Unusual Association of NF-κB Components in Tumor-Associated Macrophages (TAMs) Promotes HSPG2-Mediated Immune-Escaping Mechanism in Breast Cancer
Abstract
:1. Introduction
2. Results
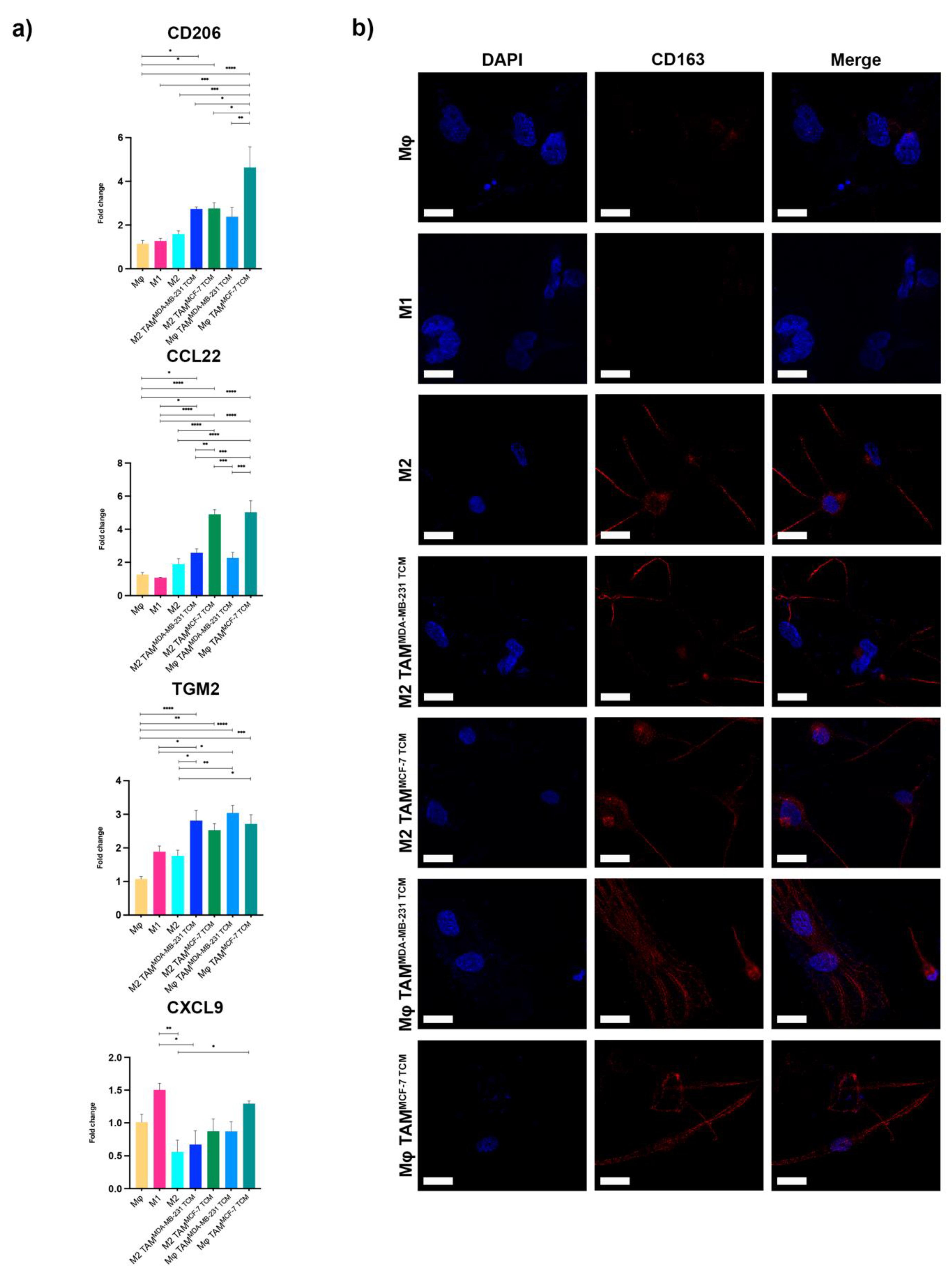
2.1. HSPG2 Overexpression in TNBC by TAM
2.2. HSPG2 Transcriptional Modulation and Unconventional NF-κB Balance in M2 Macrophages
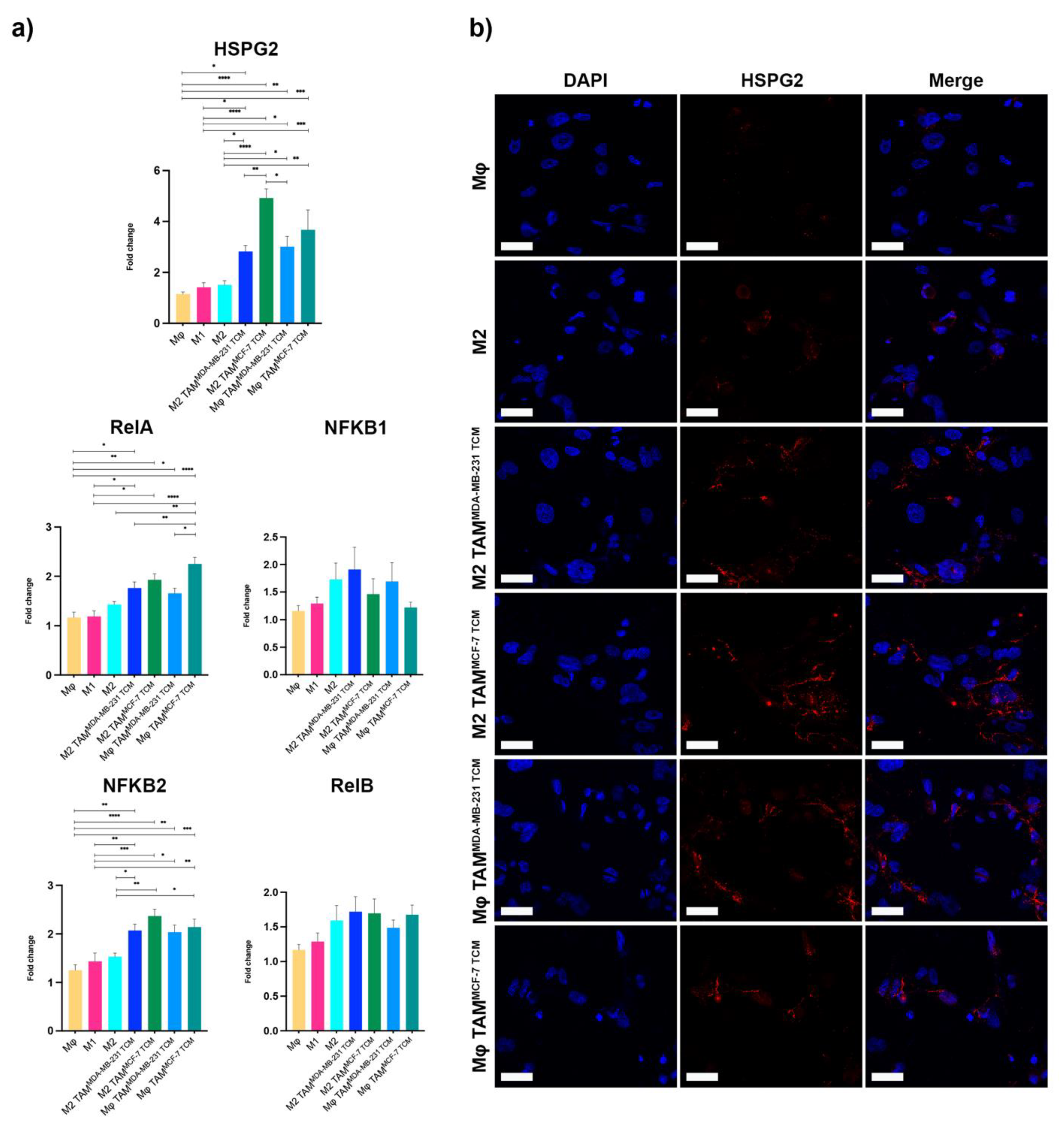
2.3. NF-κB Was Responsible for HSPG2 Expression in TAMs, Pointing to the Unexplored p65/p52 Heterodimer Formation
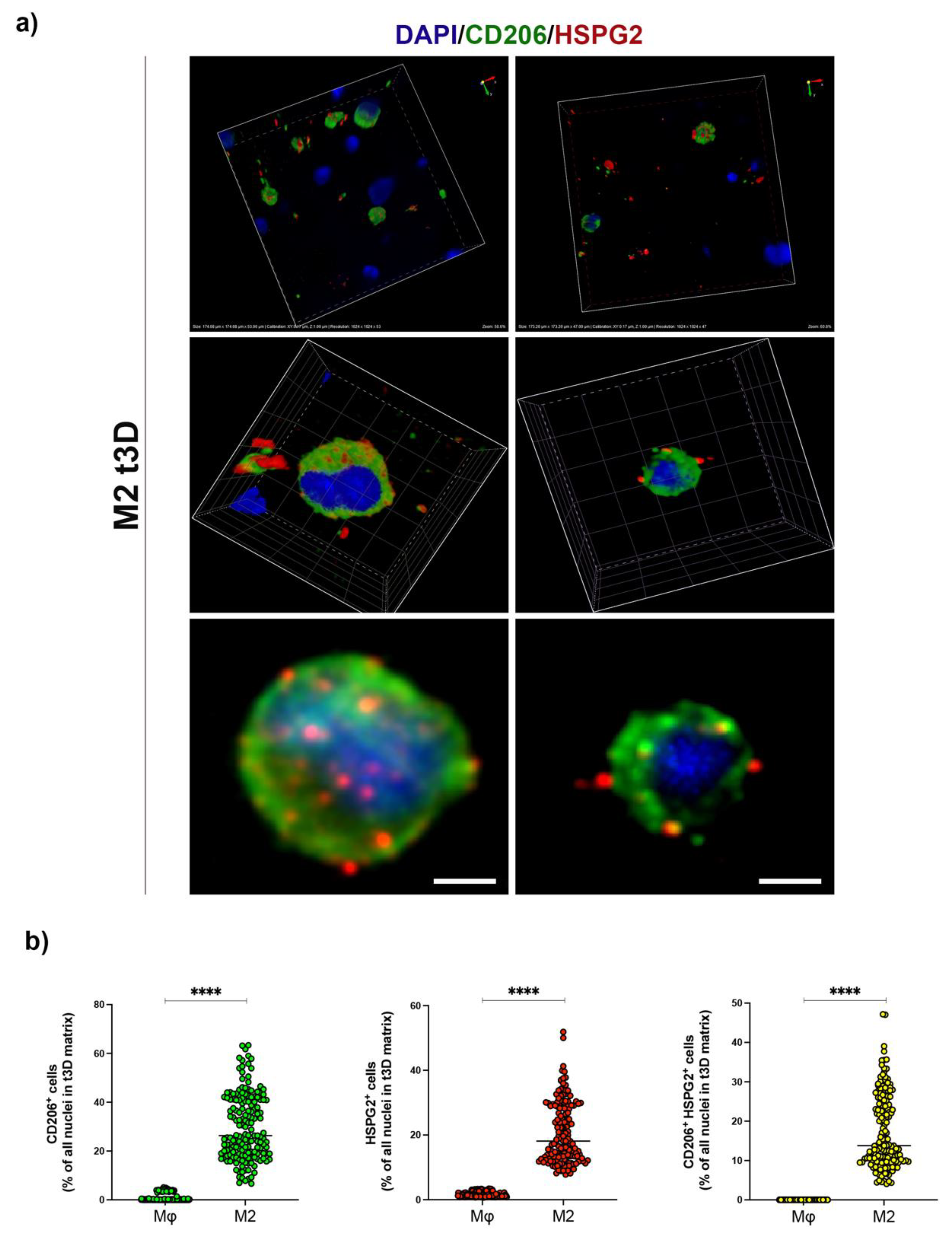
2.4. HSPG2 Was Expressed on the Surface of M2 Macrophages in the t3D Model
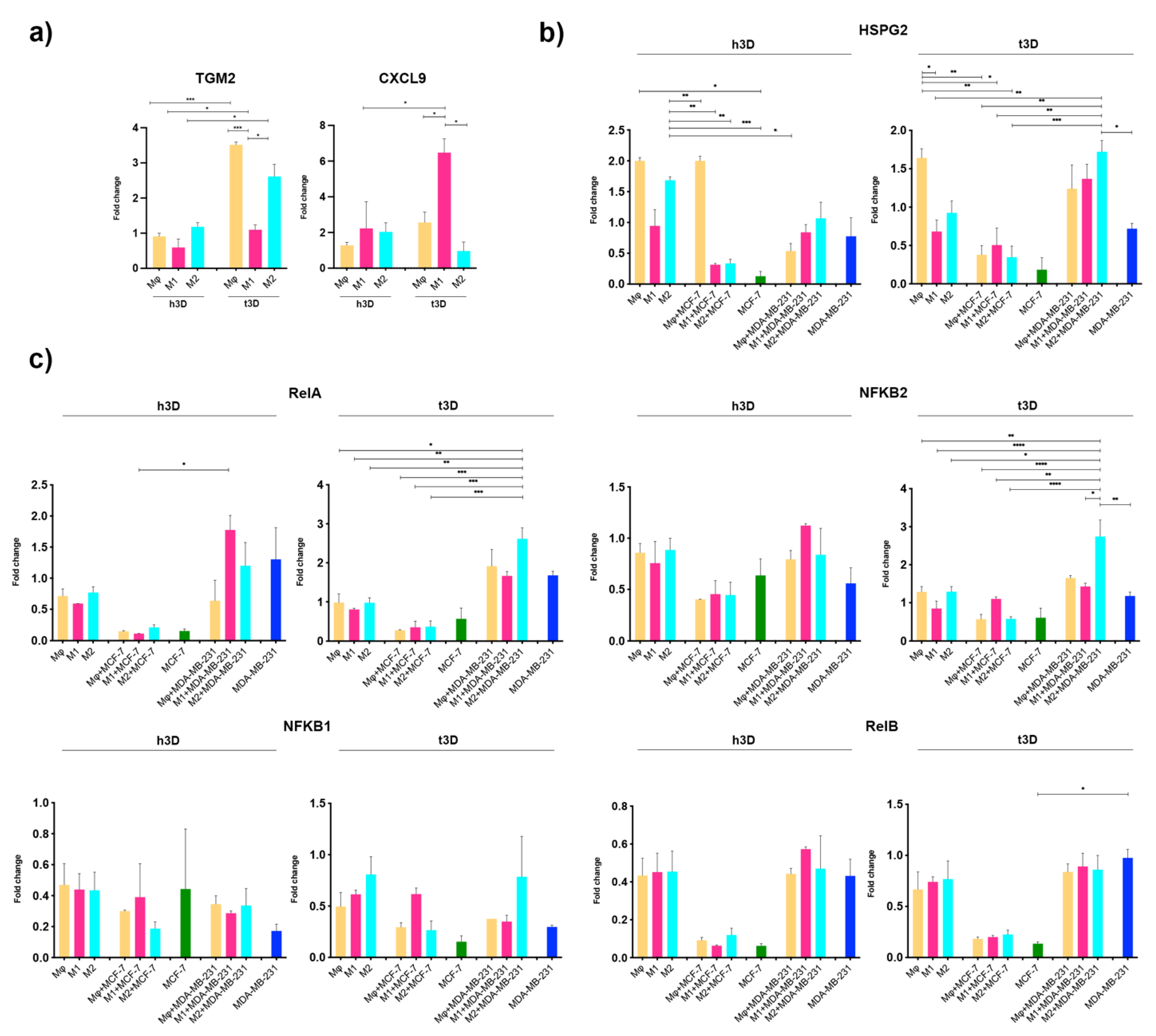
2.5. Cancer Stiffness Exacerbated HSPG2/NF-κB Dysregulation in the TNBC Model
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Hydrogel Formulation
4.3. Gene Expression Analysis
4.4. Patients and Specimens
4.5. Immunofluorescence Analysis
4.6. Image Acquisition and Processing
4.7. Co-Immunoprecipitation Assay
4.8. Nuclear-Cytoplasmic Fractionation
4.9. SDS-PAGE and Western Blots
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zuo, W.-J.; Shao, Z.-M.; Jiang, Y.-Z. Molecular Subtypes and Precision Treatment of Triple-Negative Breast Cancer. Ann. Transl. Med. 2020, 8, 499. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.M.; Pepin, A.; Thomas, R.; Biagi, T.; Stark, E.; Sparks, A.D.; Johnson, K.; Kaltman, R. Racial Disparities in Breast Cancer Hereditary Risk Assessment Referrals. J. Genet. Couns. 2020, 29, 587–593. [Google Scholar] [CrossRef]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Immunotherapy for Triple-Negative Breast Cancer: A Molecular Insight into the Microenvironment, Treatment, and Resistance. J. Natl. Cancer Cent. 2021, 1, 75–87. [Google Scholar] [CrossRef]
