Disruption of O-GlcNAcylation Homeostasis Induced Ovarian Granulosa Cell Injury in Bovine
Abstract
:1. Introduction
2. Results
2.1. Altered O-GlcNAc Levels Affect the Expression of OGT and OGA in Bovine GCs
2.2. Disruption of O-GlcNAc Cycling Affects Viability and Proliferation of GCs
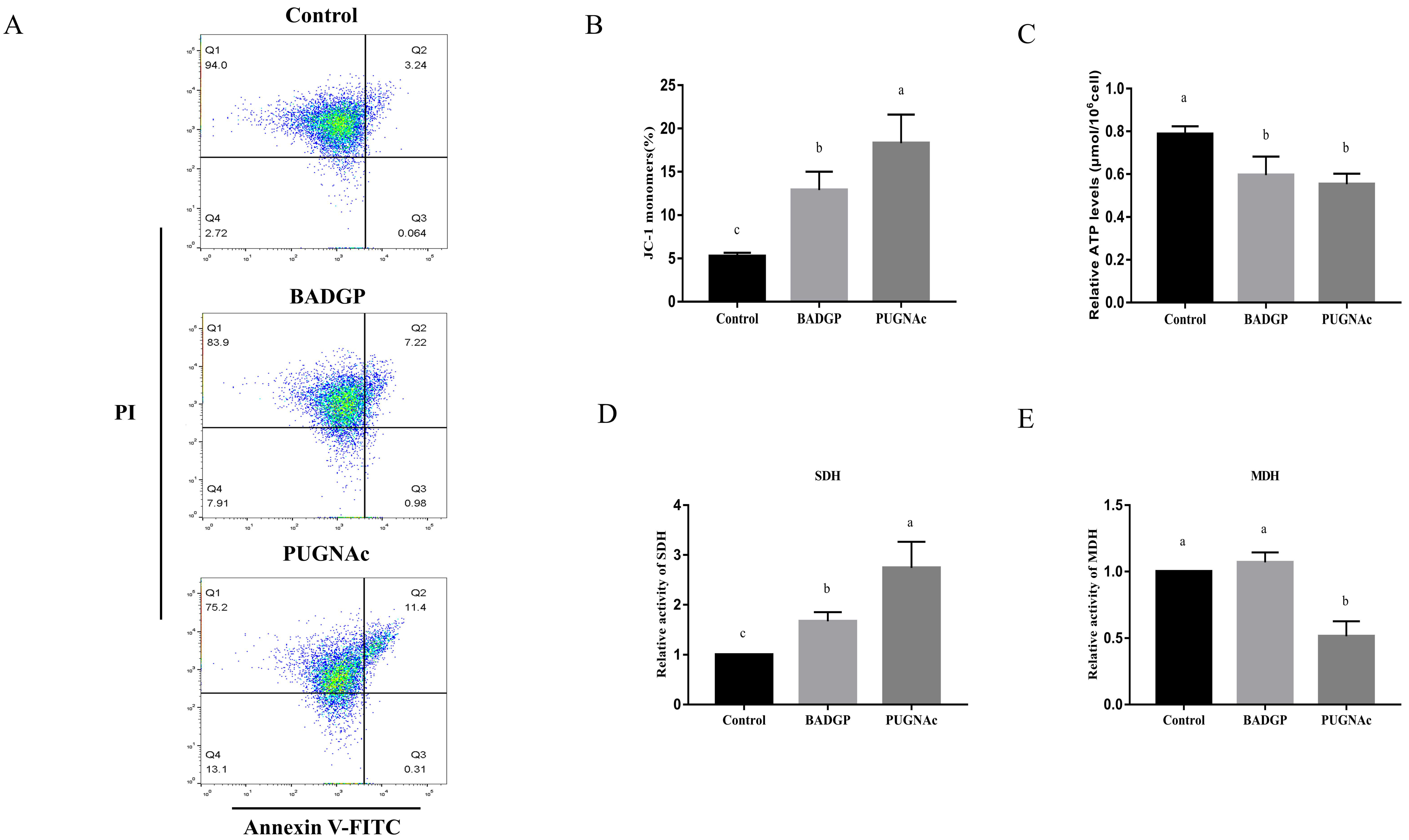
2.3. Disruption of O-GlcNAc Cycling Induces Cell Apoptosis in Bovine GCs
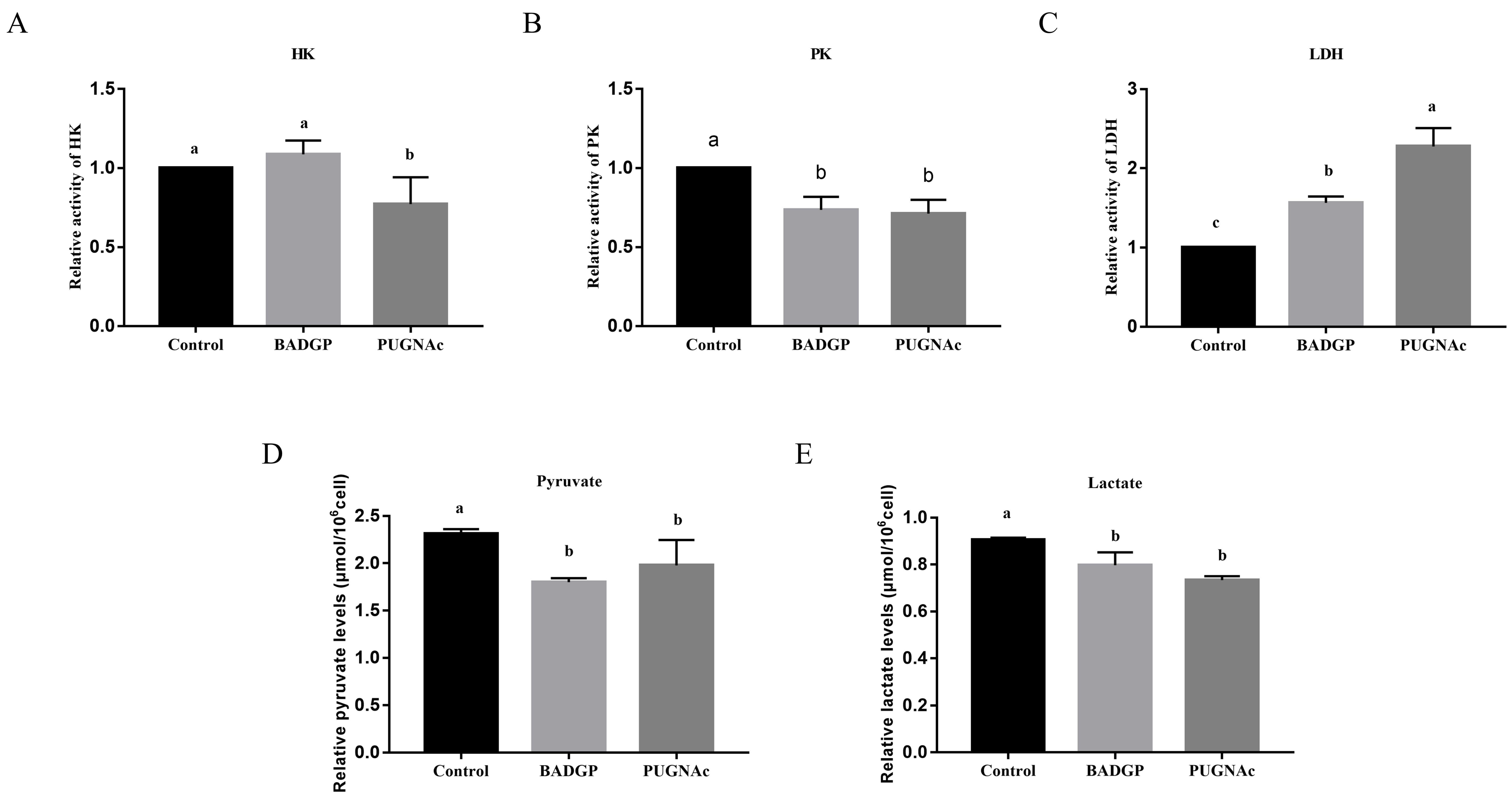
2.4. Perturbation of O-GlcNAc Cycling in Bovine GCs Reduces Glycolysis
2.5. Alteration in O-GlcNAc Levels Impairs Mitochondria Homeostasis, ATP Production, and the Tricarboxylic Acid (TCA) Cycle
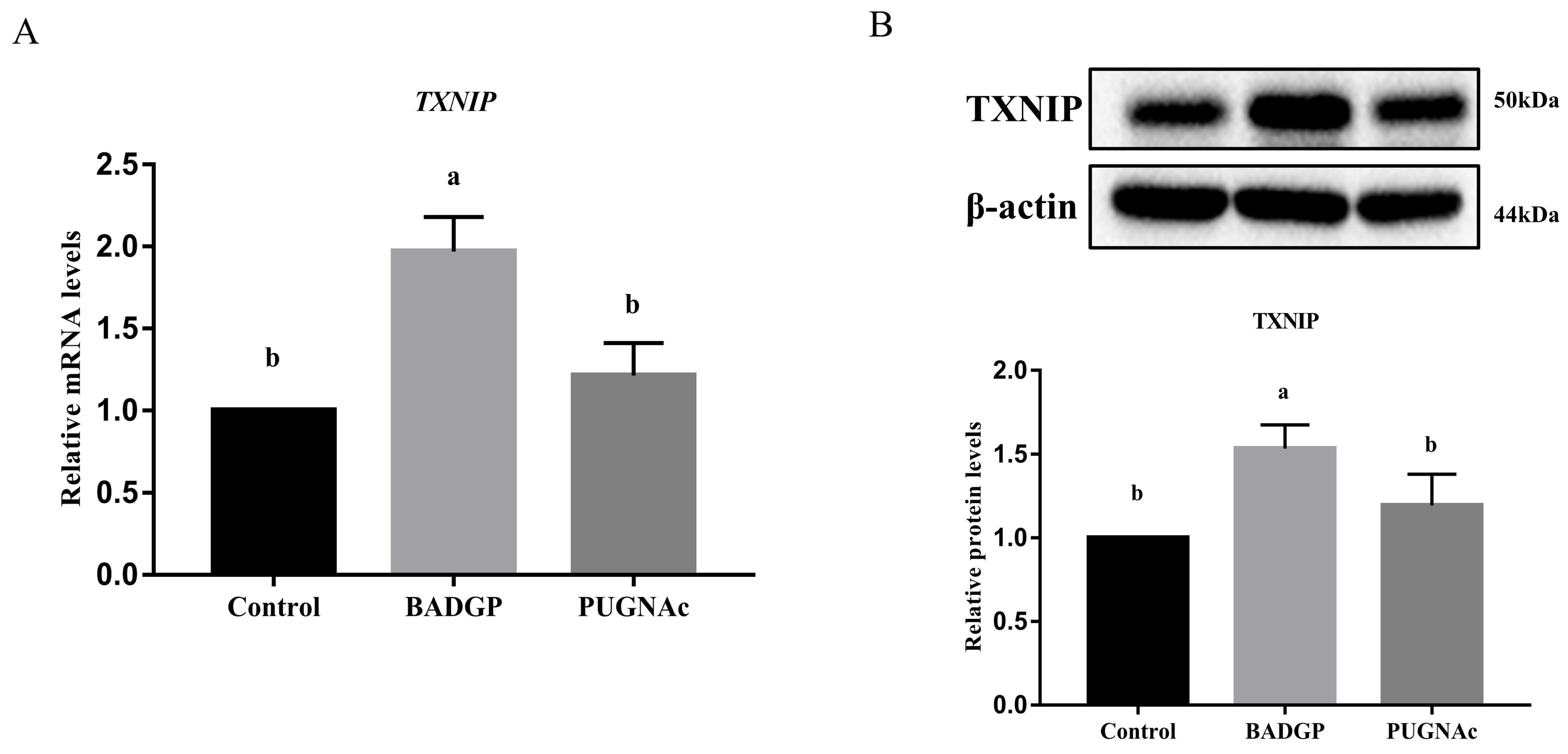
2.6. Disruption of O-GlcNAc Cycling Changes the Expression of Thioredoxin-Interacting Protein (TXNIP)
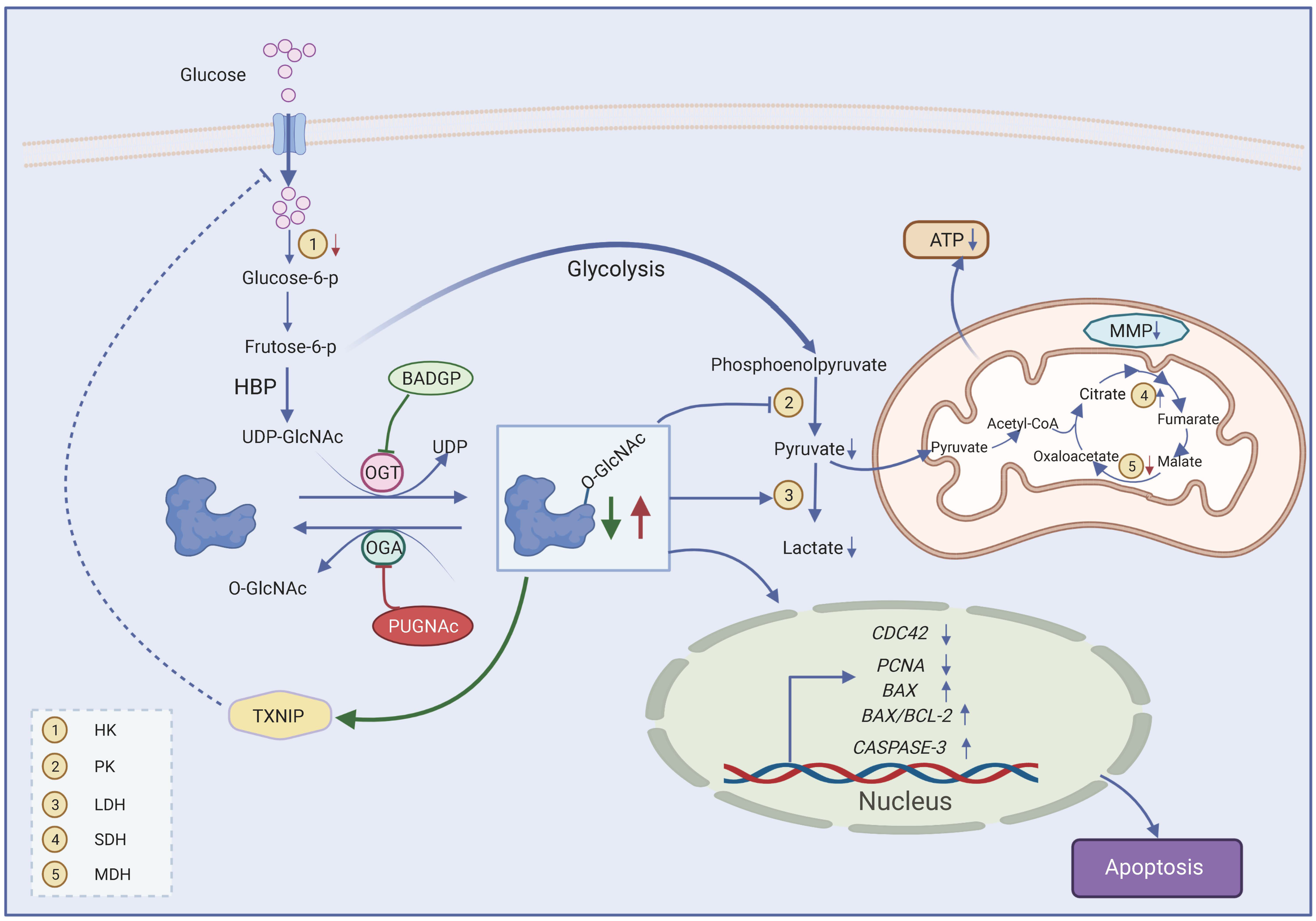
3. Discussion
4. Materials and Methods
4.1. GC Isolation and Culture
4.2. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
4.3. Protein Extraction and Western Blot Analysis
4.4. Cell Viability Analysis
4.5. Apoptotic Assay
4.6. Assessment of MMP
4.7. Enzymatic Activity Assay
4.8. Metabolites Measurement and Intracellular ATP Level Assays
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kordus, R.J.; LaVoie, H.A. Granulosa cell biomarkers to predict pregnancy in ART: Pieces to solve the puzzle. Reproduction 2017, 153, R69–R83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, Y.; Liu, H.; Pan, Z. MicroRNAs in ovarian follicular atresia and granulosa cell apoptosis. Reprod. Biol. Endocrinol. 2019, 17, 9. [Google Scholar] [CrossRef]
- Maruo, T.; Laoag-Fernandez, J.B.; Takekida, S.; Peng, X.; Deguchi, J.; Samoto, T.; Kondo, H.; Matsuo, H. Regulation of granulosa cell proliferation and apoptosis during follicular development. Gynecol. Endocrinol. 1999, 13, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.Q.; Sugiura, K.; Eppig, J.J. Mouse oocyte control of granulosa cell development and function: Paracrine regulation of cumulus cell metabolism. Semin. Reprod. Med. 2009, 27, 32–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hart, G.W.; Housley, M.P.; Slawson, C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007, 446, 1017–1022. