A Perspective on Studies of Phage DNA Packaging Dynamics
Abstract
:1. General Features of Studies of Nucleic Acid Packaging via a Procapsid
2. Phages and Phage DNA Packaging in Relation to Biomedicine: A Perspective
2.1. Phage Therapy of Infectious Disease
2.2. Metastatic Cancer
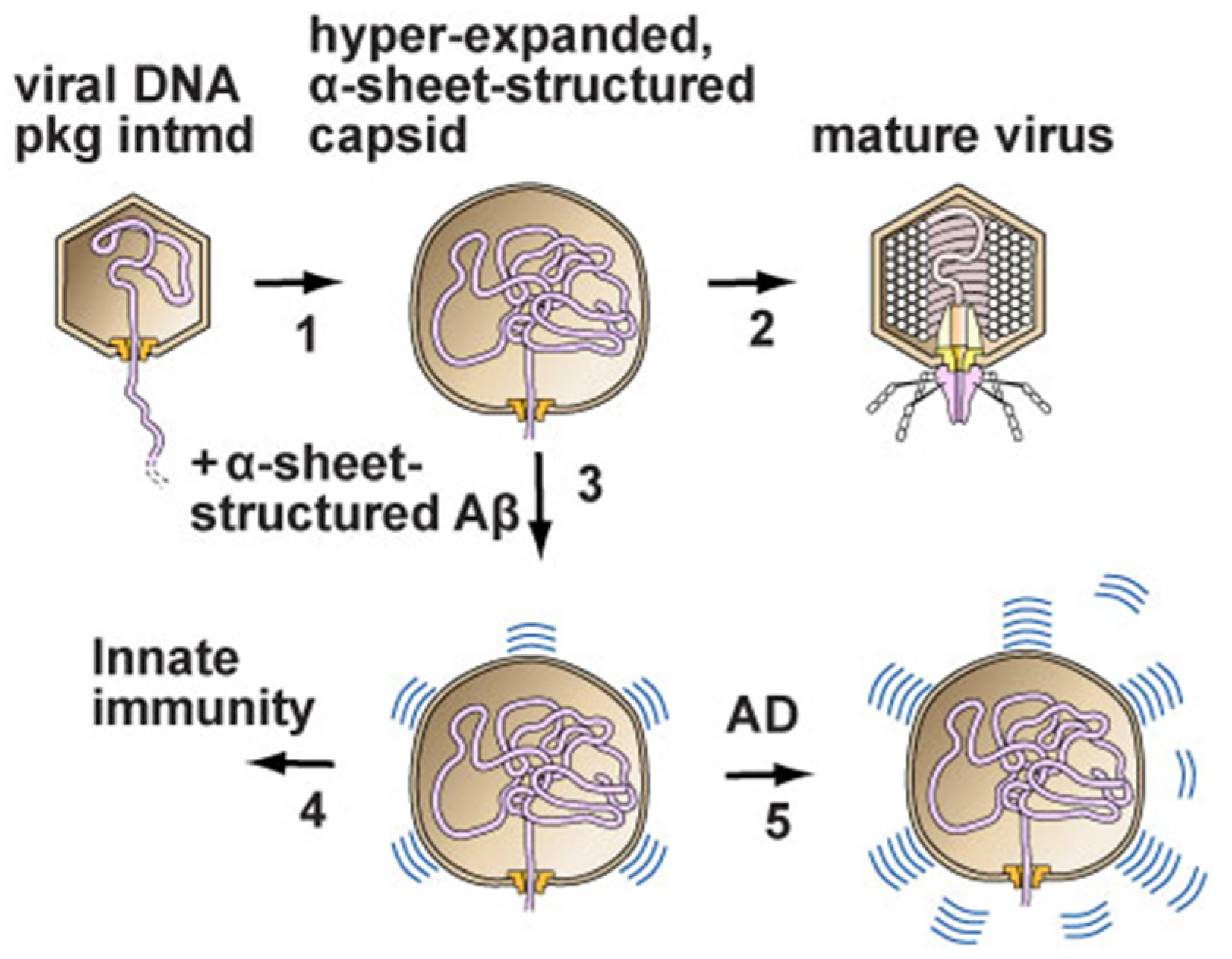
2.3. Neurodegenerative Disease
3. Strategy for Solving Key Problems of Biomedicine: Ramifications of Studies of Phages
Funding
Acknowledgments
Conflicts of Interest
References
- Murialdo, H.; Feiss, M. Enteric chromosomal islands: DNA packaging specificity and role of λ-like helper phage terminase. Viruses 2022, 14, 818. [Google Scholar] [CrossRef]
- Castillo, J.P.; Tong, A.B.; Tafoya, S.; Jardine, P.J.; Bustamante, C.A. DNA packaging motor inchworms along one strand allowing it to adapt to alternative double-helical structures. Nat. Commun. 2021, 12, 3439. [Google Scholar] [CrossRef]
- Rao, V.B.; Feiss, M. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu. Rev. Virol. 2015, 2, 351–378. [Google Scholar] [CrossRef] [Green Version]
- Black, L.W. Old, new, and widely true: The bacteriophage T4 DNA packaging mechanism. Virology 2015, 479–480, 650–656. [Google Scholar] [CrossRef] [Green Version]
- Serwer, P.; Jiang, W. Dualities in the analysis of phage DNA packaging motors. Bacteriophage 2012, 2, 239–255. [Google Scholar] [CrossRef] [Green Version]
- Casjens, S.R. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011, 9, 647–657. [Google Scholar] [CrossRef]
- Hendrix, R.W. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell 1998, 94, 147–150. [Google Scholar] [CrossRef] [Green Version]
- Fujisawa, H.; Morita, M. Phage DNA packaging. Genes Cells 1997, 2, 537–545. [Google Scholar] [CrossRef]
- Luftig, R.B.; Wood, W.B.; Okinaka, R. Bacteriophage T4 head morphogenesis: On the nature of gene 49-defective heads and their role as intermediates. J. Mol. Biol. 1971, 57, 555–573. [Google Scholar] [CrossRef]
- Kerr, C.; Sadsowski, P.D. Packaging and maturation of DNA of bacteriophage T7 in vitro. Proc. Natl. Acad. Sci. USA 1974, 71, 3545–3549. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger, E. What Is Life? Mind and Matter; Cambridge University Press: Cambridge, UK, 1968. [Google Scholar]
- Harding, C. Interview with Max Delbruck. 1979. Available online: https://oralhistories.library.caltech.edu/16/ (accessed on 21 May 2022).
- Delbrück, M. A physicist looks at biology. In Phage and the Origins of Molecular Biology; Cairns, J., Stent, G.S., Watson, J.D., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1966; pp. 9–22. [Google Scholar]
- Feynman, R.; Leighton, R.B.; Sands, M. The Feynman Lectures on Physics; Addison-Wesley: Reading, MA, USA, 1963; Volume I, Chapter 40. [Google Scholar]