- Liubomirski, Y.; Lerrer, S.; Meshel, T.; Rubinstein-Achiasaf, L.; Morein, D.; Wiemann, S.; Körner, C.; Ben-Baruch, A. Tumor-Stroma-Inflammation Networks Promote Pro-Metastatic Chemokines and Aggressiveness Characteristics in Triple-Negative Breast Cancer. Front. Immunol. 2019, 10, 757. [Google Scholar] [CrossRef] [Green Version]
- Deepak, K.G.K.; Vempati, R.; Nagaraju, G.P.; Dasari, V.R.; Nagini, S.; Rao, D.N.; Malla, R.R. Tumor Microenvironment: Challenges and Opportunities in Targeting Metastasis of Triple Negative Breast Cancer. Pharmacol. Res. 2020, 153, 104683. [Google Scholar] [CrossRef] [PubMed]
- Tower, H.; Ruppert, M.; Britt, K. The Immune Microenvironment of Breast Cancer Progression. Cancers 2019, 11, 1375. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between Cancer-Associated Fibroblasts and Immune Cells in the Tumor Microenvironment: New Findings and Future Perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef]
- Karamanos, N.K.; Piperigkou, Z.; Theocharis, A.D.; Watanabe, H.; Franchi, M.; Baud, S.; Brézillon, S.; Götte, M.; Passi, A.; Vigetti, D.; et al. Proteoglycan Chemical Diversity Drives Multifunctional Cell Regulation and Therapeutics. Chem. Rev. 2018, 118, 9152–9232. [Google Scholar] [CrossRef]
- Cruz, L.A.; Tellman, T.V.; Farach-Carson, M.C. Flipping the Molecular Switch: Influence of Perlecan and Its Modifiers in the Tumor Microenvironment. In Tumor Microenvironment; Birbrair, A., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2020; Volume 1245. [Google Scholar] [CrossRef]
- Kalscheuer, S.; Khanna, V.; Kim, H.; Li, S.; Sachdev, D.; DeCarlo, A.; Yang, D.; Panyam, J. Discovery of HSPG2 (Perlecan) as a Therapeutic Target in Triple Negative Breast Cancer. Sci. Rep. 2019, 9, 12492. [Google Scholar] [CrossRef]