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cai, Y.; Jin, J. Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell 2017, 8, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Lima, V.V.; Dela Justina, V.; Dos Passos, R.R., Jr.; Volpato, G.T.; Souto, P.C.S.; San Martin, S.; Giachini, F.R. O-GlcNAc Modification During Pregnancy: Focus on Placental Environment. Front. Physiol. 2018, 9, 1263. [Google Scholar] [CrossRef]
- Groves, J.A.; Lee, A.; Yildirir, G.; Zachara, N.E. Dynamic O-GlcNAcylation and its roles in the cellular stress response and homeostasis. Cell Stress Chaperones 2013, 18, 535–558. [Google Scholar] [CrossRef] [Green Version]
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231. [Google Scholar] [CrossRef] [Green Version]
- Bacigalupa, Z.A.; Bhadiadra, C.H.; Reginato, M.J. O-GlcNAcylation: Key regulator of glycolytic pathways. J. Bioenerg. Biomembr. 2018, 50, 189–198. [Google Scholar] [CrossRef]
- Tian, J.L.; Gomeshtapeh, F.I. Potential Roles of O-GlcNAcylation in Primary Cilia- Mediated Energy Metabolism. Biomolecules 2020, 10, 1504. [Google Scholar] [CrossRef]
- Fehl, C.; Hanover, J.A. Tools, tactics and objectives to interrogate cellular roles of O-GlcNAc in disease. Nat. Chem. Biol. 2022, 18, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Love, D.C.; Hanover, J.A. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci. STKE 2005, 2005, re13. [Google Scholar] [CrossRef] [PubMed]
- Hardiville, S.; Hart, G.W. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014, 20, 208–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Yao, R.Z.; Lian, S.; Liu, P.; Hu, Y.J.; Shi, H.Z.; Lv, H.M.; Yang, Y.Y.; Xu, B.; Li, S.Z. O-GlcNAcylation: The “stress and nutrition receptor” in cell stress response. Cell Stress Chaperones 2021, 26, 297–309. [Google Scholar] [CrossRef]
- Frank, L.A.; Sutton-McDowall, M.L.; Brown, H.M.; Russell, D.L.; Gilchrist, R.B.; Thompson, J.G. Hyperglycaemic conditions perturb mouse oocyte in vitro developmental competence via beta-O-linked glycosylation of heat shock protein 90. Hum. Reprod. 2014, 29, 1292–1303. [Google Scholar] [CrossRef] [Green Version]
- Shafi, R.; Iyer, S.P.; Ellies, L.G.; O’Donnell, N.; Marek, K.W.; Chui, D.; Hart, G.W.; Marth, J.D. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 2000, 97, 5735–5739. [Google Scholar] [CrossRef] [Green Version]
