Exploring C-to-G and A-to-Y Base Editing in Rice by Using New Vector Tools
Abstract
:1. Introduction
2. Results
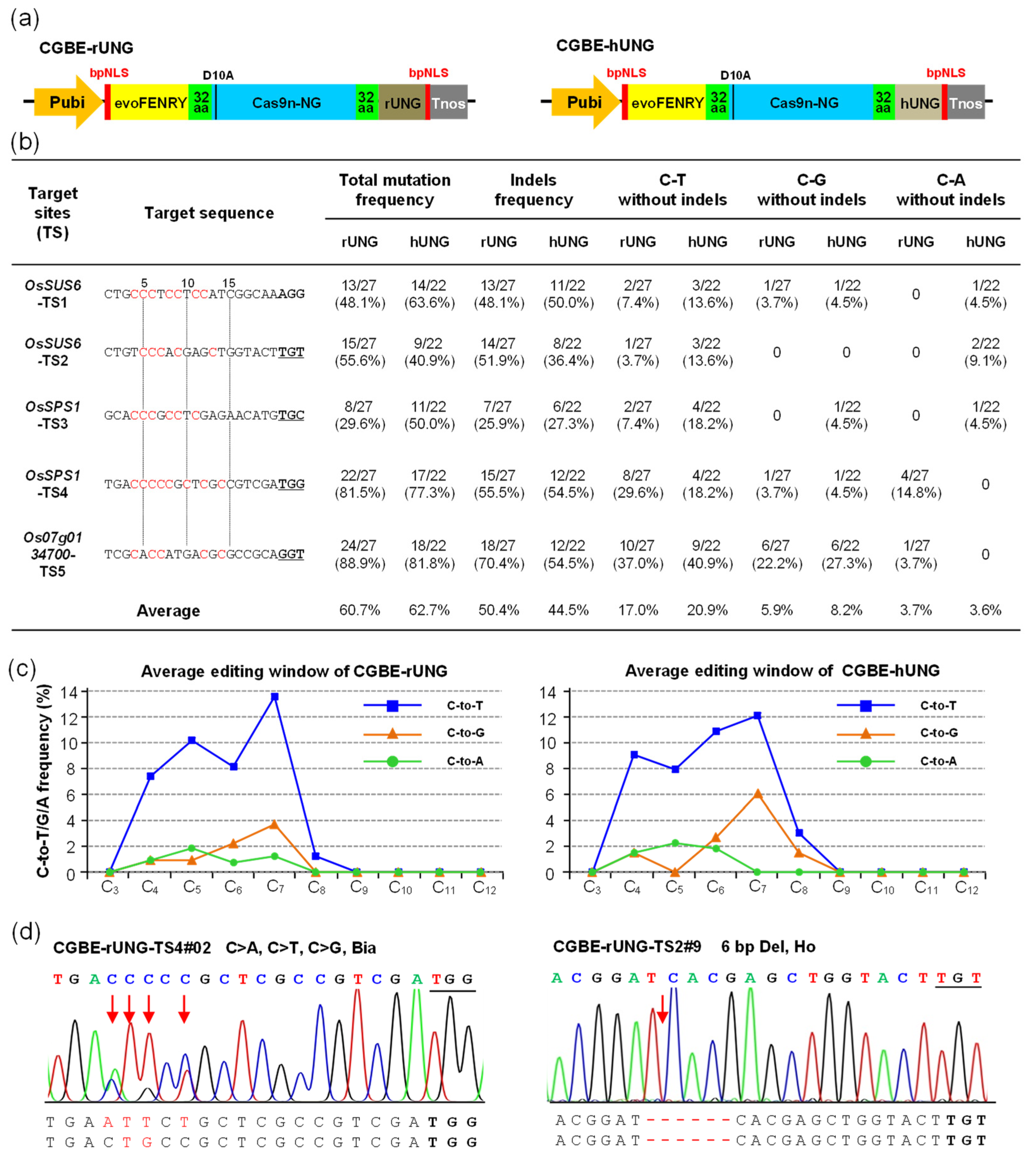
2.1. CGBE-rUNG and CGBE-hUNG Achieved C-to-G Base Editing
2.2. A-to-Y Transversions Were Not Detected by ABE8e-EndoV and ABE8e-hAAG
2.3. ABE8e-EndoV Produces Predictable Small DNA Fragment Deletions
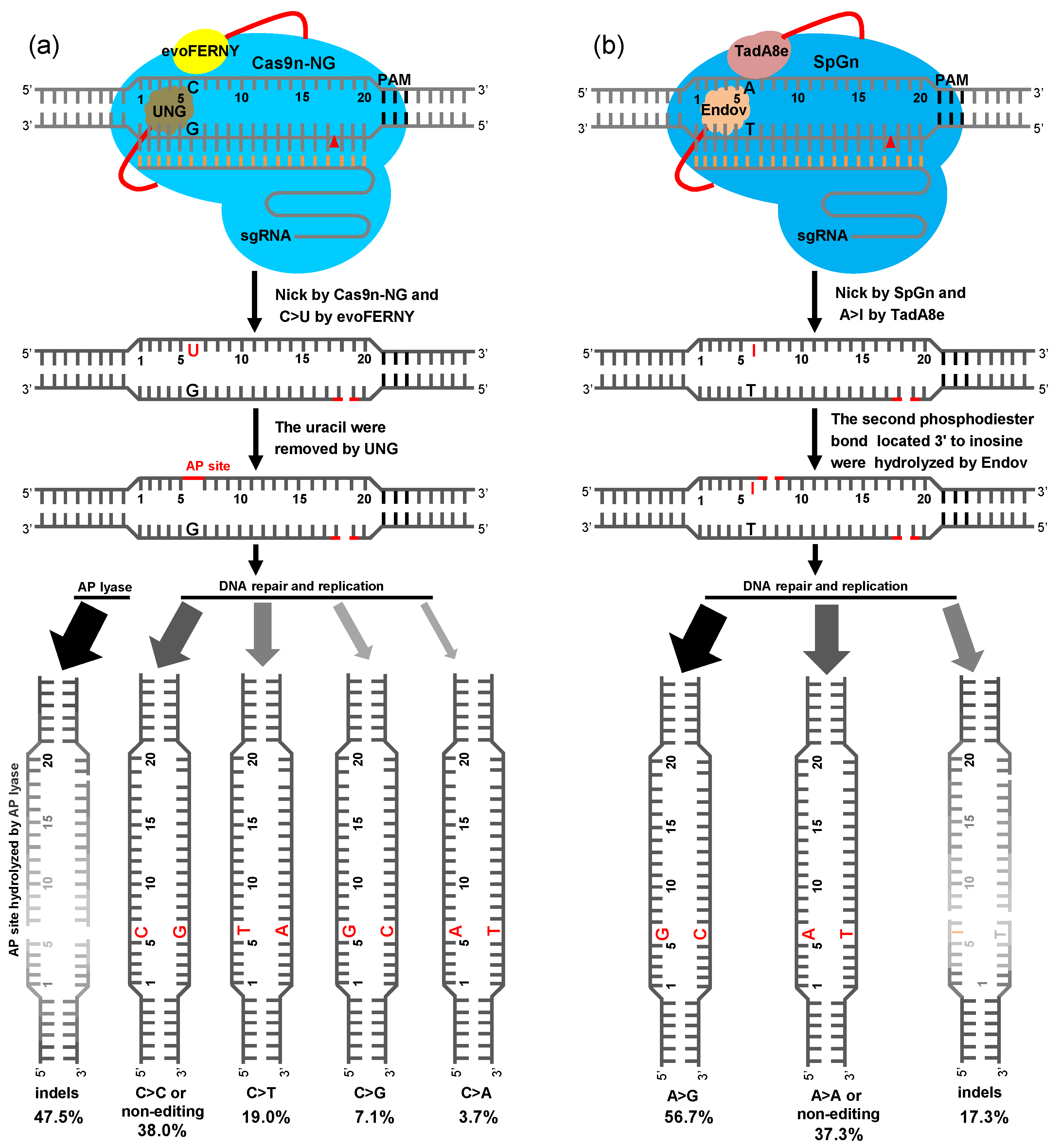
2.4. BER Pathway of Plant CGBE and ABE8e-EndoV
3. Discussion
4. Materials and Methods
4.1. Construction of Plant CGBE-rUNG, CGBE-hUNG, ABE8e-hAAG, and ABE8e-EndoV Vectors
4.2. Target Design and gRNA Expression Cassette Assembly
4.3. Genetic Transformation
4.4. On-Target and Off-Target Efficiency Analysis of T0 Rice
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Thuronyi, B.W.; Koblan, L.W.; Levy, J.M.; Yeh, W.; Zheng, C.; Newby, G.A.; Wilson, C.; Bhaumik, M.; Shubina-Oleinik, O.; Holt, J.R.; et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 2019, 37, 1070–1079. [Google Scholar] [CrossRef]
- Zeng, D.; Liu, T.; Tan, J.; Zhang, Y.; Zheng, Z.; Wang, B.; Zhou, D.; Xie, X.; Guo, M.; Liu, Y.G.; et al. PhieCBEs: Plant High-Efficiency Cytidine Base Editors with Expanded Target Range. Mol. Plant 2020, 13, 1666–1669. [Google Scholar] [CrossRef]
- Zong, Y.; Song, Q.; Li, C.; Jin, S.; Zhang, D.; Wang, Y.; Qiu, J.; Gao, C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018, 36, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Z.; Mao, Y.; Lu, Y.; Yang, R.; Tao, X.; Zhu, J.K. Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnol. J. 2019, 17, 1697–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Yang, Y.; Liu, Y.; Kang, G.; Wang, F.; Li, L.; Lv, X.; Zhao, S.; Yuan, S.; Song, J.; et al. Discriminated sgRNAs-Based SurroGate System Greatly Enhances the Screening Efficiency of Plant Base-Edited Cells. Mol. Plant 2020, 13, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.F.; Zhao, K.T.; Eton, E.; Lapinaite, A.; Newby, G.A.; Thuronyi, B.W.; Wilson, C.; Koblan, L.W.; Zeng, J.; Bauer, D.E.; et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020, 38, 883–891. [Google Scholar] [CrossRef]