- Hu, H.; Li, S.; Cui, X.; Lv, X.; Jiao, Y.; Yu, F.; Yao, H.; Song, E.; Chen, Y.; Wang, M.; et al. The Overexpression of Hypomethylated MiR-663 Induces Chemotherapy Resistance in Human Breast Cancer Cells by Targeting Heparin Sulfate Proteoglycan 2 (HSPG2). J. Biol. Chem. 2013, 288, 10973–10985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Neriah, Y.; Karin, M. Inflammation Meets Cancer, with NF-ΚB as the Matchmaker. Nat. Immunol. 2011, 12, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.R.; Grindel, B.J.; Francis, L.; Carson, D.D.; Farach-Carson, M.C. Transcriptional Activation by NFκB Increases Perlecan/HSPG2 Expression in the Desmoplastic Prostate Tumor Microenvironment. J. Cell. Biochem. 2014, 115, 1322–1333. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, G.; Wang, V.Y.-F. Origin of the Functional Distinctiveness of NF-ΚB/P52. Front. Cell Dev. Biol. 2021, 9, 764164. [Google Scholar] [CrossRef]
- Xie, J.; Li, Q.; Zhu, X.-H.; Gao, Y.; Zhao, W.-H. IGF2BP1 Promotes LPS-Induced NFκB Activation and pro-Inflammatory Cytokines Production in Human Macrophages and Monocytes. Biochem. Biophys. Res. Commun. 2019, 513, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, Q.; Yin, G.; Ding, X.; Xie, J. Ku70 and Ku80 Participate in LPS-Induced pro-Inflammatory Cytokines Production in Human Macrophages and Monocytes. Aging 2020, 12, 20432–20444. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zheng, H.; Duan, Z.; Hu, D.; Li, M.; Liu, S.; Li, Z.; Deng, X.; Wang, Z.; Tang, M.; et al. LMP1-Augmented Kappa Intron Enhancer Activity Contributes to Upregulation Expression of Ig Kappa Light Chain via NF-KappaB and AP-1 Pathways in Nasopharyngeal Carcinoma Cells. Mol. Cancer 2009, 8, 92. [Google Scholar] [CrossRef] [Green Version]
