The Transcription Factors TaTDRL and TaMYB103 Synergistically Activate the Expression of TAA1a in Wheat, Which Positively Regulates the Development of Microspore in Arabidopsis
Abstract
:1. Introduction
2. Results
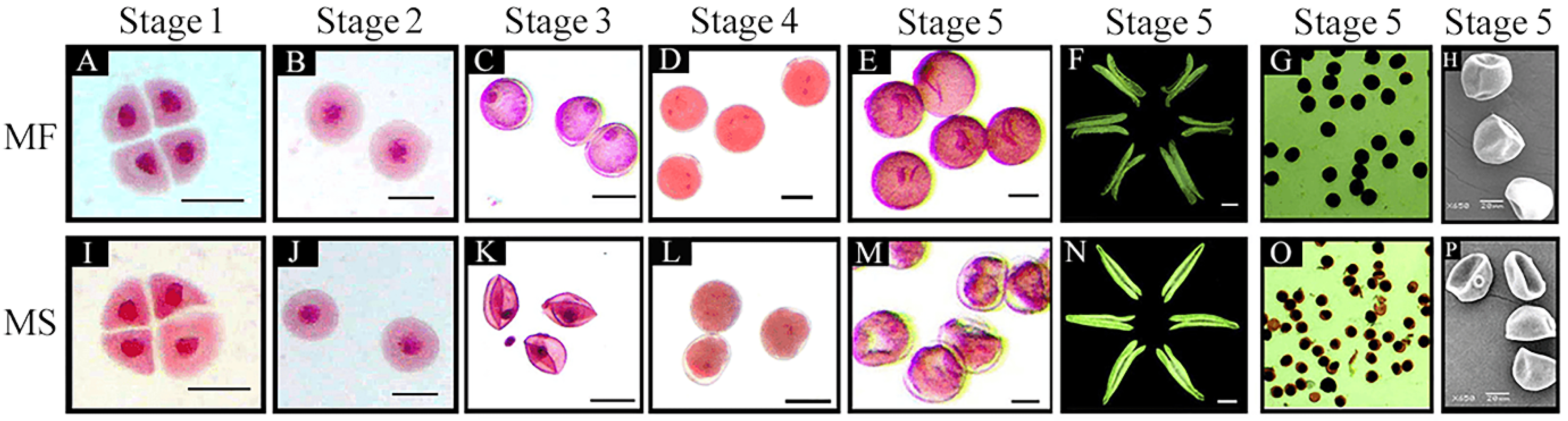
2.1. Characterization of Abnormal Anther and Microspore Development in the CMS Line