- Di Mauro, M.; Esposito, S.; Naddeo, A. When Physics Meets Biology: A Less Known Feynman. arXiv 2018, arXiv:1805.03854. [Google Scholar] [CrossRef] [Green Version]
- Feynman, R.P. Feynman Hughes Lectures, Volume 4. Available online: https://thehugheslectures.info/wp-content/uploads/lectures/FeynmanHughesLectures_Vol4.pdf (accessed on 21 May 2022).
- Caflisch, K.M.; Suh, G.A.; Patel, R. Biological challenges of phage therapy and proposed solutions: A literature review. Expert Rev. Anti-Infect. Ther. 2019, 17, 1011–1041. [Google Scholar] [CrossRef]
- Brives, C.; Pourraz, J. Phage therapy as a potential solution in the fight against AMR: Obstacles and possible futures. Palgrave Commun. 2020, 6, 100. [Google Scholar] [CrossRef]
- Melo, L.D.R.; Oliveira, H.; Pires, D.P.; Dabrowska, K.; Azeredo, J. Phage therapy efficacy: A review of the last 10 years of preclinical studies. Crit. Rev. Microbiol. 2020, 46, 78–99. [Google Scholar] [CrossRef]
- Dąbrowska, K.; Abedon, S.T. Pharmacologically aware phage therapy: Pharmacodynamic and pharmacokinetic obstacles to phage antibacterial action in animal and human bodies. Microbiol. Mol. Biol. Rev. 2019, 83, e00012–e00019. [Google Scholar] [CrossRef]
- Alsaadi, A.; Beamud, B.; Easwaran, M.; Abdelrahman, F.; El-Shibiny, A.; Alghoribi, M.F.; Domingo-Calap, P. Learning from mistakes: The role of phages in pandemics. Front. Microbiol. 2021, 12, 653107. [Google Scholar] [CrossRef]
- Nale, J.Y.; Clokie, M.R. Preclinical data and safety assessment of phage therapy in humans. Curr. Opin. Biotechnol. 2021, 68, 310–317. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef]
- Mutalik, V.K.; Arkin, A.P. A phage foundry framework to systematically develop viral countermeasures to combat antibiotic-resistant bacterial pathogens. iScience 2022, 25, 104121. [Google Scholar] [CrossRef]
- Serwer, P.; Wright, E.T.; De La Chapa, J.; Gonzales, C.B. Basics for improved use of phages for therapy. Antibiotics 2021, 10, 723. [Google Scholar] [CrossRef]
- Merril, C.R.; Biswas, B.; Carlton, R.; Jensen, N.C.; Creed, G.J.; Zullo, S.; Adhya, S. Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. USA 1996, 93, 3188–3192. [Google Scholar] [CrossRef] [Green Version]
- Cerella, C.; Radogna, F.; Dicato, M.; Diederich, M. Natural compounds as regulators of the cancer cell metabolism. Int. J. Cell Biol. 2013, 2013, 639401. [Google Scholar] [CrossRef] [Green Version]
- Li, L.Y.; Guan, Y.D.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA Repair pathways in cancer therapy and resistance. Front. Pharmacol. 2021, 11, 629266. [Google Scholar] [CrossRef]
- Amjad, M.T.; Chidharl, A.; Kasi, A. Cancer Chemotherapy. 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK564367/ (accessed on 22 May 2022).
