The Increase in the Drug Resistance of Acute Myeloid Leukemia THP-1 Cells in High-Density Cell Culture Is Associated with Inflammatory-like Activation and Anti-Apoptotic Bcl-2 Proteins
Abstract
1. Introduction
2. Results
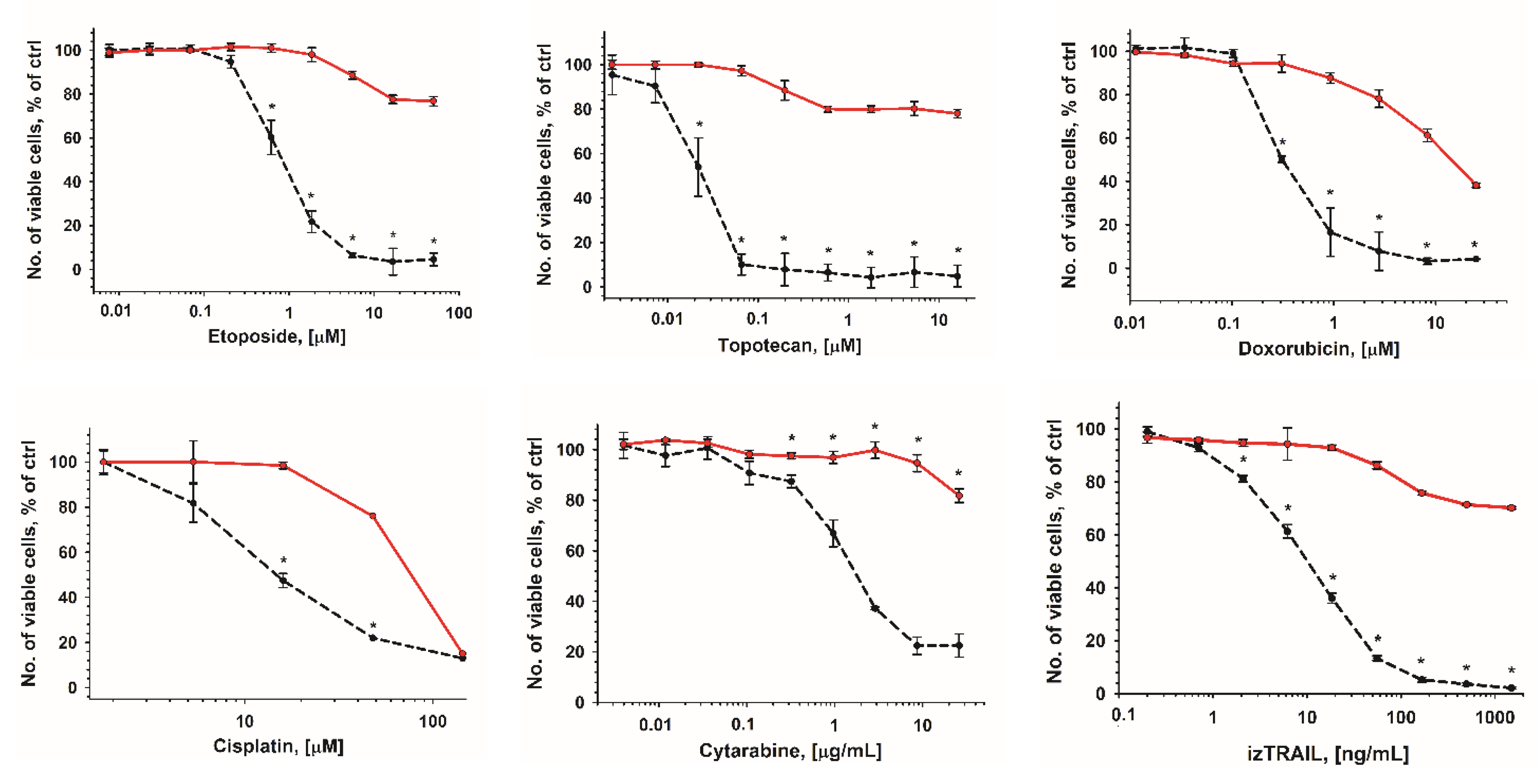
2.1. Reversible Increase in the Resistance of THP-1 Cells in High-Density Cell Cultures to DNA-Damaging Drugs and izTRAIL
2.2. The Increase in Drug Resistance of THP-1 Cells in High-Density Cultures May Depend Not Only on Culture Density
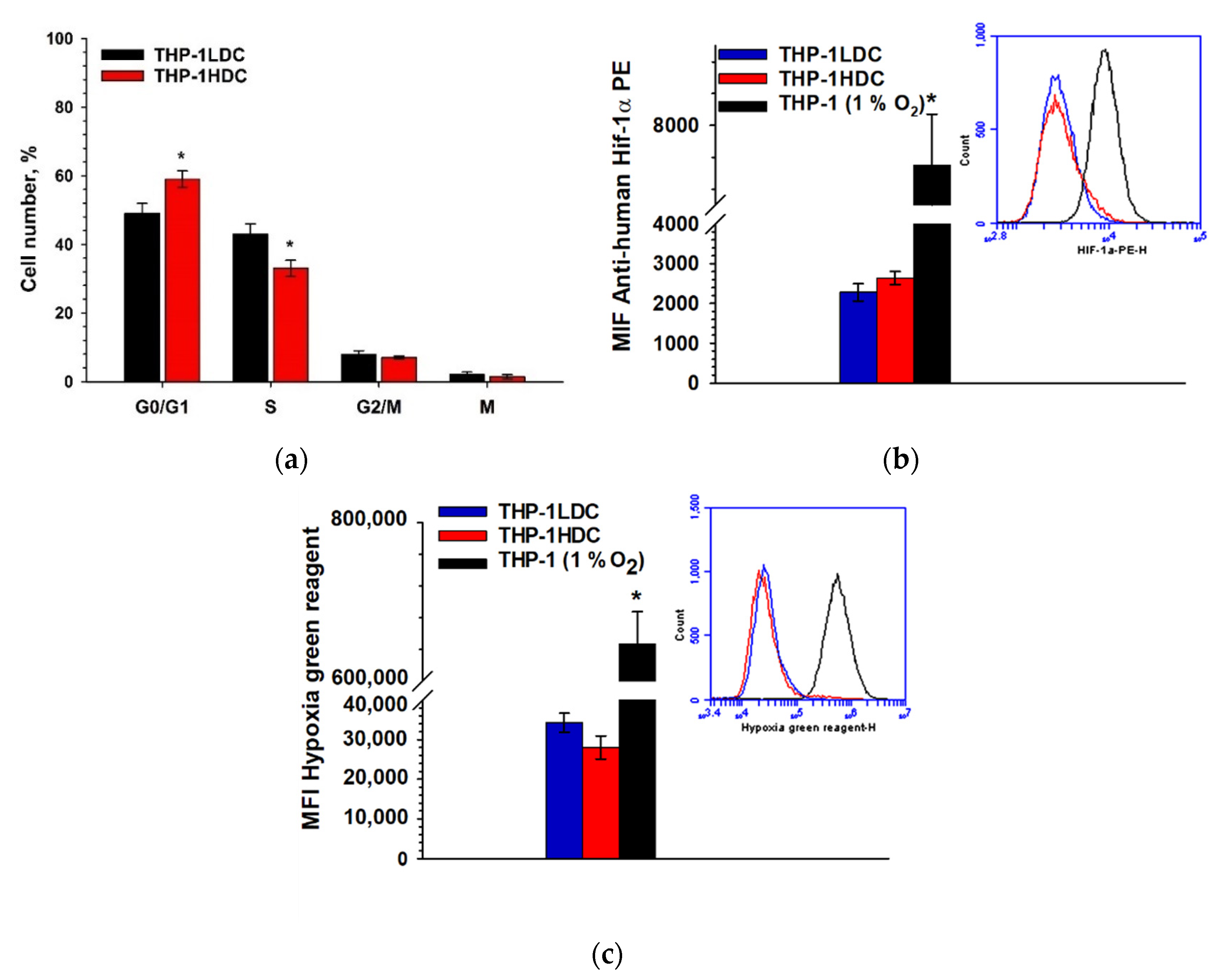
2.3. The Increase in Drug Resistance of THP-1HDC Cells Occurs While Maintaining Their Proliferative Activity and Is Not Associated with Cell Hypoxia
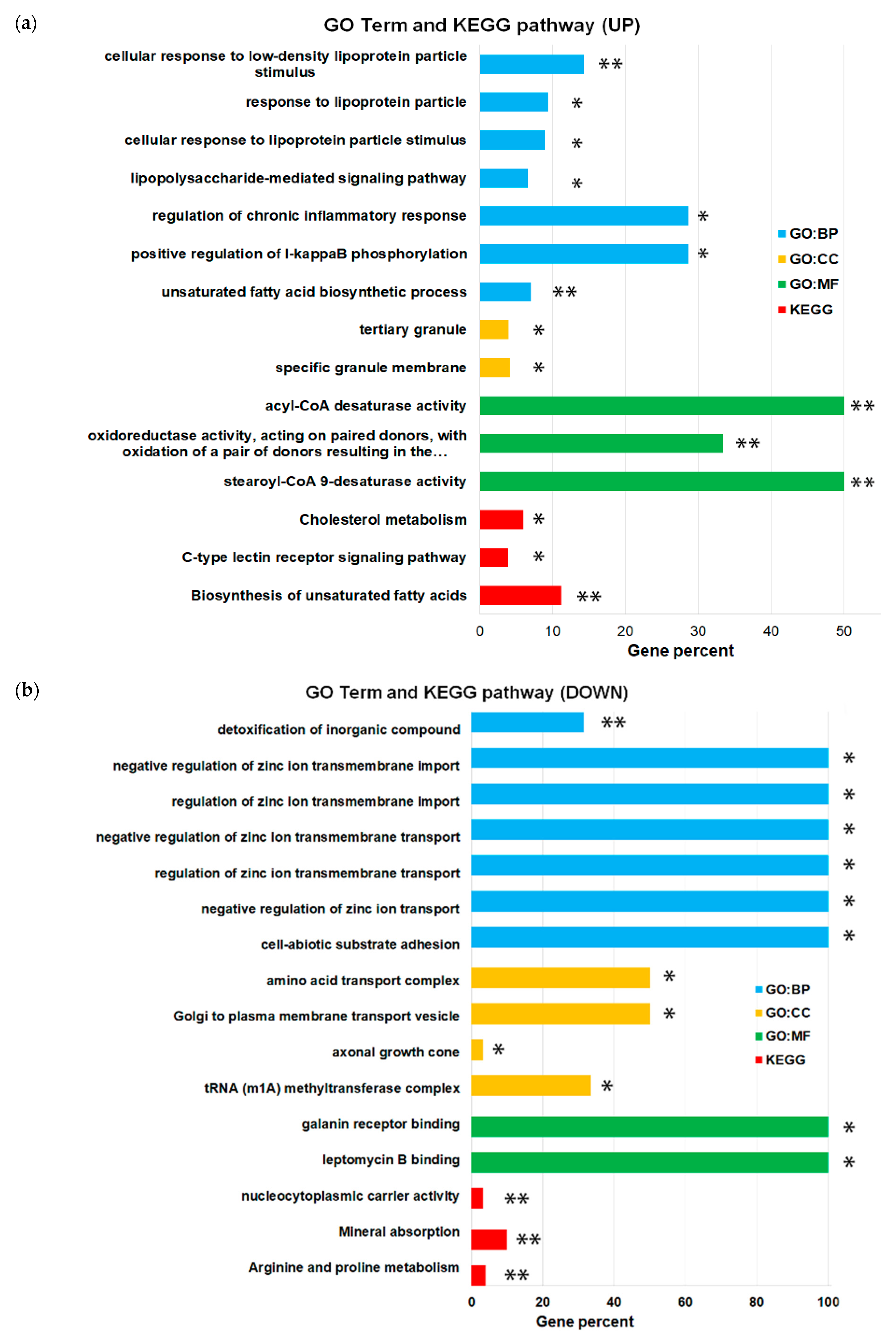
2.4. Comparative Bioinformatics Analysis of the Transcriptomes of THP-1HDC and THP-1LDC Cells
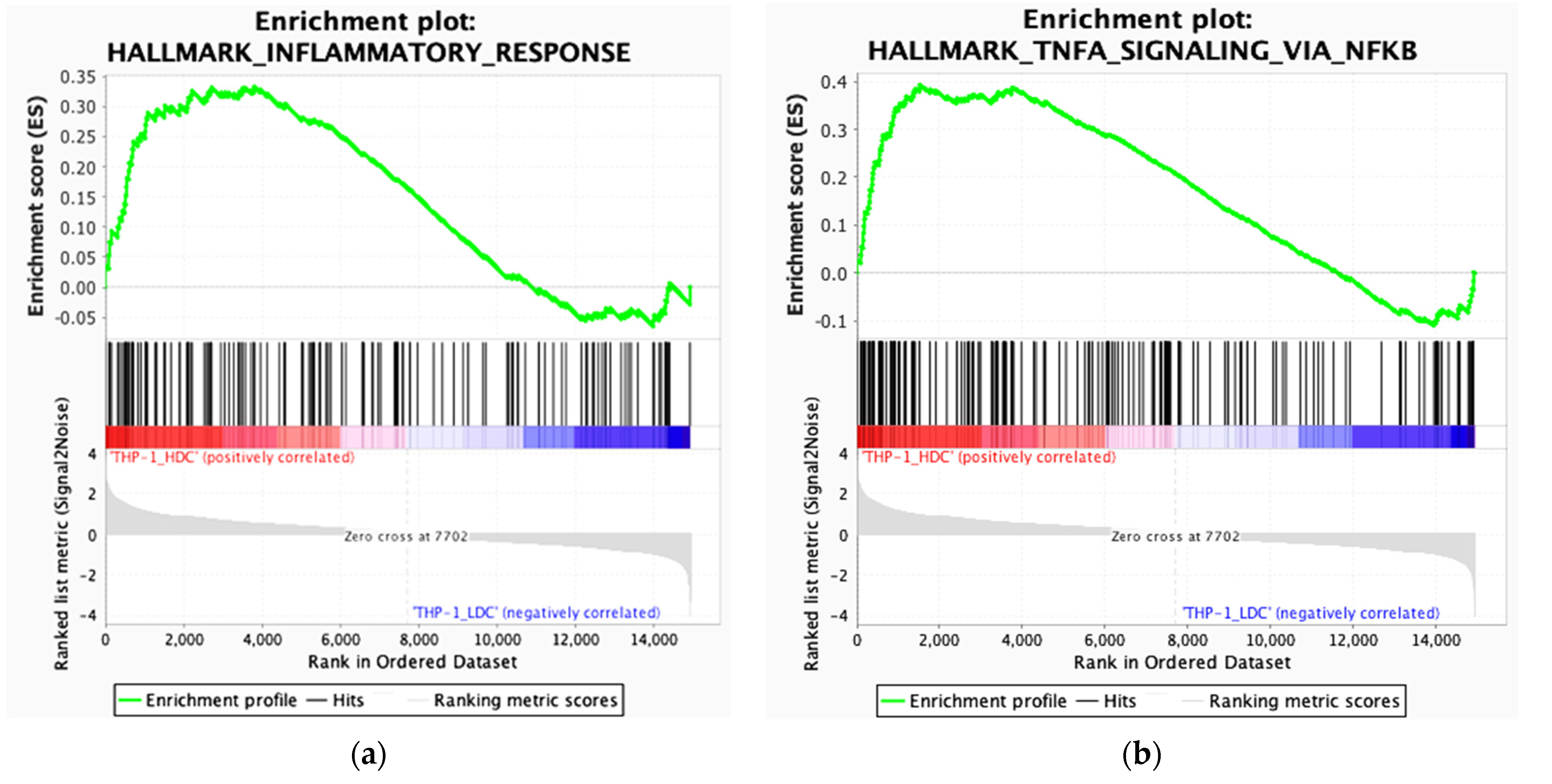
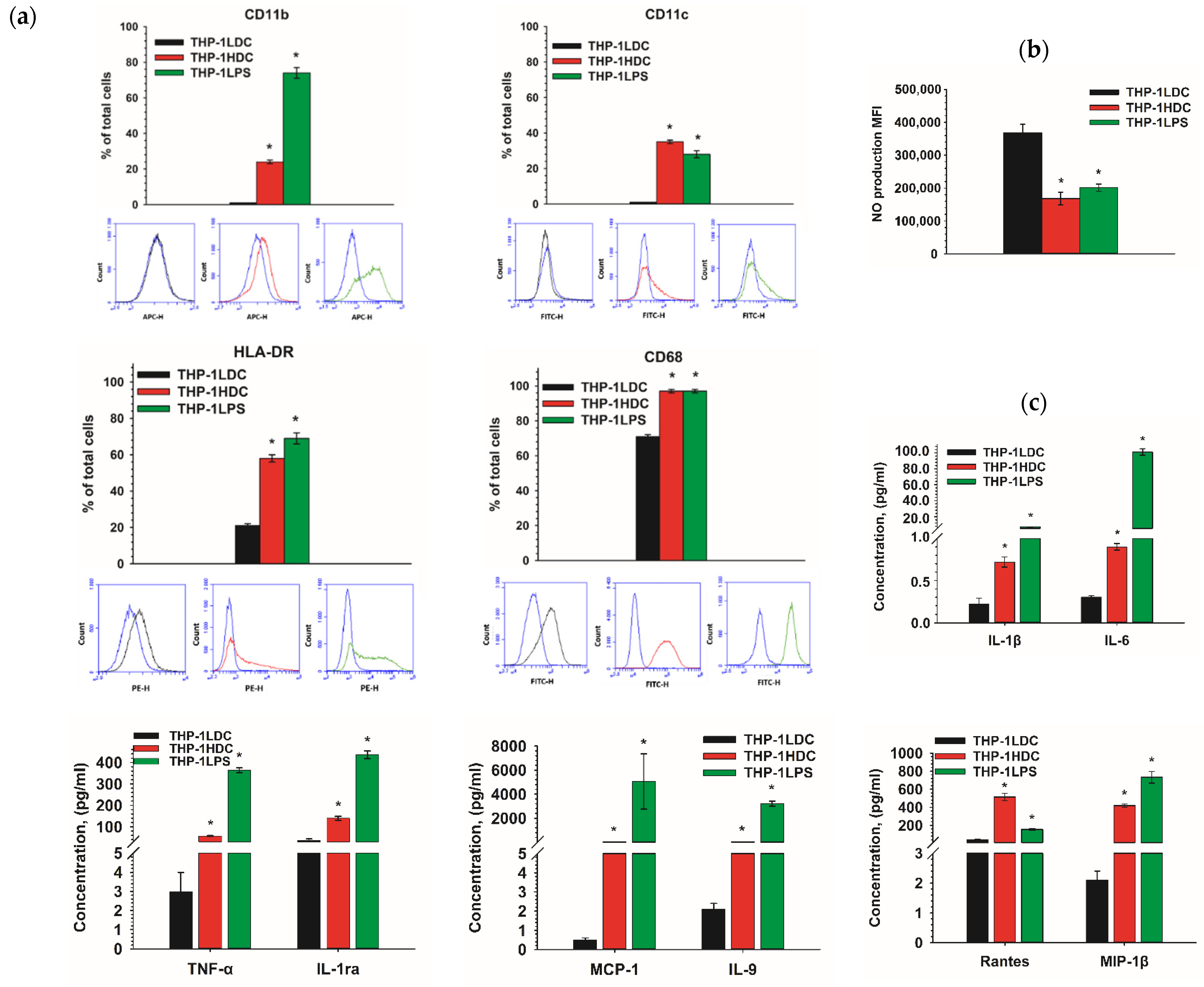
2.5. THP-1HDC Cells Demonstrate Signs of Inflammatory-like Activation
2.6. The Transcription Factor NF-kB Is Activated and the Content of Anti-Apoptotic Proteins Bcl-2 and Bcl-xL Increases in THP-1HDC Cells
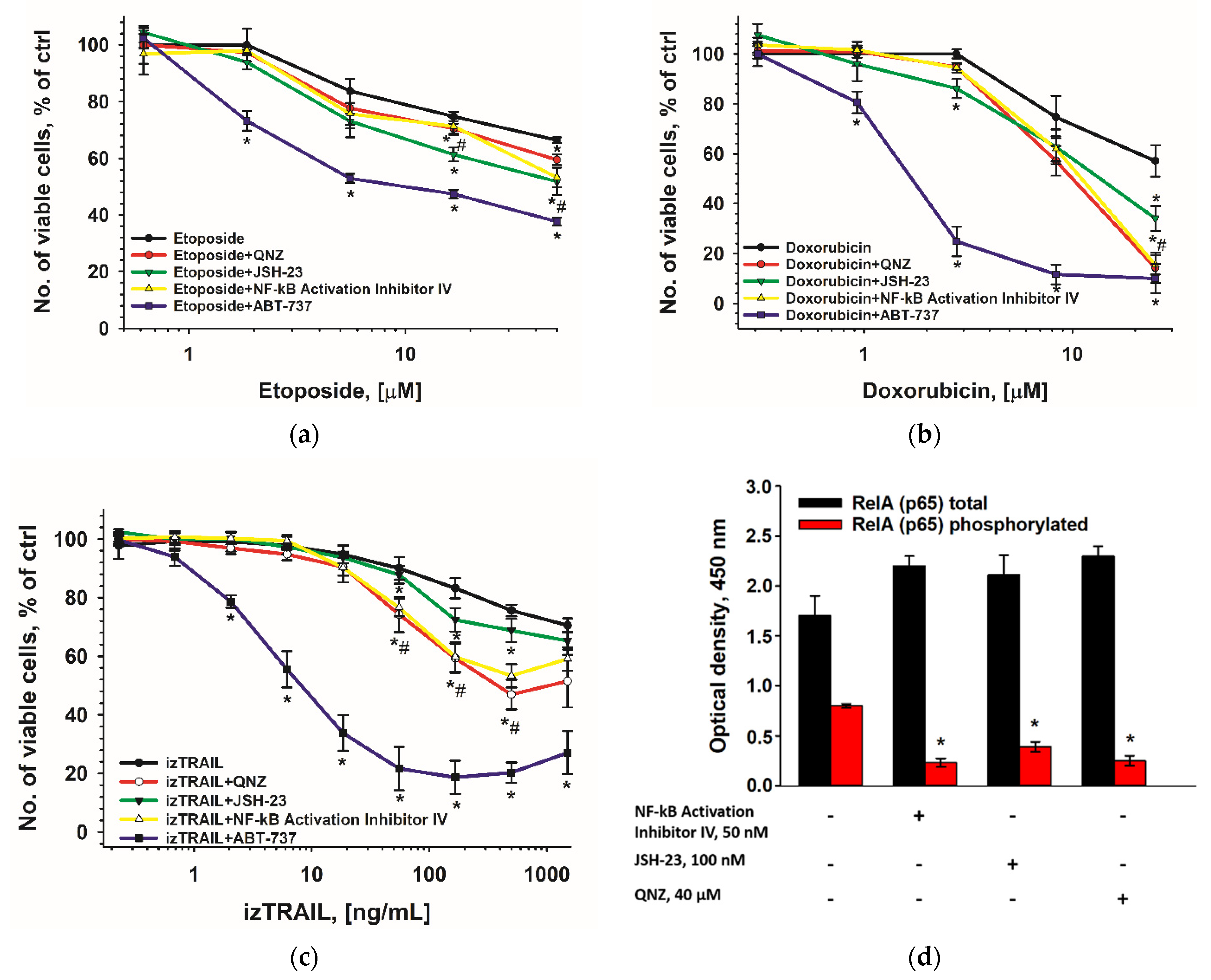
2.7. The Inhibitors of NF-kB and Anti-Apoptotic Proteins of the Bcl-2 Family Contribute to Suppression of the Increased Drug Resistance in THP-1HDC Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Protocol of izTRAIL Preparation
4.3. Cell Cultures
4.4. Cytotoxicity Assay and Drug Treatment
4.5. Cell Immunophenotyping
4.6. Cell Proliferation and Cell Viability Assays
4.7. Cell Hypoxia Assessment
4.8. Analysis of Genome-Wide Transcriptional Activity
4.9. Bioinformatics Analyses of Transcriptome
4.10. Intracellular NO Assay
4.11. Multiplex Analysis of Cytokine Production
4.12. Analysis of NF-kB Activation
4.13. Western Blotting Analysis
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sorf, A.; Sucha, S.; Morell, A.; Novotna, E.; Staud, F.; Zavrelova, A.; Visek, B.; Wsol, V.; Ceckova, M. Targeting Pharmacokinetic Drug Resistance in Acute Myeloid Leukemia Cells with CDK4/6 Inhibitors. Cancers 2020, 12, 1596. [Google Scholar] [CrossRef]
- Daver, N.; Schlenk, R.F.; Russell, N.H.; Levis, M.J. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia 2019, 33, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Montalban-Bravo, G.; Garcia-Manero, G. Novel drugs for older patients with acute myeloid leukemia. Leukemia 2015, 29, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Cucchi, D.G.J.; Groen, R.W.J.; Janssen, J.; Cloos, J. Ex vivo cultures and drug testing of primary acute myeloid leukemia samples: Current techniques and implications for experimental design and outcome. Drug Resist. Updates 2020, 53, 100730. [Google Scholar] [CrossRef] [PubMed]