- Tan, J.; Zeng, D.; Zhao, Y.; Wang, Y.; Liu, T.; Li, S.; Xue, Y.; Luo, Y.; Xie, X.; Chen, L.; et al. PhieABEs: A PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol. J. 2022, 20, 934–943. [Google Scholar] [CrossRef]
- Yan, D.; Ren, B.; Liu, L.; Yan, F.; Li, S.; Wang, G.; Sun, W.; Zhou, X.; Zhou, H. High-efficiency and multiplex adenine base editing in plants using new TadA variants. Mol. Plant 2021, 14, 722–731. [Google Scholar] [CrossRef]
- Ren, Q.; Sretenovic, S.; Liu, S.; Tang, X.; Huang, L.; He, Y.; Liu, L.; Guo, Y.; Zhong, Z.; Liu, G.; et al. PAM-less plant genome editing using a CRISPR–SpRY toolbox. Nat. Plants 2021, 7, 25–33. [Google Scholar] [CrossRef]
- Wei, C.; Wang, C.; Jia, M.; Guo, H.X.; Luo, P.Y.; Wang, M.G.; Zhu, J.K.; Zhang, H. Efficient generation of homozygous substitutions in rice in one generation utilizing an rABE8e base editor. J. Integr. Plant Biol. 2021, 63, 1595–1599. [Google Scholar] [CrossRef]
- Li, J.; Xu, R.; Qin, R.; Liu, X.; Kong, F.; Wei, P. Genome editing mediated by SpCas9 variants with broad non-canonical PAM compatibility in plants. Mol. Plant 2021, 14, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, X.; Xie, X.; Deng, L.; Zheng, H.; Pan, H.; Li, D.; Li, L.; Zhong, C. ABE8e with Polycistronic tRNA-gRNA Expression Cassette Sig-Nificantly Improves Adenine Base Editing Efficiency in Nicotiana benthamiana. Int. J. Mol. Sci. 2021, 22, 5663. [Google Scholar] [CrossRef] [PubMed]
