration of Human Colon Carcinoma Cells. Cancer Cell Int. 2015, 15, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.-H.; Tao, Z.-H.; Zhang, J.; Li, T.; Ni, C.; Xie, J.; Zhang, J.-F.; Hu, X.-C. MiRNA-21 Induces Epithelial to Mesenchymal Transition and Gemcitabine Resistance via the PTEN/AKT Pathway in Breast Cancer. Tumor Biol. 2016, 37, 7245–7254. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yan, P.; Wang, J.; Zhang, Y.; Zhang, M.; Wang, Z.; Fu, Q.; Liang, W. Clinical Significance of Tumor MiR-21, MiR-221, MiR-143, and MiR-106a as Biomarkers in Patients with Osteosarcoma. Int. J. Biol. Markers 2019, 34, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhao, J.; Tao, Y.; Guo, M.; Ya, Z.; Chen, C.; Qin, N.; Zheng, J.; Luo, J.; Xu, L. MicroRNA-21: A Promising Biomarker for the Prognosis and Diagnosis of Non-Small Cell Lung Cancer. Oncol. Lett. 2018, 16, 2777–2782. [Google Scholar] [CrossRef]
- Ji, R.; Zhang, X.; Gu, H.; Ma, J.; Wen, X.; Zhou, J.; Qian, H.; Xu, W.; Qian, J.; Lin, J. MiR-374a-5p: A New Target for Diagnosis and Drug Resistance Therapy in Gastric Cancer. Mol. Ther.—Nucleic Acids 2019, 18, 320–331. [Google Scholar] [CrossRef] [Green Version]
- Son, D.; Kim, Y.; Lim, S.; Kang, H.-G.; Kim, D.-H.; Park, J.W.; Cheong, W.; Kong, H.K.; Han, W.; Park, W.-Y.; et al. MiR-374a-5p Promotes Tumor Progression by Targeting ARRB1 in Triple Negative Breast Cancer. Cancer Lett. 2019, 454, 224–233. [Google Scholar] [CrossRef]
- Lian, F.; Cui, Y.; Zhou, C.; Gao, K.; Wu, L. Identification of a Plasma Four-MicroRNA Panel as Potential Noninvasive Biomarker for Osteosarcoma. PLoS ONE 2015, 10, e0121499. [Google Scholar] [CrossRef]
- Ren, L.; Chen, H.; Song, J.; Chen, X.; Lin, C.; Zhang, X.; Hou, N.; Pan, J.; Zhou, Z.; Wang, L.; et al. MiR-454-3p-Mediated Wnt/β-Catenin Signaling Antagonists Suppression Promotes Breast Cancer Metastasis. Theranostics 2019, 9, 449–465. [Google Scholar] [CrossRef]
- Song, Y.; Guo, Q.; Gao, S.; Hua, K. MiR-454-3p Promotes Proliferation and Induces Apoptosis in Human Cervical Cancer Cells by Targeting TRIM3. Biochem. Biophys. Res. Commun. 2019, 516, 872–879. [Google Scholar] [CrossRef]
- Liao, H.; Liang, Y.; Kang, L.; Xiao, Y.; Yu, T.; Wan, R. MiR-454-3p Inhibits Non-Small Cell Lung Cancer Cell Proliferation and Metastasis by Targeting TGFB2. Oncol. Rep. 2021, 45, 67. [Google Scholar] [CrossRef]
- Yan, A.; Wang, C.; Zheng, L.; Zhou, J.; Zhang, Y. MicroRNA-454-3p Inhibits Cell Proliferation and Invasion in Esophageal Cancer by Targeting Insulin-like Growth Factor 2 MRNA-Binding Protein 1. Oncol. Lett. 2020, 20, 359. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, G.; Zheng, W.; Xue, Q.; Wei, D.; Zheng, Y.; Yuan, J. MiR-454-3p and MiR-374b-5p Suppress Migration and Invasion of Bladder Cancer Cells through Targetting ZEB2. Biosci. Rep. 2018, 38, BSR20181436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, J.; Yu, H.; Xie, P.; Liu, W.; Wang, K.; Ni, H. MiR-454-3p Exerts Tumor-Suppressive Functions by down-Regulation of NFATc2 in Glioblastoma. Gene 2019, 710, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Hui, X.; Zhang, S.; Wang, Y. MiR-454-3p Suppresses Cell Migration and Invasion by Targeting CPEB1 in Human Glioblastoma. Mol. Med. Rep. 2018, 18, 3965–3972. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Li, Y.; Chen, G.; Wang, F.; Shen, Z.; Zhou, R. Overexpression of MiR-15b-5p Promotes Gastric Cancer Metastasis by Regulating PAQR3. Oncol. Rep. 2017, 38, 352–358. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Zhang, N.; Zhao, S.; Chen, X.; Li, F.; Tao, X. MiR-221-3p and MiR-15b-5p Promote Cell Proliferation and Invasion by Targeting Axin2 in Liver Cancer. Oncol. Lett. 2019, 18, 6491–6500. [Google Scholar] [CrossRef] [Green Version]