- Chawla, M.; Mukherjee, T.; Deka, A.; Chatterjee, B.; Sarkar, U.A.; Singh, A.K.; Kedia, S.; Lum, J.; Dhillon, M.K.; Banoth, B.; et al. An Epithelial Nfkb2 Pathway Exacerbates Intestinal Inflammation by Supplementing Latent RelA Dimers to the Canonical NF-ΚB Module. Proc. Natl. Acad. Sci. USA 2021, 118, e2024828118. [Google Scholar] [CrossRef]
- Saraiva, D.P.; Matias, A.T.; Braga, S.; Jacinto, A.; Cabral, M.G. Establishment of a 3D Co-Culture With MDA-MB-231 Breast Cancer Cell Line and Patient-Derived Immune Cells for Application in the Development of Immunotherapies. Front. Oncol. 2020, 10, 1543. [Google Scholar] [CrossRef]
- Benner, B.; Scarberry, L.; Suarez-Kelly, L.P.; Duggan, M.C.; Campbell, A.R.; Smith, E.; Lapurga, G.; Jiang, K.; Butchar, J.P.; Tridandapani, S.; et al. Generation of Monocyte-Derived Tumor-Associated Macrophages Using Tumor-Conditioned Media Provides a Novel Method to Study Tumor-Associated Macrophages in Vitro. J. Immunother. Cancer 2019, 7, 140. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Yang, L.; Yue, D.; Cao, L.; Li, L.; Wang, D.; Ping, Y.; Shen, Z.; Zheng, Y.; Wang, L.; et al. Macrophage-Derived CCL22 Promotes an Immunosuppressive Tumor Microenvironment via IL-8 in Malignant Pleural Effusion. Cancer Lett. 2019, 452, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.-Y.; Whitelock, J.M.; Williams, H.; Kim, H.N.; Medbury, H.J.; Lord, M.S. Macrophages Bind LDL Using Heparan Sulfate and the Perlecan Protein Core. J. Biol. Chem. 2021, 296, 100520. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Guo, Z.; Cheng, J.; Sun, B.; Yang, J.; Li, H.; Wu, S.; Dong, F.; Yan, X. Differential Metabolic Responses in Breast Cancer Cell Lines to Acidosis and Lactic Acidosis Revealed by Stable Isotope Assisted Metabolomics. Sci. Rep. 2020, 10, 21967. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, C.; Roldan, M.; Ruedas-Rama, M.J.; Orte, A.; Martin, M. Breast Cancer Cell Subtypes Display Different Metabolic Phenotypes That Correlate with Their Clinical Classification. Biology 2021, 10, 1267. [Google Scholar] [CrossRef] [PubMed]