2.2. Isolation and Sequence Analysis of the TAA1a Promoter
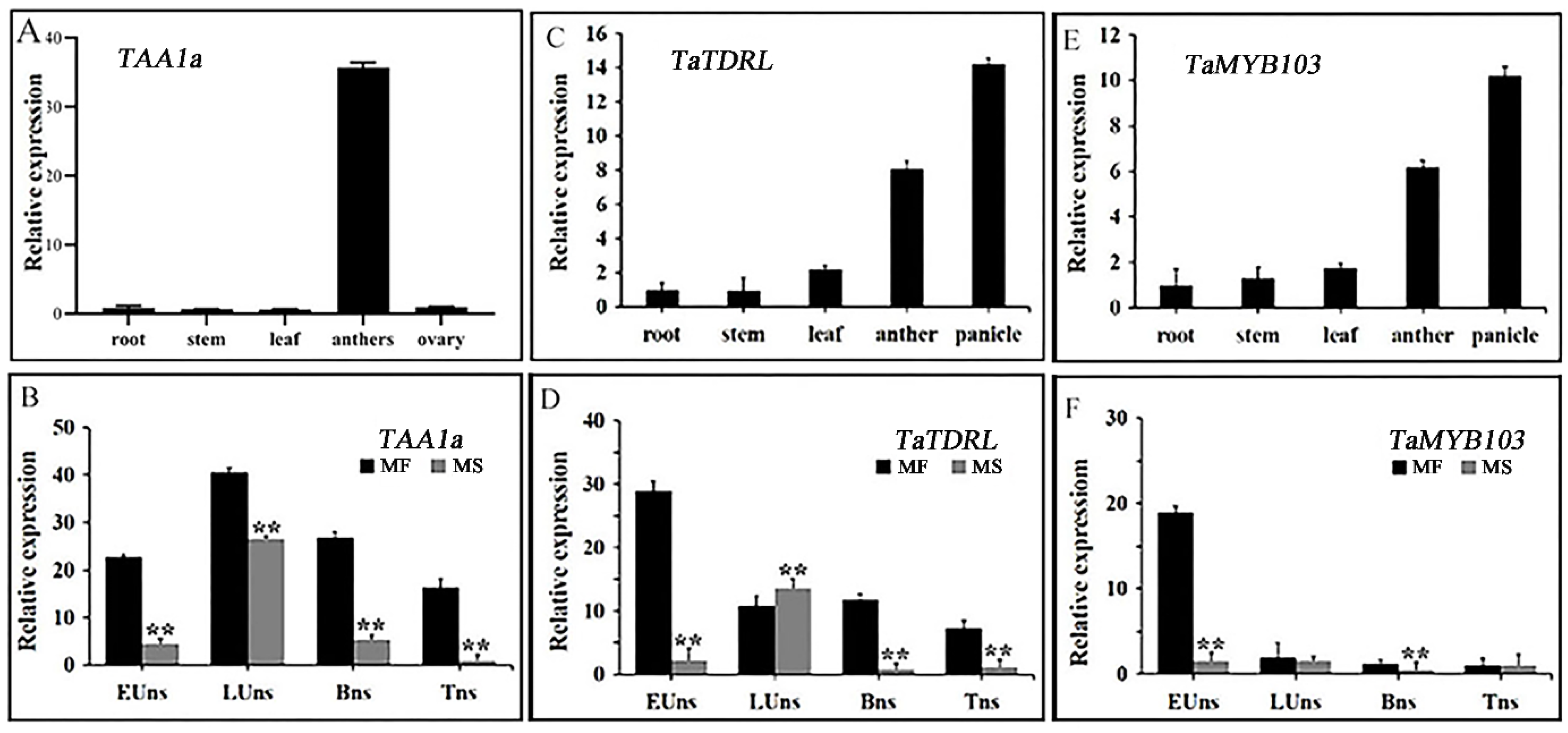
2.3. Expression Patterns of the Transcription Factors TaTDRL, TaMYB03, and TAA1a in Wheat Anthers
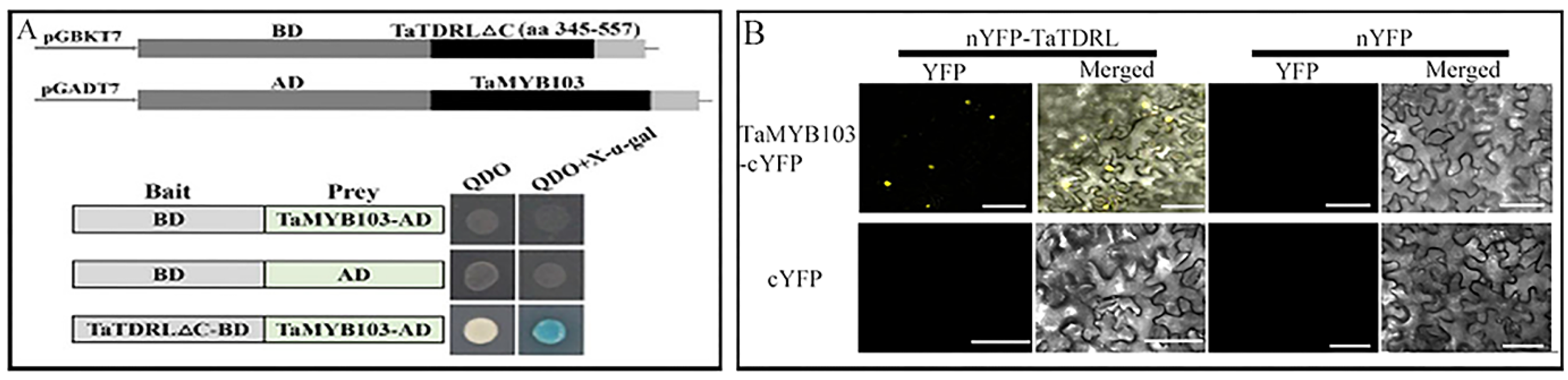
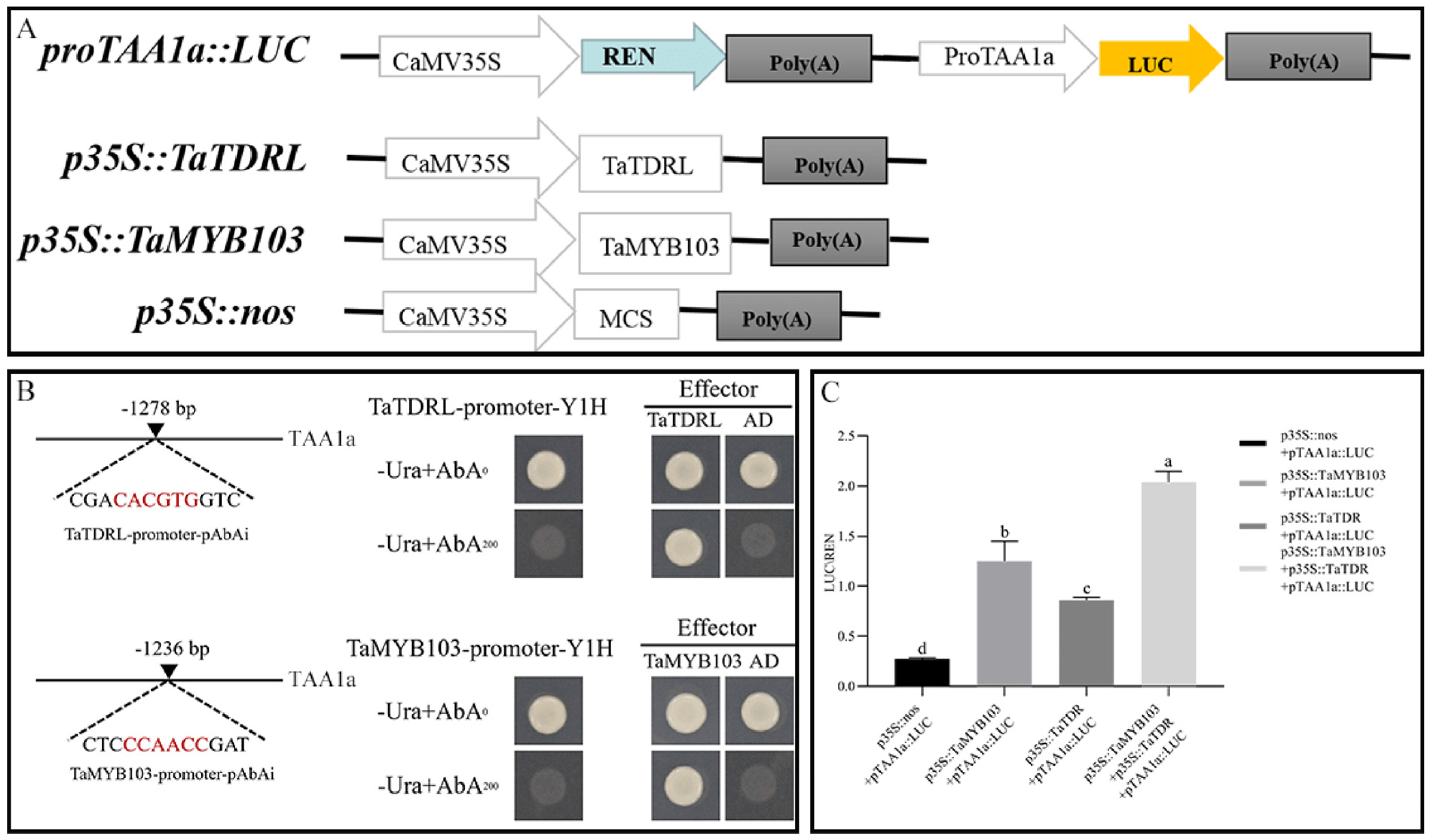
2.4. TaTDRL and TaMYB103 Synergistically Activate the Expression of TAA1a