- Weng, Y.; Shen, Y.; He, Y.; Pan, X.; Xu, J.; Jiang, Y.; Zhang, Q.; Wang, S.; Kong, F.; Zhao, S.; et al. The MiR-15b-5p/PDK4 Axis Regulates Osteosarcoma Proliferation through Modulation of the Warburg Effect. Biochem. Biophys. Res. Commun. 2018, 503, 2749–2757. [Google Scholar] [CrossRef]
- Courtney, K.D.; Corcoran, R.B.; Engelman, J.A. The PI3K Pathway As Drug Target in Human Cancer. J. Clin. Oncol. 2010, 28, 1075–1083. [Google Scholar] [CrossRef] [Green Version]
- Willems, L.; Tamburini, J.; Chapuis, N.; Lacombe, C.; Mayeux, P.; Bouscary, D. PI3K and MTOR Signaling Pathways in Cancer: New Data on Targeted Therapies. Curr. Oncol. Rep. 2012, 14, 129–138. [Google Scholar] [CrossRef]
- Gutierrez, A.; Snyder, E.L.; Marino-Enriquez, A.; Zhang, Y.-X.; Sioletic, S.; Kozakewich, E.; Grebliunaite, R.; Ou, W.; Sicinska, E.; Raut, C.P.; et al. Aberrant AKT Activation Drives Well-Differentiated Liposarcoma. Proc. Natl. Acad. Sci. USA 2011, 108, 16386–16391. [Google Scholar] [CrossRef] [Green Version]
- Demicco, E.G.; Torres, K.E.; Ghadimi, M.P.; Colombo, C.; Bolshakov, S.; Hoffman, A.; Peng, T.; Bovée, J.V.M.G.; Wang, W.-L.; Lev, D.; et al. Involvement of the PI3K/Akt Pathway in Myxoid/Round Cell Liposarcoma. Mod. Pathol. 2012, 25, 212–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Lopez-Marquez, H.; Fan, K.C.; Choy, E.; Cote, G.; Harmon, D.; Nielsen, G.P.; Yang, C.; Zhang, C.; Mankin, H.; et al. Synergistic Effects of Targeted PI3K Signaling Inhibition and Chemotherapy in Liposarcoma. PLoS ONE 2014, 9, e93996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codenotti, S.; Mansoury, W.; Pinardi, L.; Monti, E.; Marampon, F.; Fanzani, A. Animal Models of Well-Differentiated/Dedifferentiated Liposarcoma: Utility and Limitations. OncoTargets Ther. 2019, 12, 5257–5268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lejeune, D.; Dumoutier, L.; Constantinescu, S.; Kruijer, W.; Schuringa, J.J.; Renauld, J.-C. Interleukin-22 (IL-22) Activates the JAK/STAT, ERK, JNK, and P38 MAP Kinase Pathways in a Rat Hepatoma Cell Line: Pathways That Are Shared with and Distinct from Il-10. J. Biol. Chem. 2002, 277, 33676–33682. [Google Scholar] [CrossRef] [Green Version]

| MDM2 FISH Results | MFS (n = 16) | UPS (n = 18) |
|---|---|---|
| Amplified (positive) | 0 | 0 |
| Non-amplified (negative) | 13 | 11 |
| Data not available | 3 | 7 |
| Upregulated | Downregulated | |
|---|---|---|
| In liposarcoma (including WLDPS and DDLPS) compared to lipoma | miR-196a-5p | miR-144-3p miR-144-5p miR-451a |
| In DDLPS compared to WDLPS | miR-21-5p miR-454-3p miR-374a-5p miR-15b-5p | miR-193a-5p miR-423-5p |
| Pathway | Key Components | p-Value | q-Value |
|---|---|---|---|
| Cardiac EGF pathway | GPCR, Phospholipase C, PKC Delta, MAPK | 1.00 × 10−4 | 1.49 × 10−2 |