- Veillet, F.; Perrot, L.; Guyon-Debast, A.; Kermarrec, M.; Chauvin, L.; Chauvin, J.; Gallois, J.; Mazier, M.; Nogué, F. Expanding the CRISPR Toolbox in P. patens Using SpCas9-NG Variant and Application for Gene and Base Editing in Solanaceae Crops. Int. J. Mol. Sci. 2020, 21, 1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.; Guyon-Debast, A.; Chauvin, J.; Nogué, F.; Mazier, M. Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Qin, R.; Li, J.; Liao, S.; Shan, T.; Xu, R.; Wu, D.; Wei, P. A CRISPR-Cas9-mediated domain-specific base-editing screen enables functional assessment of ACCase variants in rice. Plant Biotechnol. J. 2020, 18, 1845–1847. [Google Scholar] [CrossRef] [Green Version]
- Zeng, D.; Liu, T.; Ma, X.; Wang, B.; Zheng, Z.; Zhang, Y.; Xie, X.; Yang, B.; Zhao, Z.; Zhu, Q.; et al. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5′UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 2020, 18, 2385–2387. [Google Scholar] [CrossRef]
- Khan, Z.A.; Kumar, R.; Dasgupta, I. CRISPR/Cas-Mediated Resistance against Viruses in Plants. Int. J. Mol. Sci. 2022, 23, 2303. [Google Scholar] [CrossRef]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N. Control of Bacterial Diseases of Banana Using CRISPR/Cas-Based Gene Editing. Int. J. Mol. Sci. 2022, 23, 3619. [Google Scholar] [CrossRef]
- Kantor, A.; McClements, M.; MacLaren, R. CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int. J. Mol. Sci. 2020, 21, 6240. [Google Scholar] [CrossRef]
- Zhao, D.; Li, J.; Li, S.; Xin, X.; Hu, M.; Price, M.A.; Rosser, S.J.; Bi, C.; Zhang, X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021, 39, 35–40. [Google Scholar] [CrossRef]
- Kurt, I.C.; Zhou, R.; Iyer, S.; Garcia, S.P.; Miller, B.R.; Langner, L.M.; Grünewald, J.; Joung, J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021, 39, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Sretenovic, S.; Liu, S.; Li, G.; Cheng, Y.; Fan, T.; Xu, Y.; Zhou, J.; Zheng, X.; Coleman, G.; Zhang, Y.; et al. Exploring C-To-G Base Editing in Rice, Tomato, and Poplar. Front. Genome Ed. 2021, 3, 756766. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Shen, R.; Li, Z.; Yao, Q.; Zhang, X.; Zhong, D.; Tan, X.; Song, M.; Han, H.; Zhu, J.K.; et al. Efficient C-to-G editing in rice using an optimized base editor. Plant Biotechnol. J. 2022, 20, 1238–1240. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I. Diversity of Endonuclease V: From DNA Repair to RNA Editing. Biomolecules 2015, 5, 2194–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedglin, M.; O’Brien, P.J. Human Alkyladenine DNA Glycosylase Employs a Processive Search for DNA Damage. Biochemistry 2008, 47, 11434–11445. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Samara, N.L.; Kuraoka, I.; Yang, W. Evolution of Inosine-Specific Endonuclease V from Bacterial DNase to Eukaryotic RNase. Mol. Cell 2019, 76, 44–56. [Google Scholar] [CrossRef]
- Kiyonari, S.; Egashira, Y.; Ishino, S.; Ishino, Y. Biochemical characterization of endonuclease V from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biochem. 2014, 155, 325–333. [Google Scholar] [CrossRef]
- Dalhus, B.; Alseth, I.; Bjørås, M. Structural basis for incision at deaminated adenines in DNA and RNA by endonuclease V. Prog. Biophys. Mol. Biol. 2015, 117, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zeng, D.; Zheng, Z.; Lin, Z.; Xue, Y.; Li, T.; Xie, X.; Ma, G.; Liu, Y.G.; Zhu, Q. The ScCas9++ variant expands the CRISPR toolbox for genome editing in plants. J. Integr. Plant Biol. 2021, 63, 1611–1619. [Google Scholar] [CrossRef]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y.G. CRISPR-GE: A Convenient Software Toolkit for CRISPR-Based Genome Editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zong, Y.; Lin, Q.; Zhang, H.; Chai, Z.; Zhang, D.; Chen, K.; Qiu, J.L.; Gao, C. Precise, predictable multi-nucleotide deletions in rice and wheat using APOBEC-Cas9. Nat. Biotechnol. 