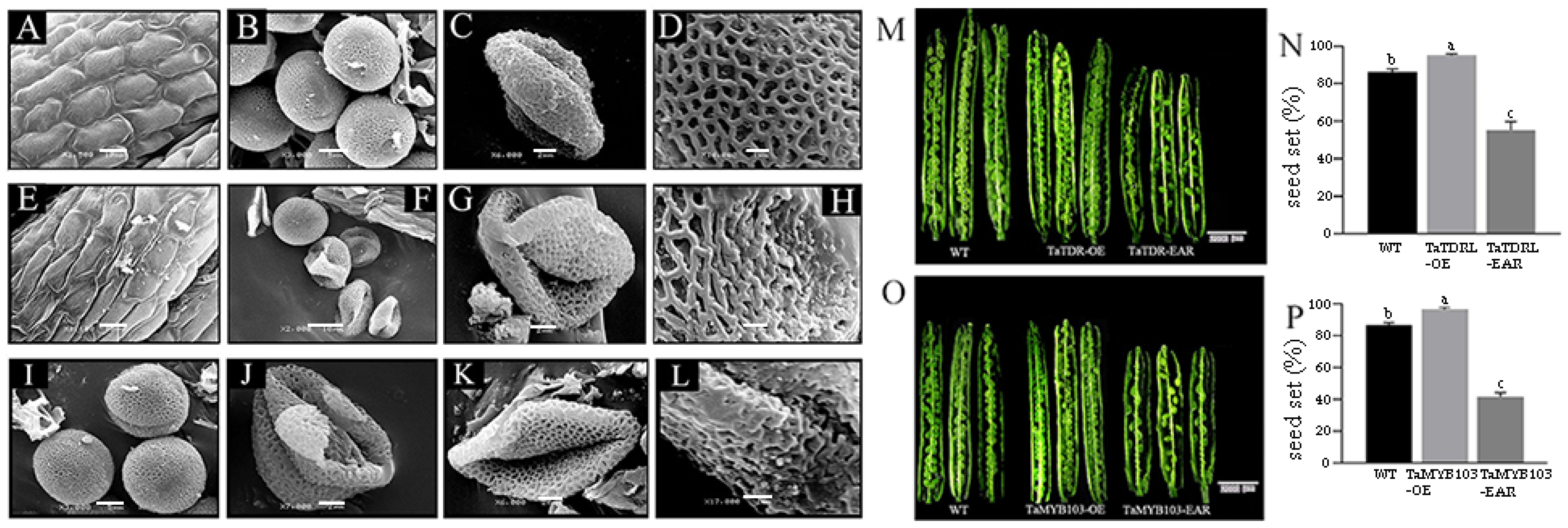
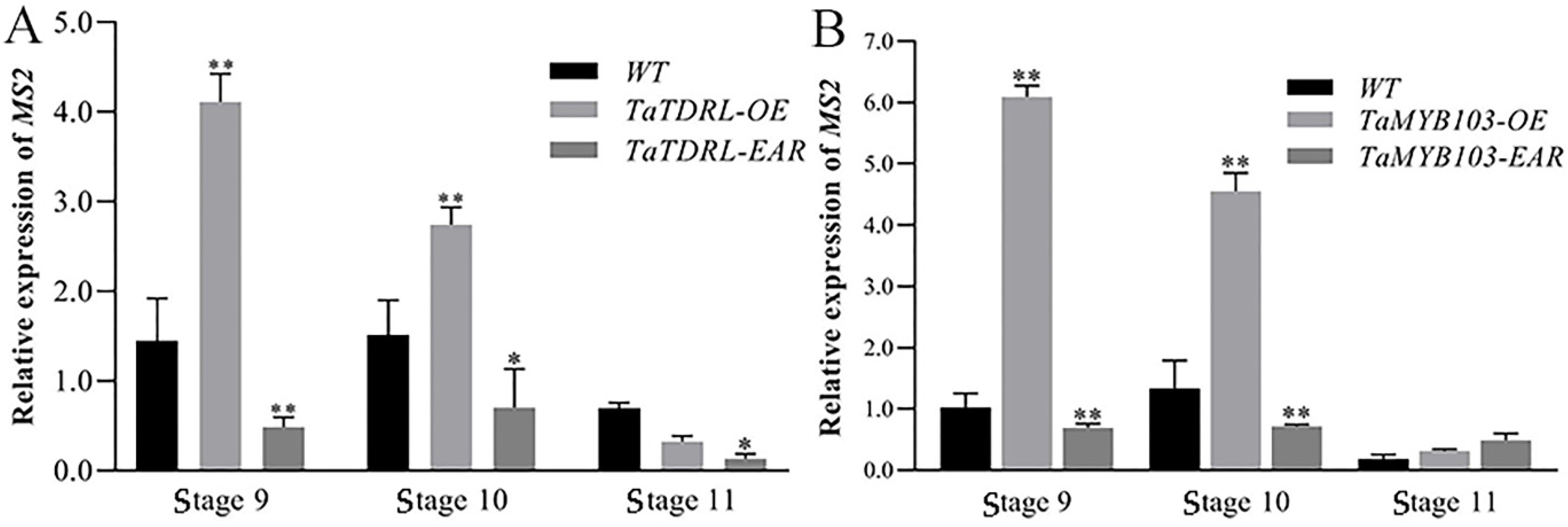
2.5. Pollen Development and the Expression of MS2 Were Dysregulated in TaTDRL-EAR and TaMYB103-EAR Plants
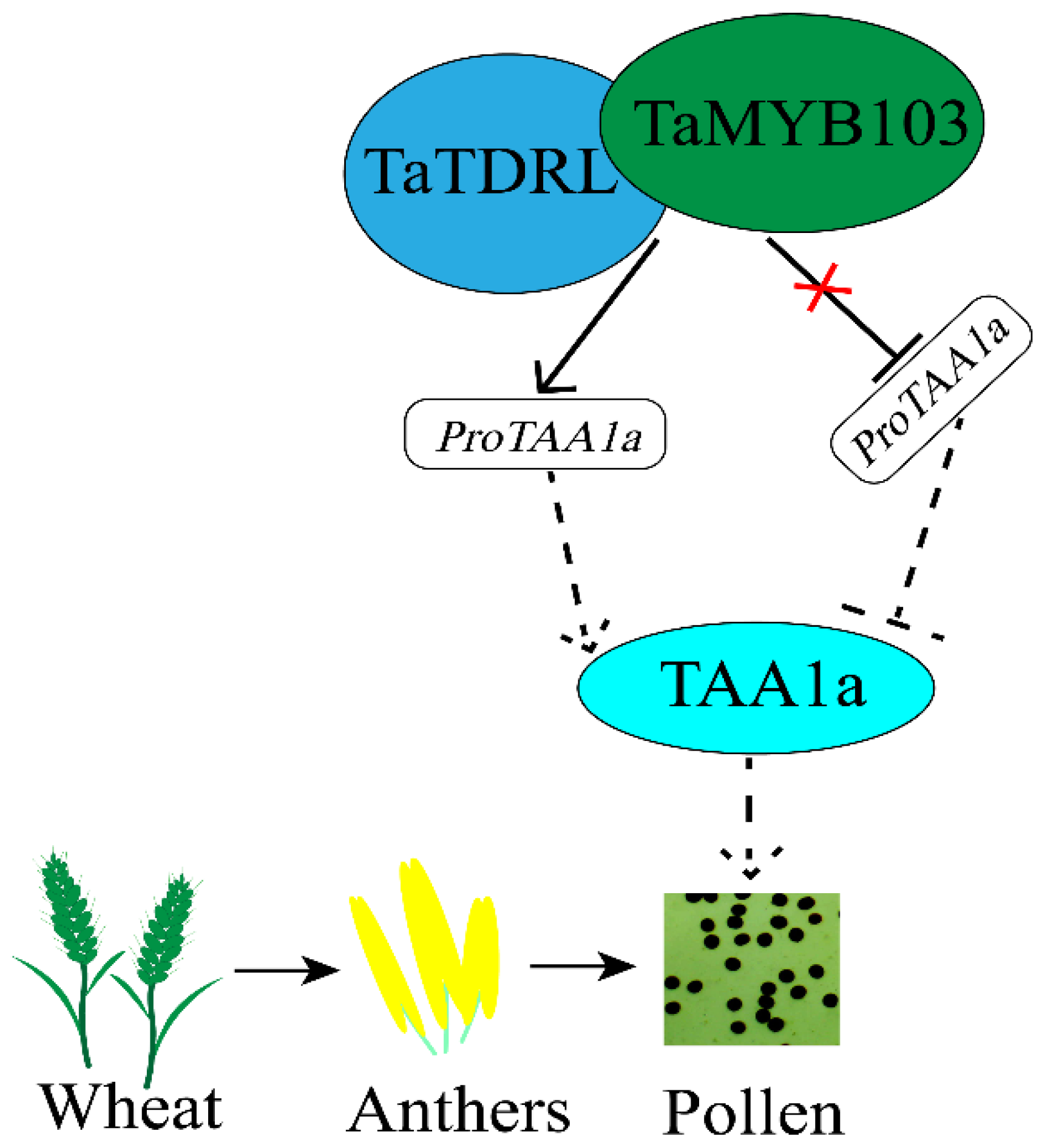
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Isolation of the TAA1a Promoter and Bioinformatics Analysis