- Muha, V.; Authier, F.; Szoke-Kovacs, Z.; Johnson, S.; Gallagher, J.; McNeilly, A.; McCrimmon, R.J.; Teboul, L.; van Aalten, D.M.F. Loss of O-GlcNAcase catalytic activity leads to defects in mouse embryogenesis. J. Biol. Chem. 2021, 296, 100439. [Google Scholar] [CrossRef]
- Slawson, C.; Duncan, F.E. Sweet action: The dynamics of O-GlcNAcylation during meiosis in mouse oocytes. Mol. Reprod. Dev. 2015, 82, 915. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.T.; Romar, R.; Pavone, M.E.; Soriano-Ubeda, C.; Zhang, J.; Slawson, C.; Duncan, F.E. Disruption of O-GlcNAc homeostasis during mammalian oocyte meiotic maturation impacts fertilization. Mol. Reprod. Dev. 2019, 86, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Ruane, P.T.; Tan, C.M.J.; Adlam, D.J.; Kimber, S.J.; Brison, D.R.; Aplin, J.D.; Westwood, M. Protein O-GlcNAcylation Promotes Trophoblast Differentiation at Implantation. Cells 2020, 9, 2246. [Google Scholar] [CrossRef] [PubMed]
- Sutton-McDowall, M.L.; Gilchrist, R.B.; Thompson, J.G. The pivotal role of glucose metabolism in determining oocyte developmental competence. Reproduction 2010, 139, 685–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maucieri, A.M.; Townson, D.H. Evidence and manipulation of O-GlcNAcylation in granulosa cells of bovine antral folliclesdagger. Biol. Reprod. 2021, 104, 914–923. [Google Scholar] [CrossRef]
- Pantaleon, M.; Tan, H.Y.; Kafer, G.R.; Kaye, P.L. Toxic effects of hyperglycemia are mediated by the hexosamine signaling pathway and o-linked glycosylation in early mouse embryos. Biol. Reprod. 2010, 82, 751–758. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Tan, E.P.; VandenHull, N.J.; Peterson, K.R.; Slawson, C. O-GlcNAcase Expression is Sensitive to Changes in O-GlcNAc Homeostasis. Front. Endocrinol. 2014, 5, 206. [Google Scholar] [CrossRef] [Green Version]
- Nagy, T.; Fisi, V.; Frank, D.; Katai, E.; Nagy, Z.; Miseta, A. Hyperglycemia-Induced Aberrant Cell Proliferation; A Metabolic Challenge Mediated by Protein O-GlcNAc Modification. Cells 2019, 8, 999. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.H.; Liao, C.C.; Chen, M.Y.; Chou, T.Y. Feedback Regulation of O-GlcNAc Transferase through Translation Control to Maintain Intracellular O-GlcNAc Homeostasis. Int. J. Mol. Sci. 2021, 22, 3463. [Google Scholar] [CrossRef]