- Bolandi, S.M.; Pakjoo, M.; Beigi, P.; Kiani, M.; Allahgholipour, A.; Goudarzi, N.; Khorashad, J.S.; Eiring, A.M. A Role for the Bone Marrow Microenvironment in Drug Resistance of Acute Myeloid Leukemia. Cells 2021, 10, 2833. [Google Scholar] [CrossRef]
- Auberger, P.; Tamburini-Bonnefoy, J.; Puissant, A. Drug Resistance in Hematological Malignancies. Int. J. Mol. Sci. 2020, 21, 6091. [Google Scholar] [CrossRef]
- Grosse-Wilde, A.; Voloshanenko, O.; Bailey, S.L.; Longton, G.M.; Schaefer, U.; Csernok, A.I.; Schutz, G.; Greiner, E.F.; Kemp, C.J.; Walczak, H. TRAIL-R deficiency in mice enhances lymph node metastasis without affecting primary tumor development. J. Clin. Investig. 2008, 118, 100–110. [Google Scholar] [CrossRef]
- Dimberg, L.Y.; Anderson, C.K.; Camidge, R.; Behbakht, K.; Thorburn, A.; Ford, H.L. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 2013, 32, 1341–1350. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005, 12, 228–237. [Google Scholar] [CrossRef]
- Cheng, J.; Hylander, B.L.; Baer, M.R.; Chen, X.; Repasky, E.A. Multiple mechanisms underlie resistance of leukemia cells to Apo2 Ligand/TRAIL. Mol. Cancer Ther. 2006, 5, 1844–1853. [Google Scholar] [CrossRef][Green Version]
- Zhang, J.; Gu, Y.; Chen, B. Mechanisms of drug resistance in acute myeloid leukemia. OncoTargets Ther. 2019, 12, 1937–1945. [Google Scholar] [CrossRef] [PubMed]
- Bakker, E.; Qattan, M.; Mutti, L.; Demonacos, C.; Krstic-Demonacos, M. The role of microenvironment and immunity in drug response in leukemia. Biochim. Biophys. Acta 2016, 1863, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Jones, V.S.; Huang, R.Y.; Chen, L.P.; Chen, Z.S.; Fu, L.; Huang, R.P. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochim. Biophys. Acta 2016, 1865, 255–265. [Google Scholar] [CrossRef]
- Giles, F.J.; Krawczyk, J.; O’Dwyer, M.; Swords, R.; Freeman, C. The role of inflammation in leukaemia. Adv. Exp. Med. Biol. 2014, 816, 335–360. [Google Scholar] [CrossRef]
- Binder, S.; Luciano, M.; Horejs-Hoeck, J. The cytokine network in acute myeloid leukemia (AML): A focus on pro- and anti-inflammatory mediators. Cytokine Growth Factor Rev. 2018, 43, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Miao, H.; Zhang, Y.; Li, W.; Li, Z.; Zhou, Y.; Zhao, L.; Guo, Q. Bone marrow microenvironment confers imatinib resistance to chronic myelogenous leukemia and oroxylin A reverses the resistance by suppressing Stat3 pathway. Arch. Toxicol. 2015, 89, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Rajput, D.; Naval, R.; Yadav, K.; Tungaria, A.; Behari, S. Bilateral proptosis and bitemporal swelling: A rare manifestation of acute myeloid leukemia. J. Pediatric Neurosci. 2010, 5, 68–71. [Google Scholar] [CrossRef]