| Caspase pathway | Caspase cascade | 1.00 × 10−4 | 1.49 × 10−2 |
| AMB2 neutrophils pathway | PI3K, Akt, MAPK | 1.00 × 10−4 | 1.49 × 10−2 |
| RAP1 signaling | PI3K, Akt, MAPK | 1.00 × 10−4 | 1.49 × 10−2 |
| Insulin glucose pathway | PI3K, Akt | 1.00 × 10−4 | 1.49 × 10−2 |
| PI3KCI AKT pathway | PI3K, Akt | 2.00 × 10−4 | 2.13 × 10−2 |
| PKB mediated events | Akt | 2.00 × 10−4 | 2.13 × 10−2 |
| Integrin alphaIIB beta3 signaling | thrombin, ADP, collagen, fibrinogen and thrombospondin, PTK2/FAK | 3.00 × 10−4 | 2.49 × 10−2 |
| Costimulation by the CD28 family | CD28, CTLA4, ICOS, PD1 and BTLA receptors, PI3K, Akt | 4.00 × 10−4 | 2.49 × 10−2 |
| CD40 pathway | JAK/STAT, NF-kappaB, MAPK, P38, JNK | 4.00 × 10−4 | 2.49 × 10−2 |
| Regulation of the actin cytoskeleton by Rho GTPases | Rho, ROCK | 4.00 × 10−4 | 2.49 × 10−2 |
| P38 MAPK pathway | MKK3, MKK6, P38 | 5.00 × 10−4 | 2.49 × 10−2 |
| IL2 PI3K pathway | IL2, PI3K, Akt | 5.00 × 10−4 | 2.49 × 10−2 |
| Lysophospholipid pathway | Rho, PI3K, Akt, MAPK, PLC, cAMP | 5.00 × 10−4 | 2.49 × 10−2 |
| Telomerase pathway | ATM, ATR, TERT, Akt | 5.00 × 10−4 | 2.49 × 10−2 |
| Chemical pathway | cytochrome c, AIF, caspase cascade | 6.00 × 10−4 | 2.49 × 10−2 |
| ECM regulators | Components Regulating remodeling of the extra-cellular matrix | 6.00 × 10−4 | 2.49 × 10−2 |
| HNF3B pathway | Regulated by PI3K, MAPK. Regulates cancer | 6.00 × 10−4 | 2.49 × 10−2 |
| Lipoma (n = 34) | WDLPS 1 (n = 24) | DDLPS 2 (n = 20) | MFS 3 (n = 16) | UPS 4 (n = 18) | |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 23 | 17 | 13 | 5 | 10 |
| Female | 11 | 7 | 7 | 11 | 8 |
| Age (median, range) | 51.5 (5–81) | 59 (42–87) | 64 (39–85) | 67 (36–94) | 60.5 (26–94) |
| Race | |||||
| Chinese | 25 | 15 | 14 | 15 | 12 |
| Malay | 4 | 3 | 3 | 1 | 2 |
| Indian | 2 | 0 | 0 | 0 | 0 |
| Others 5 | 3 | 6 | 3 | 0 | 4 |
| Anatomic site | |||||
| Extremities 6 | 3 | 6 | 6 | 9 | 8 |
| Central body sites 7 | 24 | 16 | 13 | 5 | 10 |
| Others 8 | 7 | 2 | 1 | 2 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, H.M.; Cheng, H.; Tang, Y.C.; Leong, S.M.; Teo, P.Y.; Lee, C.K.; Lee, V.K.M.; Hue, S.S.-S. MicroRNAs as Potential Biomarkers in the Differential Diagnosis of Lipomatous Tumors and Their Mimics. Int. J. Mol. Sci. 2022, 23, 7804. https://doi.org/10.3390/ijms23147804
Tan HM, Cheng H, Tang YC, Leong SM, Teo PY, Lee CK, Lee VKM, Hue SS-S. MicroRNAs as Potential Biomarkers in the Differential Diagnosis of Lipomatous Tumors and Their Mimics. International Journal of Molecular Sciences. 2022; 23(14):7804. https://doi.org/10.3390/ijms23147804
Chicago/Turabian StyleTan, Hui Min, He Cheng, Yew Chung Tang, Sai Mun Leong, Poh Yin Teo, Chi Kuen Lee, Victor Kwan Min Lee, and Susan Swee-Shan Hue. 2022. "MicroRNAs as Potential Biomarkers in the Differential Diagnosis of Lipomatous Tumors and Their Mimics" International Journal of Molecular Sciences 23, no. 14: 7804. https://doi.org/10.3390/ijms23147804
APA StyleTan, H. M., Cheng, H., Tang, Y. C., Leong, S. M., Teo, P. Y., Lee, C. K., Lee, V. K. M., & Hue, S. S.-S. (2022). MicroRNAs as Potential Biomarkers in the Differential Diagnosis of Lipomatous Tumors and Their Mimics. International Journal of Molecular Sciences, 23(14), 7804. https://doi.org/10.3390/ijms23147804