- Piersma, B.; Hayward, M.-K.; Weaver, V.M. Fibrosis and Cancer: A Strained Relationship. Biochim. Biophys. Acta BBA-Rev. Cancer 2020, 1873, 188356. [Google Scholar] [CrossRef]
- Chirivì, M.; Maiullari, F.; Milan, M.; Presutti, D.; Cordiglieri, C.; Crosti, M.; Sarnicola, M.L.; Soluri, A.; Volpi, M.; Święszkowski, W.; et al. Tumor Extracellular Matrix Stiffness Promptly Modulates the Phenotype and Gene Expression of Infiltrating T Lymphocytes. Int. J. Mol. Sci. 2021, 22, 5862. [Google Scholar] [CrossRef]
- van der Leun, A.M.; Thommen, D.S.; Schumacher, T.N. CD8+ T Cell States in Human Cancer: Insights from Single-Cell Analysis. Nat. Rev. Cancer 2020, 20, 218–232. [Google Scholar] [CrossRef]
- Choi, J.; Gyamfi, J.; Jang, H.; Koo, J.S. The Role of Tumor-Associated Macrophage in Breast Cancer Biology. Histol. Histopathol. 2018, 33, 133–145. [Google Scholar] [CrossRef]
- Jung, K.Y.; Cho, S.W.; Kim, Y.A.; Kim, D.; Oh, B.-C.; Park, D.J.; Park, Y.J. Cancers with Higher Density of Tumor-Associated Macrophages Were Associated with Poor Survival Rates. J. Pathol. Transl. Med. 2015, 49, 318–324. [Google Scholar] [CrossRef] [Green Version]
- McMahon, M.; Ye, S.; Izzard, L.; Dlugolenski, D.; Tripp, R.A.; Bean, A.G.D.; McCulloch, D.R.; Stambas, J. ADAMTS5 Is a Critical Regulator of Virus-Specific T Cell Immunity. PLoS Biol. 2016, 14, e1002580. [Google Scholar] [CrossRef] [Green Version]