- Hagiya, A.; Vaidya, P.; Khedro, T.; Yaghmour, B.; Siddiqi, I.; Yaghmour, G. Bone Marrow Features in Patients With Acute Myeloid Leukemia Treated With Novel Targeted Isocitrate Dehydrogenase 1/2 Inhibitors. World J. Oncol. 2019, 10, 226–230. [Google Scholar] [CrossRef]
- Vucetic, M.; Daher, B.; Cassim, S.; Meira, W.; Pouyssegur, J. Together we stand, apart we fall: How cell-to-cell contact/interplay provides resistance to ferroptosis. Cell Death Dis. 2020, 11, 789. [Google Scholar] [CrossRef]
- Gujral, T.S.; Kirschner, M.W. Hippo pathway mediates resistance to cytotoxic drugs. Proc. Natl. Acad. Sci. USA 2017, 114, E3729–E3738. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Choi, N.; Kim, K.; Koo, H.J.; Choi, J.; Kim, H.N. Chemoresistance of Cancer Cells: Requirements of Tumor Microenvironment-mimicking In Vitro Models in Anti-Cancer Drug Development. Theranostics 2018, 8, 5259–5275. [Google Scholar] [CrossRef] [PubMed]
- Navratilova, J.; Jungova, P.; Vanhara, P.; Preisler, J.; Kanicky, V.; Smarda, J. Copper ions regulate cytotoxicity of disulfiram to myeloid leukemia cells. Int. J. Mol. Med. 2009, 24, 661–670. [Google Scholar] [CrossRef]
- Lavi, O.; Greene, J.M.; Levy, D.; Gottesman, M.M. The role of cell density and intratumoral heterogeneity in multidrug resistance. Cancer Res. 2013, 73, 7168–7175. [Google Scholar] [CrossRef] [PubMed]
- Grantab, R.; Sivananthan, S.; Tannock, I.F. The penetration of anticancer drugs through tumor tissue as a function of cellular adhesion and packing density of tumor cells. Cancer Res. 2006, 66, 1033–1039. [Google Scholar] [CrossRef]
- Sabelstrom, H.; Quigley, D.A.; Fenster, T.; Foster, D.J.; Fuchshuber, C.A.M.; Saxena, S.; Yuan, E.; Li, N.; Paterno, F.; Phillips, J.J.; et al. High density is a property of slow-cycling and treatment-resistant human glioblastoma cells. Exp. Cell Res. 2019, 378, 76–86. [Google Scholar] [CrossRef]
- He, Y.; Zhu, Q.; Chen, M.; Huang, Q.; Wang, W.; Li, Q.; Huang, Y.; Di, W. The changing 50% inhibitory concentration (IC50) of cisplatin: A pilot study on the artifacts of the MTT assay and the precise measurement of density-dependent chemoresistance in ovarian cancer. Oncotarget 2016, 7, 70803–70821. [Google Scholar] [CrossRef]
- Pearl Mizrahi, S.; Gefen, O.; Simon, I.; Balaban, N.Q. Persistence to anti-cancer treatments in the stationary to proliferating transition. Cell Cycle 2016, 15, 3442–3453. [Google Scholar] [CrossRef]