- Abramowitz, L.K.; Harly, C.; Das, A.; Bhandoola, A.; Hanover, J.A. Blocked O-GlcNAc cycling disrupts mouse hematopoeitic stem cell maintenance and early T cell development. Sci. Rep. 2019, 9, 12569. [Google Scholar] [CrossRef]
- Keembiyehetty, C.; Love, D.C.; Harwood, K.R.; Gavrilova, O.; Comly, M.E.; Hanover, J.A. Conditional knock-out reveals a requirement for O-linked N-Acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J. Biol. Chem. 2015, 290, 7097–7113. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Zhao, X.; Chen, J.; Qu, W.; Huang, X.; Wang, M.; Shao, Z.; Shu, Q.; Li, X. O-GlcNAc transferase Ogt regulates embryonic neuronal development through modulating Wnt/β-catenin signaling. Hum. Mol. Genet. 2021, 31, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Decourcelle, A.; Loison, I.; Baldini, S.; Leprince, D.; Dehennaut, V. Evidence of a compensatory regulation of colonic O-GlcNAc transferase and O-GlcNAcase expression in response to disruption of O-GlcNAc homeostasis. Biochem. Biophys. Res. Commun. 2020, 521, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, Y.; Gao, Y.; Li, Q.; Cao, Y.; Shen, Y.; Chen, P.; Yan, J.; Li, J. Vitamin E regulates bovine granulosa cell apoptosis via NRF2-mediated defence mechanism by activating PI3K/AKT and ERK1/2 signalling pathways. Reprod. Domest. Anim. 2021, 56, 1066–1084. [Google Scholar] [CrossRef]
- O’Donnell, N.; Zachara, N.E.; Hart, G.W.; Marth, J.D. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell Biol. 2004, 24, 1680–1690. [Google Scholar]
- Jang, H.; Kim, T.W.; Yoon, S.; Choi, S.Y.; Kang, T.W.; Kim, S.Y.; Kwon, Y.W.; Cho, E.J.; Youn, H.D. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 2012, 11, 62–74. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.R.; Song, M.; Lee, H.; Jeon, Y.; Choi, E.J.; Jang, H.J.; Moon, H.Y.; Byun, H.Y.; Kim, E.K.; Kim, D.H.; et al. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 2012, 11, 439–448. [Google Scholar] [CrossRef] [Green Version]