- Schirrmacher, V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment (Review). Int. J. Oncol. 2019, 54, 407–419. [Google Scholar]
- Lodovichi, S.; Cervelli, T.; Pellicioli, A.; Galli, A. Inhibition of DNA repair in cancer therapy: Toward a multi-target approach. Int. J. Mol. Sci. 2020, 21, 6684. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, P.K. DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Sig. Transduct. Target. Ther. 2021, 6, 254. [Google Scholar] [CrossRef]
- Ramos, A.; Sadeghi, S.; Tabatabaeian, H. Battling chemoresistance in cancer: Root causes and strategies to uproot them. Int. J. Mol. Sci. 2021, 22, 9451. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Zheng, M.; Huang, J.; Tong, A.; Yang, H. Oncolytic viruses for cancer therapy: Barriers and recent advances. Mol. Ther. Oncolytics 2019, 15, 234–247. [Google Scholar] [CrossRef] [Green Version]
- Azizian, K.; Pustokhina, I.; Ghanavati, R.; Hamblin, M.; Amini, A.; Kouhsari, E. The potential use of theranostic bacteria in cancer. J. Cell. Physiol. 2021, 236, 4184–4194. [Google Scholar] [CrossRef]
- Sedighi, M.; Zahedi Bialvaei, A.; Hamblin, M.R.; Ohadi, E.; Asadi, A.; Halajzadeh, M.; Lohrasbi, V.; Mohammadzadeh, N.; Amiriani, T.; Krutova, M.; et al. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. 2019, 8, 3167–3181. [Google Scholar] [CrossRef]
- Forbes, N.S.; Coffin, R.S.; Deng, L.; Evgin, L.; Fiering, S.; Giacalone, M.; Gravekamp, C.; Gulley, J.L.; Gunn, H.; Hoffman, R.M.; et al. White paper on microbial anti-cancer therapy and prevention. J. Immunother. Cancer 2018, 6, 78. [Google Scholar] [CrossRef]
- Liu, P.; Chen, G.; Zhang, J. A review of liposomes as a drug delivery system: Current status of approved products, regulatory environments, and future perspectives. Molecules 2022, 27, 1372. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef]
- Danhier, F. To exploit the tumor microenvironment: Since the EPR effect fails in the clinic, what is the future of nanomedicine? J. Control. Release 2016, 244, 108–121. [Google Scholar] [CrossRef]
- Barenholz, Y. Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Wu, J. The enhanced permeability and retention (EPR) effect: The significance of the concept and methods to enhance its application. J. Pers. Med. 2021, 11, 771. [Google Scholar] [CrossRef]
- Mahato, R.; Tai, W.; Cheng, K. Prodrugs for improving tumor targetability and efficiency. Adv. Drug Deliv. Rev. 2011, 63, 659–670. [Google Scholar] [CrossRef] [Green Version]
- Souza, C.; Pellosi, D.S.; Tedesco, A.C. Prodrugs for targeted cancer therapy. Expert Rev. Anticancer Ther. 2019, 19, 483–502. [Google Scholar] [CrossRef]
- Abraham, S.A.; Waterhouse, D.N.; Mayer, L.D.; Cullis, P.R.; Madden, T.D.; Bally, M.B. The liposomal formulation of doxorubicin. Meth. Enzymol. 2005, 39, 71–96. [Google Scholar]
- Griess, G.A.; Khan, S.A.; Serwer, P. Variation of the permeability of bacteriophage T4: Analysis by use of a protein-specific probe for the T4 interior. Biopolymers 1991, 31, 11–21. [Google Scholar] [CrossRef]
- Serwer, P.; Wright, E.T.; Gonzales, C.B. Phage capsids as gated, long-persistence, uniform drug delivery vehicles. In Current and Future Aspects of Nanomedicine; Khalil, I., Ed.; InTech Open: Rijeka, Croatia, 2020; Available online: https://www.intechopen.com/online-first/phage-capsids-as-gated-long-persistence-uniform-drug-delivery-vehicles (accessed on 23 May 2022).