- Menegakis, A.; Klompmaker, R.; Vennin, C.; Arbusa, A.; Damen, M.; van den Broek, B.; Zips, D.; van Rheenen, J.; Krenning, L.; Medema, R.H. Resistance of Hypoxic Cells to Ionizing Radiation Is Mediated in Part via Hypoxia-Induced Quiescence. Cells 2021, 10, 610. [Google Scholar] [CrossRef]
- Xu, P.; Wang, M.; Jiang, Y.; Ouyang, J.; Chen, B. The association between expression of hypoxia inducible factor-1α and multi-drug resistance of acute myeloid leukemia. Transl. Cancer Res. 2017, 6, 198–205. [Google Scholar] [CrossRef]
- Viatour, P.; Merville, M.P.; Bours, V.; Chariot, A. Phosphorylation of NF-kappaB and IkappaB proteins: Implications in cancer and inflammation. Trends Biochem. Sci. 2005, 30, 43–52. [Google Scholar] [CrossRef]
- Solakivi, T.; Kunnas, T.; Jaakkola, O.; Renko, J.; Lehtimaki, T.; Nikkari, S.T. Delta-6-desaturase gene polymorphism is associated with lipoprotein oxidation in vitro. Lipids Health Dis. 2013, 12, 80. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Greene, E.R.; Huang, S.; Serhan, C.N.; Panigrahy, D. Regulation of inflammation in cancer by eicosanoids. Prostaglandins Other Lipid Mediat. 2011, 96, 27–36. [Google Scholar] [CrossRef]
- Calder, P.C. Polyunsaturated fatty acids and inflammatory processes: New twists in an old tale. Biochimie 2009, 91, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Shinkai, H. Leptomycin B reduces matrix metalloproteinase-9 expression and suppresses cutaneous inflammation. J. Investig. Dermatol. 2005, 124, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.C.; Munoz-Najar, U.; Paik, J.H.; Claffey, K.; Yoshida, M.; Hla, T. Leptomycin B, an inhibitor of the nuclear export receptor CRM1, inhibits COX-2 expression. J. Biol. Chem. 2003, 278, 2773–2776. [Google Scholar] [CrossRef]
- Kimura, T.; Hashimoto, I.; Nagase, T.; Fujisawa, J. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-alpha1 mRNA. J. Cell Sci. 2004, 117, 2259–2270. [Google Scholar] [CrossRef]
- Ghosh, C.C.; Vu, H.Y.; Mujo, T.; Vancurova, I. Analysis of nucleocytoplasmic shuttling of NF kappa B proteins in human leukocytes. Methods Mol. Biol. 2008, 457, 279–292. [Google Scholar] [CrossRef]
- Koller, A.; Brunner, S.M.; Bianchini, R.; Ramspacher, A.; Emberger, M.; Locker, F.; Schlager, S.; Kofler, B. Galanin is a potent modulator of cytokine and chemokine expression in human macrophages. Sci. Rep. 2019, 9, 7237. [Google Scholar] [CrossRef]
- Jarosz, M.; Olbert, M.; Wyszogrodzka, G.; Mlyniec, K.; Librowski, T. Antioxidant and anti-inflammatory effects of zinc. Zinc-dependent NF-kappaB signaling. Inflammopharmacology 2017, 25, 11–24. [Google Scholar] [CrossRef]