- Sangaletti, S.; Chiodoni, C.; Tripodo, C.; Colombo, M.P. Common Extracellular Matrix Regulation of Myeloid Cell Activity in the Bone Marrow and Tumor Microenvironments. Cancer Immunol. Immunother. CII 2017, 66, 1059–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jachetti, E.; Caputo, S.; Mazzoleni, S.; Brambillasca, C.S.; Parigi, S.M.; Grioni, M.; Piras, I.S.; Restuccia, U.; Calcinotto, A.; Freschi, M.; et al. Tenascin-C Protects Cancer Stem-like Cells from Immune Surveillance by Arresting T-Cell Activation. Cancer Res. 2015, 75, 2095–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theocharis, A.D.; Skandalis, S.S.; Tzanakakis, G.N.; Karamanos, N.K. Proteoglycans in Health and Disease: Novel Roles for Proteoglycans in Malignancy and Their Pharmacological Targeting. FEBS J. 2010, 277, 3904–3923. [Google Scholar] [CrossRef] [PubMed]
- Mongiat, M.; Taylor, K.; Otto, J.; Aho, S.; Uitto, J.; Whitelock, J.M.; Iozzo, R.V. The Protein Core of the Proteoglycan Perlecan Binds Specifically to Fibroblast Growth Factor-7. J. Biol. Chem. 2000, 275, 7095–7100. [Google Scholar] [CrossRef] [Green Version]
- Elgundi, Z.; Papanicolaou, M.; Major, G.; Cox, T.R.; Melrose, J.; Whitelock, J.M.; Farrugia, B.L. Cancer Metastasis: The Role of the Extracellular Matrix and the Heparan Sulfate Proteoglycan Perlecan. Front. Oncol. 2020, 9, 1482. [Google Scholar] [CrossRef] [Green Version]
- Witherel, C.E.; Sao, K.; Brisson, B.K.; Han, B.; Volk, S.W.; Petrie, R.J.; Han, L.; Spiller, K.L. Regulation of Extracellular Matrix Assembly and Structure by Hybrid M1/M2 Macrophages. Biomaterials 2021, 269, 120667. [Google Scholar] [CrossRef]
- Brasil da Costa, F.H.; Lewis, M.S.; Truong, A.; Carson, D.D.; Farach-Carson, M.C. SULF1 Suppresses Wnt3A-Driven Growth of Bone Metastatic Prostate Cancer in Perlecan-Modified 3D Cancer-Stroma-Macrophage Triculture Models. PLoS ONE 2020, 15, e0230354. [Google Scholar] [CrossRef]