- Majewska, J.; Beta, W.; Lecion, D.; Hodyra-Stefaniak, K.; Kłopot, A.; Kaźmierczak, Z.; Miernikiewicz, P.; Piotrowicz, A.; Ciekot, J.; Owczarek, B.; et al. Oral application of T4 phage induces weak antibody production in the gut and in the blood. Viruses 2015, 7, 4783–4799. [Google Scholar] [CrossRef]
- Majewska, J.; Kaźmierczak, Z.; Lahutta, K.; Lecion, D.; Szymczak, A.; Miernikiewicz, P.; Drapała, J.; Harhala, M.; Marek-Bukowiec, K.; Jędruchniewicz, N.; et al. Induction of phage-specific antibodies by two therapeutic staphylococcal bacteriophages administered per os. Front. Immunol. 2019, 10, 2607. [Google Scholar] [CrossRef]
- Dugger, B.N.; Dickson, D.W. Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 2017, 9, a028035. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.A.; Leyns, C.E.G.; Holtzman, D.M. Intercellular spread of protein aggregates in neurodegenerative disease. Annu. Rev. Cell Dev. Biol. 2018, 34, 545–568. [Google Scholar] [CrossRef]
- Miller, J.H.; Das, V. Potential for treatment of neurodegenerative diseases with natural products or synthetic compounds that stabilize microtubules. Curr. Pharm. Des. 2020, 26, 4362–4372. [Google Scholar] [CrossRef]
- Steinhilb, M.L.; Khanna, M. Animal models of neurodegenerative disease: Recent advances in fly highlight innovative approaches to drug discovery. Front. Mol. Neurosci. 2022, 15, 883358. [Google Scholar]
- Serwer, P.; Wright, E.T. A protein assembly hypothesis for population-specific decrease in dementia with time. Biophysica 2021, 1, 15–21. [Google Scholar] [CrossRef]
- Serwer, P.; Wright, E.T.; Hunter, B. Additions to alpha-sheet based hypotheses for the cause of Alzheimer’s disease. J. Alzheimer’s Dis. 2022, 1–10. [Google Scholar] [CrossRef]
- Itzhaki, R.F. Corroboration of a major role for herpes simplex virus type 1 in Alzheimer’s disease. Front. Aging Neurosci. 2018, 10, 324. [Google Scholar] [CrossRef] [Green Version]
- Itzhaki, R.F.; Golde, T.E.; Heneka, M.T.; Readhead, B. Do infections have a role in the pathogenesis of Alzheimer disease? Nat. Rev. Neurol. 2020, 16, 193–197. [Google Scholar] [CrossRef]
- Studier, F.W. Relationships among different strains of T7 and among T7-related bacteriophages. Virology 1979, 95, 70–84. [Google Scholar] [CrossRef]
- Serwer, P.; Watson, R.H.; Hayes, S.J.; Allen, J.L. Comparison of the physical properties and assembly pathways of the related bacteriophages T7, T3 and phi II. J. Mol. Biol. 1983, 170, 447–469. [Google Scholar] [CrossRef]
- Fang, P.A.; Wright, E.T.; Weintraub, S.T.; Hakala, K.; Wu, W.; Serwer, P.; Jiang, W. Visualization of bacteriophage T3 capsids with DNA incompletely packaged in vivo. J. Mol. Biol. 2008, 384, 1384–1399. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serwer, P. A Perspective on Studies of Phage DNA Packaging Dynamics. Int. J. Mol. Sci. 2022, 23, 7854. https://doi.org/10.3390/ijms23147854
Serwer P. A Perspective on Studies of Phage DNA Packaging Dynamics. International Journal of Molecular Sciences. 2022; 23(14):7854. https://doi.org/10.3390/ijms23147854
Chicago/Turabian StyleSerwer, Philip. 2022. "A Perspective on Studies of Phage DNA Packaging Dynamics" International Journal of Molecular Sciences 23, no. 14: 7854. https://doi.org/10.3390/ijms23147854
APA StyleSerwer, P. (2022). A Perspective on Studies of Phage DNA Packaging Dynamics. International Journal of Molecular Sciences, 23(14), 7854. https://doi.org/10.3390/ijms23147854