- Zelova, H.; Hosek, J. TNF-alpha signalling and inflammation: Interactions between old acquaintances. Inflamm. Res. 2013, 62, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Tomar, D.; Sripada, L.; Prajapati, P.; Singh, R.; Singh, A.K.; Singh, R. Nucleo-cytoplasmic trafficking of TRIM8, a novel oncogene, is involved in positive regulation of TNF induced NF-kappaB pathway. PLoS ONE 2012, 7, e48662. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Biehl, A.; Gadina, M.; Hasni, S.; Schwartz, D.M. JAK-STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs 2017, 77, 521–546. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Tripathi, P.; Kashyap, L.; Singh, V. The role of nitric oxide in inflammatory reactions. FEMS Immunol. Med. Microbiol. 2007, 51, 443–452. [Google Scholar] [CrossRef]
- Gudowska-Sawczuk, M.; Mroczko, B. The Role of Nuclear Factor Kappa B (NF-kappaB) in Development and Treatment of COVID-19: Review. Int. J. Mol. Sci. 2022, 23, 5283. [Google Scholar] [CrossRef] [PubMed]
- Poma, P. NF-kappaB and Disease. Int. J. Mol. Sci. 2020, 21, 9181. [Google Scholar] [CrossRef]
- Banjara, S.; Suraweera, C.D.; Hinds, M.G.; Kvansakul, M. The Bcl-2 Family: Ancient Origins, Conserved Structures, and Divergent Mechanisms. Biomolecules 2020, 10, 128. [Google Scholar] [CrossRef]
- Ganesan, S.; Mathews, V.; Vyas, N. Microenvironment and drug resistance in acute myeloid leukemia: Do we know enough? Int. J. Cancer 2022, 150, 1401–1411. [Google Scholar] [CrossRef]
- Tabe, Y.; Konopleva, M. Role of Microenvironment in Resistance to Therapy in AML. Curr. Hematol. Malig. Rep. 2015, 10, 96–103. [Google Scholar] [CrossRef]
- Masquelier, M.; Vitols, S. Drastic effect of cell density on the cytotoxicity of daunorubicin and cytosine arabinoside. Biochem. Pharmacol. 2004, 67, 1639–1646. [Google Scholar] [CrossRef]
- Takemura, Y.; Kobayashi, H.; Miyachi, H.; Hayashi, K.; Sekiguchi, S.; Ohnuma, T. The influence of tumor cell density on cellular accumulation of doxorubicin or cisplatin in vitro. Cancer Chemother. Pharmacol. 1991, 27, 417–422. [Google Scholar] [CrossRef]
- Jeon, K.H.; Park, S.; Jang, H.J.; Hwang, S.Y.; Shrestha, A.; Lee, E.S.; Kwon, Y. AK-I-190, a New Catalytic Inhibitor of Topoisomerase II with Anti-Proliferative and Pro-Apoptotic Activity on Androgen-Negative Prostate Cancer Cells. Int. J. Mol. Sci. 2021, 22, 11246. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.C.; Chiang, S.F.; Yang, P.C.; Ke, T.W.; Chen, T.W.; Hu, C.H.; Huang, Y.W.; Chang, H.Y.; Chen, W.T.; Chao, K.S.C. Immunogenic Cell Death by the Novel Topoisomerase I Inhibitor TLC388 Enhances the Therapeutic Efficacy of Radiotherapy. Cancers 2021, 13, 1218. [Google Scholar] [CrossRef] [PubMed]