- Slawson, C.; Copeland, R.J.; Hart, G.W. O-GlcNAc signaling: A metabolic link between diabetes and cancer? Trends Biochem. Sci. 2010, 35, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, M.; Yu, R.K. O-linked beta-N-acetylglucosaminylation in mouse embryonic neural precursor cells. J. Neurosci. Res. 2009, 87, 3535–3545. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Wu, C.; Hart, G.W. Analytical and Biochemical Perspectives of Protein O-GlcNAcylation. Chem. Rev. 2021, 121, 1513–1581. [Google Scholar] [CrossRef]
- Gloster, T.M.; Vocadlo, D.J. Mechanism, Structure, and Inhibition of O-GlcNAc Processing Enzymes. Curr. Signal Transduct. Ther. 2010, 5, 74–91. [Google Scholar] [CrossRef] [Green Version]
- Bai, L.; Wang, W.; Xiang, Y.; Wang, S.; Wan, S.; Zhu, Y. Aberrant elevation of GDF8 impairs granulosa cell glucose metabolism via upregulating SERPINE1 expression in patients with PCOS. Mol. Ther. Nucleic Acids 2021, 23, 294–309. [Google Scholar] [CrossRef]
- Chahal, N.; Geethadevi, A.; Kaur, S.; Lakra, R.; Nagendra, A.; Shrivastav, T.G.; De Pascali, F.; Reiter, E.; Crepieux, P.; Devi, M.G.; et al. Direct impact of gonadotropins on glucose uptake and storage in preovulatory granulosa cells: Implications in the pathogenesis of polycystic ovary syndrome. Metabolism 2021, 115, 154458. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, K.; Pei, L.; Hu, X.; Cai, Y.; Ding, J.; Li, D.; Han, X.; Wu, J. The mechanisms of mitochondrial dysfunction and glucose intake decrease induced by Microcystin-LR in ovarian granulosa cells. Ecotoxicol. Environ. Saf. 2021, 212, 111931. [Google Scholar] [CrossRef]
- Pan, Y.; Zhu, J.; Lv, Q.; Shi, D.; Yang, S.; Xing, Q.; Zhang, R.; Cheng, J.; Deng, Y. Follicle-stimulating hormone regulates glycolysis of water buffalo follicular granulosa cells through AMPK/SIRT1 signalling pathway. Reprod. Domest. Anim. 2022, 57, 185–195. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, W.; Wang, Z.; Zheng, N.; Yuan, F.; Li, B.; Li, X.; Deng, L.; Lin, M.; Chen, X.; et al. Enhanced glycolysis in granulosa cells promotes the activation of primordial follicles through mTOR signaling. Cell Death Dis. 2022, 13, 87. [Google Scholar] [CrossRef]