- Vennin, C.; Mélénec, P.; Rouet, R.; Nobis, M.; Cazet, A.S.; Murphy, K.J.; Herrmann, D.; Reed, D.A.; Lucas, M.C.; Warren, S.C.; et al. CAF Hierarchy Driven by Pancreatic Cancer Cell P53-Status Creates a pro-Metastatic and Chemoresistant Environment via Perlecan. Nat. Commun. 2019, 10, 3637. [Google Scholar] [CrossRef] [Green Version]
- Afik, R.; Zigmond, E.; Vugman, M.; Klepfish, M.; Shimshoni, E.; Pasmanik-Chor, M.; Shenoy, A.; Bassat, E.; Halpern, Z.; Geiger, T.; et al. Tumor Macrophages Are Pivotal Constructors of Tumor Collagenous Matrix. J. Exp. Med. 2016, 213, 2315–2331. [Google Scholar] [CrossRef]
- Zhang, T.; Jia, Y.; Yu, Y.; Zhang, B.; Xu, F.; Guo, H. Targeting the Tumor Biophysical Microenvironment to Reduce Resistance to Immunotherapy. Adv. Drug Deliv. Rev. 2022, 186, 114319. [Google Scholar] [CrossRef]
- Salinas-Vera, Y.M.; Valdés, J.; Pérez-Navarro, Y.; Mandujano-Lazaro, G.; Marchat, L.A.; Ramos-Payán, R.; Nuñez-Olvera, S.I.; Pérez-Plascencia, C.; López-Camarillo, C. Three-Dimensional 3D Culture Models in Gynecological and Breast Cancer Research. Front. Oncol. 2022, 12, 826113. [Google Scholar] [CrossRef] [PubMed]


| Gene Symbol | Sense-Forward Primer | Antisense-Reverse Primer |
|---|---|---|
| GAPDH | TCTTTTGCGTCGCCAGCCGAG | TGACCAGGCGCCCAATACGAC |
| HSPG2 | TCGGCCATGAGTCCTTCTAC | GTGTATCGCAACTTCCCACC |
| RelA | ATCCCATCTTTGACAATCGTGC | CTGGTCCCGTGAAATACACCTC |
| NFKB1 | GTGGAGCACGACAAC | GGTGTGGTTCCATCG |
| NFKB2 | TGAAGCCAGTCATCT | CCTGCTGTCTTGTCC |
| RelB | CATTGAGCGGAAGATTCAAC | GCAGCTCTGATGTGTTTGTG |
| CD206 | CGAGGAAGAGGTTCGGTTCACC | GCAATCCCGGTTCTCATGGC |
| TGM2 | GCAGTGACTTTGACGTCTTTGCCC | GTAGCTGTTGATAACTGGCTCCACG |
| CCL22 | GCCAACATGGAAGACAGCGT | TTATCCCTGAAGGTTAGCAACACCA |
| CXCL9 | GCTGGTTCTGATTGGAGTGC | GAAGGGCTTGGGGCAAATTG |