- Serhan, N.; Mouchel, P.L.; Medina, P.; Segala, G.; Mougel, A.; Saland, E.; Rives, A.; Lamaziere, A.; Despres, G.; Sarry, J.E.; et al. Dendrogenin A synergizes with Cytarabine to Kill Acute Myeloid Leukemia Cells In Vitro and In Vivo. Cancers 2020, 12, 1725. [Google Scholar] [CrossRef] [PubMed]
- Baguley, B.C.; Drummond, C.J.; Chen, Y.Y.; Finlay, G.J. DNA-Binding Anticancer Drugs: One Target, Two Actions. Molecules 2021, 26, 552. [Google Scholar] [CrossRef]
- Tsafa, E.; Bentayebi, K.; Topanurak, S.; Yata, T.; Przystal, J.; Fongmoon, D.; Hajji, N.; Waramit, S.; Suwan, K.; Hajitou, A. Doxorubicin Improves Cancer Cell Targeting by Filamentous Phage Gene Delivery Vectors. Int. J. Mol. Sci. 2020, 21, 7867. [Google Scholar] [CrossRef]
- Montagner, D.; Tolan, D.; Andriollo, E.; Gandin, V.; Marzano, C. A Pt(IV) Prodrug Combining Chlorambucil and Cisplatin: A Dual-Acting Weapon for Targeting DNA in Cancer Cells. Int. J. Mol. Sci. 2018, 19, 3775. [Google Scholar] [CrossRef]
- Zhang, Z.; Patel, S.B.; King, M.R. Micelle-in-Liposomes for Sustained Delivery of Anticancer Agents That Promote Potent TRAIL-Induced Cancer Cell Apoptosis. Molecules 2020, 26, 157. [Google Scholar] [CrossRef]
- Valkov, N.I.; Gump, J.L.; Engel, R.; Sullivan, D.M. Cell density-dependent VP-16 sensitivity of leukaemic cells is accompanied by the translocation of topoisomerase IIalpha from the nucleus to the cytoplasm. Br. J. Haematol. 2000, 108, 331–345. [Google Scholar] [CrossRef]
- Cheon, H.; Borden, E.C.; Stark, G.R. Interferons and their stimulated genes in the tumor microenvironment. Semin. Oncol. 2014, 41, 156–173. [Google Scholar] [CrossRef]
- Nuan-Aliman, S.; Bordereaux, D.; Thieblemont, C.; Baud, V. The Alternative RelB NF-kB Subunit Exerts a Critical Survival Function upon Metabolic Stress in Diffuse Large B-Cell Lymphoma-Derived Cells. Biomedicines 2022, 10, 348. [Google Scholar] [CrossRef] [PubMed]
- Kaltschmidt, B.; Witte, K.E.; Greiner, J.F.W.; Weissinger, F.; Kaltschmidt, C. Targeting NF-kappaB Signaling in Cancer Stem Cells: A Narrative Review. Biomedicines 2022, 10, 261. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.; Banerjee, S.; Mondal, A.; Chakraborty, U.; Pumarol, J.; Croley, C.R.; Bishayee, A. Targeting the JAK/STAT Signaling Pathway Using Phytocompounds for Cancer Prevention and Therapy. Cells 2020, 9, 1451. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Role of JAK/STAT3 Signaling in the Regulation of Metastasis, the Transition of Cancer Stem Cells, and Chemoresistance of Cancer by Epithelial-Mesenchymal Transition. Cells 2020, 9, 217. [Google Scholar] [CrossRef]