- Yi, W.; Clark, P.M.; Mason, D.E.; Keenan, M.C.; Hill, C.; Goddard, W.A., 3rd; Peters, E.C.; Driggers, E.M.; Hsieh-Wilson, L.C. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012, 337, 975–980. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, J.; Jin, X.; Zhang, D.; Li, D.; Hao, F.; Feng, Y.; Gu, S.; Meng, F.; Tian, M.; et al. O-GlcNAcylation destabilizes the active tetrameric PKM2 to promote the Warburg effect. Proc. Natl. Acad. Sci. USA 2017, 114, 13732–13737. [Google Scholar] [CrossRef] [Green Version]
- Singh, J.P.; Qian, K.; Lee, J.S.; Zhou, J.; Han, X.; Zhang, B.; Ong, Q.; Ni, W.; Jiang, M.; Ruan, H.B.; et al. O-GlcNAcase targets pyruvate kinase M2 to regulate tumor growth. Oncogene 2020, 39, 560–573. [Google Scholar] [CrossRef]
- Cao, W.; Cao, J.; Huang, J.; Yao, J.; Yan, G.; Xu, H.; Yang, P. Discovery and confirmation of O-GlcNAcylated proteins in rat liver mitochondria by combination of mass spectrometry and immunological methods. PLoS ONE 2013, 8, e76399. [Google Scholar] [CrossRef]
- Ma, J.; Banerjee, P.; Whelan, S.A.; Liu, T.; Wei, A.C.; Ramirez-Correa, G.; McComb, M.E.; Costello, C.E.; O’Rourke, B.; Murphy, A.; et al. Comparative Proteomics Reveals Dysregulated Mitochondrial O-GlcNAcylation in Diabetic Hearts. J. Proteome Res. 2016, 15, 2254–2264. [Google Scholar] [CrossRef]
- Ferrer, C.M.; Lynch, T.P.; Sodi, V.L.; Falcone, J.N.; Schwab, L.P.; Peacock, D.L.; Vocadlo, D.J.; Seagroves, T.N.; Reginato, M.J. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell 2014, 54, 820–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, E.P.; Villar, M.T.; Lezi, E.; Lu, J.; Selfridge, J.E.; Artigues, A.; Swerdlow, R.H.; Slawson, C. Altering O-linked beta-N-acetylglucosamine cycling disrupts mitochondrial function. J. Biol. Chem. 2014, 289, 14719–14730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akinbiyi, E.O.; Abramowitz, L.K.; Bauer, B.L.; Stoll, M.S.K.; Hoppel, C.L.; Hsiao, C.P.; Hanover, J.A.; Mears, J.A. Blocked O-GlcNAc cycling alters mitochondrial morphology, function, and mass. Sci. Rep. 2021, 11, 22106. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, N.; Haseeb, M.; Kim, M.S.; Choi, S. Role of Thioredoxin-Interacting Protein in Diseases and Its Therapeutic Outlook. Int. J. Mol. Sci. 2021, 22, 2754. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, Q.; Gong, L.; Xu, H.; Liu, B.; Fang, X.; Yu, D.; Li, L.; Wei, T.; Wang, Y.; et al. C-terminal truncated HBx initiates hepatocarcinogenesis by downregulating TXNIP and reprogramming glucose metabolism. Oncogene 2021, 40, 1147–1161. [Google Scholar] [CrossRef]