2020, 38, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Li, J.; Wang, Q.; Xu, Z.; Sun, L.; Alariqi, M.; Manghwar, H.; Wang, G.; Li, B.; Ding, X.; et al. High-efficient and precise base editing of C•G to T•A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 18, 45–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koblan, L.W.; Arbab, M.; Shen, M.W.; Hussmann, J.A.; Anzalone, A.V.; Doman, J.L.; Newby, G.A.; Yang, D.; Mok, B.; Replogle, J.M.; et al. Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat. Biotechnol. 2021, 39, 1414–1425. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Tan, J.; Wang, Y.; Chen, S.; Lin, Z.; Zhao, Y.; Xue, Y.; Luo, Y.; Liu, Y.; Zhu, Q. An Efficient Marker Gene Excision Strategy Based on CRISPR/Cas9-Mediated Homology-Directed Repair in Rice. Int. J. Mol. Sci. 2022, 23, 1588. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, D.; Zheng, Z.; Liu, Y.; Liu, T.; Li, T.; Liu, J.; Luo, Q.; Xue, Y.; Li, S.; Chai, N.; et al. Exploring C-to-G and A-to-Y Base Editing in Rice by Using New Vector Tools. Int. J. Mol. Sci. 2022, 23, 7990. https://doi.org/10.3390/ijms23147990
Zeng D, Zheng Z, Liu Y, Liu T, Li T, Liu J, Luo Q, Xue Y, Li S, Chai N, et al. Exploring C-to-G and A-to-Y Base Editing in Rice by Using New Vector Tools. International Journal of Molecular Sciences. 2022; 23(14):7990. https://doi.org/10.3390/ijms23147990
Chicago/Turabian StyleZeng, Dongchang, Zhiye Zheng, Yuxin Liu, Taoli Liu, Tie Li, Jianhong Liu, Qiyu Luo, Yang Xue, Shengting Li, Nan Chai, and et al. 2022. "Exploring C-to-G and A-to-Y Base Editing in Rice by Using New Vector Tools" International Journal of Molecular Sciences 23, no. 14: 7990. https://doi.org/10.3390/ijms23147990
APA StyleZeng, D., Zheng, Z., Liu, Y., Liu, T., Li, T., Liu, J., Luo, Q., Xue, Y., Li, S., Chai, N., Yu, S., Xie, X., Liu, Y.-G., & Zhu, Q. (2022). Exploring C-to-G and A-to-Y Base Editing in Rice by Using New Vector Tools. International Journal of Molecular Sciences, 23(14), 7990. https://doi.org/10.3390/ijms23147990