- Scholz, C.C.; Cavadas, M.A.; Tambuwala, M.M.; Hams, E.; Rodriguez, J.; von Kriegsheim, A.; Cotter, P.; Bruning, U.; Fallon, P.G.; Cheong, A.; et al. Regulation of IL-1beta-induced NF-kappaB by hydroxylases links key hypoxic and inflammatory signaling pathways. Proc. Natl. Acad. Sci. USA 2013, 110, 18490–18495. [Google Scholar] [CrossRef]
- Shah, A.; Plaza-Sirvent, C.; Weinert, S.; Buchbinder, J.H.; Lavrik, I.N.; Mertens, P.R.; Schmitz, I.; Lindquist, J.A. YB-1 Mediates TNF-Induced Pro-Survival Signaling by Regulating NF-kappaB Activation. Cancers 2020, 12, 2188. [Google Scholar] [CrossRef]
- Haugen, J.; Chandyo, R.K.; Brokstad, K.A.; Mathisen, M.; Ulak, M.; Basnet, S.; Valentiner-Branth, P.; Strand, T.A. Cytokine Concentrations in Plasma from Children with Severe and Non-Severe Community Acquired Pneumonia. PLoS ONE 2015, 10, e0138978. [Google Scholar] [CrossRef]
- Conti, P.; DiGioacchino, M. MCP-1 and RANTES are mediators of acute and chronic inflammation. Allergy Asthma Proc. 2001, 22, 133–137. [Google Scholar] [CrossRef]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef]
- Shimabukuro-Vornhagen, A.; Godel, P.; Subklewe, M.; Stemmler, H.J.; Schlosser, H.A.; Schlaak, M.; Kochanek, M.; Boll, B.; von Bergwelt-Baildon, M.S. Cytokine release syndrome. J. Immunother. Cancer 2018, 6, 56. [Google Scholar] [CrossRef]
- Stucki, A.; Rivier, A.S.; Gikic, M.; Monai, N.; Schapira, M.; Spertini, O. Endothelial cell activation by myeloblasts: Molecular mechanisms of leukostasis and leukemic cell dissemination. Blood 2001, 97, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Recher, C. Clinical Implications of Inflammation in Acute Myeloid Leukemia. Front. Oncol. 2021, 11, 623952. [Google Scholar] [CrossRef] [PubMed]
- Fadeev, R.; Chekanov, A.; Solovieva, M.; Bezborodova, O.; Nemtsova, E.; Dolgikh, N.; Fadeeva, I.; Senotov, A.; Kobyakova, M.; Evstratova, Y.; et al. Improved Anticancer Effect of Recombinant Protein izTRAIL Combined with Sorafenib and Peptide iRGD. Int. J. Mol. Sci. 2019, 20, 525. [Google Scholar] [CrossRef] [PubMed]
- Lavogina, D.; Lust, H.; Tahk, M.J.; Laasfeld, T.; Vellama, H.; Nasirova, N.; Vardja, M.; Eskla, K.L.; Salumets, A.; Rinken, A.; et al. Revisiting the Resazurin-Based Sensing of Cellular Viability: Widening the Application Horizon. Biosensors 2022, 12, 196. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, Y.V.; Kobyakova, M.I.; Senotov, A.S.; Lomovsky, A.I.; Minaychev, V.V.; Fadeeva, I.S.; Shtatnova, D.Y.; Krasnov, K.S.; Zvyagina, A.I.; Akatov, V.S.; et al. Macrophage-like THP-1 Cells Derived from High-Density Cell Culture Are Resistant to TRAIL-Induced Cell Death via Down-Regulation of Death-Receptors DR4 and DR5. Biomolecules 2022, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Rasul, A.; Bao, R.; Malhi, M.; Zhao, B.; Tsuji, I.; Li, J.; Li, X. Induction of apoptosis by costunolide in bladder cancer cells is mediated through ROS generation and mitochondrial dysfunction. Molecules 2013, 18, 1418–1433. [Google Scholar] [CrossRef]
- Ge, X.; Son, W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Udhaya Kumar, S.; Thirumal Kumar, D.; Bithia, R.; Sankar, S.; Magesh, R.; Sidenna, M.; George Priya Doss, C.; Zayed, H. Analysis of Differentially Expressed Genes and Molecular Pathways in Familial Hypercholesterolemia Involved in Atherosclerosis: A Systematic and Bioinformatics Approach. Front. Genet. 2020, 11, 734. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobyakova, M.; Lomovskaya, Y.; Senotov, A.; Lomovsky, A.; Minaychev, V.; Fadeeva, I.; Shtatnova, D.; Krasnov, K.; Zvyagina, A.; Odinokova, I.; et al. The Increase in the Drug Resistance of Acute Myeloid Leukemia THP-1 Cells in High-Density Cell Culture Is Associated with Inflammatory-like Activation and Anti-Apoptotic Bcl-2 Proteins. Int. J. Mol. Sci. 2022, 23, 7881. https://doi.org/10.3390/ijms23147881
Kobyakova M, Lomovskaya Y, Senotov A, Lomovsky A, Minaychev V, Fadeeva I, Shtatnova D, Krasnov K, Zvyagina A, Odinokova I, et al. The Increase in the Drug Resistance of Acute Myeloid Leukemia THP-1 Cells in High-Density Cell Culture Is Associated with Inflammatory-like Activation and Anti-Apoptotic Bcl-2 Proteins. International Journal of Molecular Sciences. 2022; 23(14):7881. https://doi.org/10.3390/ijms23147881
Chicago/Turabian StyleKobyakova, Margarita, Yana Lomovskaya, Anatoly Senotov, Alexey Lomovsky, Vladislav Minaychev, Irina Fadeeva, Daria Shtatnova, Kirill Krasnov, Alena Zvyagina, Irina Odinokova, and et al. 2022. "The Increase in the Drug Resistance of Acute Myeloid Leukemia THP-1 Cells in High-Density Cell Culture Is Associated with Inflammatory-like Activation and Anti-Apoptotic Bcl-2 Proteins" International Journal of Molecular Sciences 23, no. 14: 7881. https://doi.org/10.3390/ijms23147881
APA StyleKobyakova, M., Lomovskaya, Y., Senotov, A., Lomovsky, A., Minaychev, V., Fadeeva, I., Shtatnova, D., Krasnov, K., Zvyagina, A., Odinokova, I., Akatov, V., & Fadeev, R. (2022). The Increase in the Drug Resistance of Acute Myeloid Leukemia THP-1 Cells in High-Density Cell Culture Is Associated with Inflammatory-like Activation and Anti-Apoptotic Bcl-2 Proteins. International Journal of Molecular Sciences, 23(14), 7881. https://doi.org/10.3390/ijms23147881