- Filhoulaud, G.; Benhamed, F.; Pagesy, P.; Bonner, C.; Fardini, Y.; Ilias, A.; Movassat, J.; Burnol, A.F.; Guilmeau, S.; Kerr-Conte, J.; et al. O-GlcNacylation Links TxNIP to Inflammasome Activation in Pancreatic beta Cells. Front. Endocrinol. 2019, 10, 291. [Google Scholar] [CrossRef] [Green Version]
- Pagesy, P.; Tachet, C.; Mostefa-Kara, A.; Larger, E.; Issad, T. Increased OGA Expression and Activity in Leukocytes from Patients with Diabetes: Correlation with Inflammation Markers. Exp. Clin. Endocrinol. Diabetes 2019, 127, 517–523. [Google Scholar] [CrossRef]
- Hong, S.Y.; Hagen, T. 2-Deoxyglucose induces the expression of thioredoxin interacting protein (TXNIP) by increasing O-GlcNAcylation-Implications for targeting the Warburg effect in cancer cells. Biochem. Biophys. Res. Commun. 2015, 465, 838–844. [Google Scholar] [CrossRef]
- Xu, H.; Khan, A.; Zhao, S.; Wang, H.; Zou, H.; Pang, Y.; Zhu, H. Effects of Inhibin A on Apoptosis and Proliferation of Bovine Granulosa Cells. Animals 2020, 10, 367. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.W.; Jiang, X.L.; Wang, Y.C.; Wang, Y.Y.; Hao, H.S.; Zhao, S.J.; Du, W.H.; Zhao, X.M.; Wang, L.; Zhu, H.B. Melatonin protects against paraquat-induced damage during in vitro maturation of bovine oocytes. J. Pineal Res. 2019, 66, e12532. [Google Scholar] [CrossRef]
- Jiang, X.; Pang, Y.; Zhao, S.; Hao, H.; Zhao, X.; Du, W.; Wang, Y.; Zhu, H. Thioredoxin-interacting protein regulates glucose metabolism and improves the intracellular redox state in bovine oocytes during in vitro maturation. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E405–E416. [Google Scholar] [CrossRef] [PubMed]



| Gene | Primer | Sequence | Accession No. |
|---|---|---|---|
| GAPDH | Forward | 5′-GGGTCATCATCTCTGCACCT-3′ | NM_001034034 |
| Reverse | 5′-GGTCATAAGTCCCTCCACGA-3′ | ||
| OGT | Forward | 5′-AGGGTTCGAAGGCTGTAACTG-3′ | NM_001098070 |