| IL-6 | TTCTCCACAAGCGCCTTC | AGAGGTGAGTGGCTGTCTGT |
| IL-1b | CCTGAGCTCGCCAGTGAAA | TCCTGGAAGGAGCACTTCATCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Paolis, V.; Maiullari, F.; Chirivì, M.; Milan, M.; Cordiglieri, C.; Pagano, F.; La Manna, A.R.; De Falco, E.; Bearzi, C.; Rizzi, R.; et al. Unusual Association of NF-κB Components in Tumor-Associated Macrophages (TAMs) Promotes HSPG2-Mediated Immune-Escaping Mechanism in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 7902. https://doi.org/10.3390/ijms23147902
De Paolis V, Maiullari F, Chirivì M, Milan M, Cordiglieri C, Pagano F, La Manna AR, De Falco E, Bearzi C, Rizzi R, et al. Unusual Association of NF-κB Components in Tumor-Associated Macrophages (TAMs) Promotes HSPG2-Mediated Immune-Escaping Mechanism in Breast Cancer. International Journal of Molecular Sciences. 2022; 23(14):7902. https://doi.org/10.3390/ijms23147902
Chicago/Turabian StyleDe Paolis, Veronica, Fabio Maiullari, Maila Chirivì, Marika Milan, Chiara Cordiglieri, Francesca Pagano, Alessandra Rita La Manna, Elena De Falco, Claudia Bearzi, Roberto Rizzi, and et al. 2022. "Unusual Association of NF-κB Components in Tumor-Associated Macrophages (TAMs) Promotes HSPG2-Mediated Immune-Escaping Mechanism in Breast Cancer" International Journal of Molecular Sciences 23, no. 14: 7902. https://doi.org/10.3390/ijms23147902
APA StyleDe Paolis, V., Maiullari, F., Chirivì, M., Milan, M., Cordiglieri, C., Pagano, F., La Manna, A. R., De Falco, E., Bearzi, C., Rizzi, R., & Parisi, C. (2022). Unusual Association of NF-κB Components in Tumor-Associated Macrophages (TAMs) Promotes HSPG2-Mediated Immune-Escaping Mechanism in Breast Cancer. International Journal of Molecular Sciences, 23(14), 7902. https://doi.org/10.3390/ijms23147902