| Reverse | 5′-ACTCAGCTAACCCTGTGCTG-3′ | ||
| OGA | Forward | 5′-TTGAAGAATGGCGGTCACGA-3′ | NM_001206448 |
| Reverse | 5′-TGACTACGACACCCTAACCAC-3′ | ||
| CDC42 | Forward | 5′-GTTGTTGTGGGTGATGGTGC-3′ | NM_001046332 |
| Reverse | 5′-TCCCCACCAATCATAACTGT-3′ | ||
| PCNA | Forward | 5′-GTCCAGGGCTCCATCTTGAA-3′ | NM_001034494 |
| Reverse | 5′-CAAGGAGACATGAGACGAGT-3′ | ||
| CCND2 | Forward | 5′-TGACCGCTGAGAAGTTATGC-3′ | NM_001076372 |
| Reverse | 5′-CGCCAGGTTCCATTTCAACT-3′ | ||
| BAX | Forward | 5′-GGCTGGACATTGGACTTCCTTC-3′ | NM_173894 |
| Reverse | 5′-TGGTCACTGTCTGCCATGTGG-3′ | ||
| BCL-2 | Forward | 5′-GAGTCGGATCGCAACTTGGA-3′ | NM_001077486 |
| Reverse | 5′-CTCTCGGCTGCTGCATTGT-3′ | ||
| CASPASE-3 | Forward | 5′-TACTTGGGAAGGTGTGAGAAAACTAA-3′ | NM_001077840 |
| Reverse | 5′-AACCCGTCTCCCTTTATATTGCT-3′ | ||
| TXNIP | Forward | 5′-TGGACTACTGGGTGAAGGCT-3′ | NM_001101875 |
| Reverse | 5′-AGCTGACACAGGTTCCAGTAAAT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.-F.; Feng, Z.-Q.; Sun, Y.-W.; Zhao, S.-J.; Zou, H.-Y.; Hao, H.-S.; Du, W.-H.; Zhao, X.-M.; Zhu, H.-B.; Pang, Y.-W. Disruption of O-GlcNAcylation Homeostasis Induced Ovarian Granulosa Cell Injury in Bovine. Int. J. Mol. Sci. 2022, 23, 7815. https://doi.org/10.3390/ijms23147815
Wang T-F, Feng Z-Q, Sun Y-W, Zhao S-J, Zou H-Y, Hao H-S, Du W-H, Zhao X-M, Zhu H-B, Pang Y-W. Disruption of O-GlcNAcylation Homeostasis Induced Ovarian Granulosa Cell Injury in Bovine. International Journal of Molecular Sciences. 2022; 23(14):7815. https://doi.org/10.3390/ijms23147815
Chicago/Turabian StyleWang, Teng-Fei, Zhi-Qiang Feng, Ya-Wen Sun, Shan-Jiang Zhao, Hui-Ying Zou, Hai-Sheng Hao, Wei-Hua Du, Xue-Ming Zhao, Hua-Bin Zhu, and Yun-Wei Pang. 2022. "Disruption of O-GlcNAcylation Homeostasis Induced Ovarian Granulosa Cell Injury in Bovine" International Journal of Molecular Sciences 23, no. 14: 7815. https://doi.org/10.3390/ijms23147815
APA StyleWang, T.-F., Feng, Z.-Q., Sun, Y.-W., Zhao, S.-J., Zou, H.-Y., Hao, H.-S., Du, W.-H., Zhao, X.-M., Zhu, H.-B., & Pang, Y.-W. (2022). Disruption of O-GlcNAcylation Homeostasis Induced Ovarian Granulosa Cell Injury in Bovine. International Journal of Molecular Sciences, 23(14), 7815. https://doi.org/10.3390/ijms23147815