4.3. Expression Analysis
4.4. Transformation of Arabidopsis thaliana
4.5. Analysis of Protein Subcellular Localization
4.6. Yeast One-Hybrid Assay
4.7. Yeast Two-Hybrid Assay
4.8. BiFC
4.9. Dual-Luciferase Assay
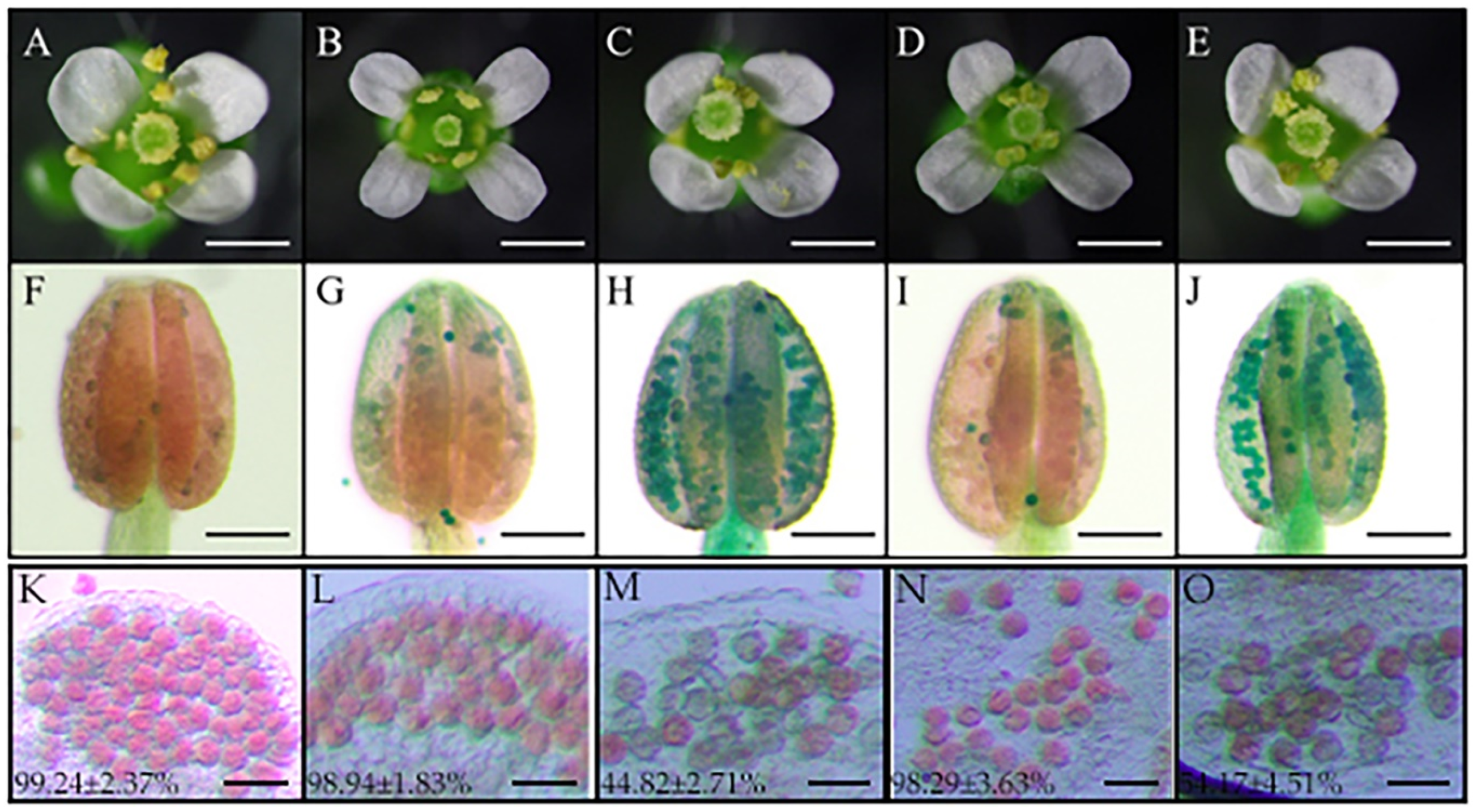
4.10. Phenotypic Characterization and Microspore Analysis of Wheat and Arabidopsis thaliana
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubcovsky, J.; Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, B.S.; Appels, R.; Botha-Oberholster, A.M.; Buell, C.R.; Bennetzen, J.L.; Chalhoub, B.; Chumley, F.; Dvorak, J.; Iwanaga, M.; Keller, B.; et al. A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics 2004, 168, 1087–1096. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.K.; Mir, R.R.; Mohan, A.; Kumar, J. Wheat genomics: Present status and future prospects. Int. J. Plant Genom. 2008, 2008, 896451. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, J.; Scott, P.; Brinton, J.; Mestre, T.C.; Bush, M.; Del Blanco, A.; Dubcovsky, J.; Uauy, C. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. TAG Theor. Appl. Genet. 2016, 129, 1099–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, T.C.; Zhang, X.K.; Yin, G.H.; Wang, L.N.; Han, Y.L.; Chen, L.; Huang, F.; Tang, J.W.; Xia, X.C.; He, Z.H. Genetic gains in grain yield, net photosynthesis and stomatal conductance achieved in Henan Province of China between 1981 and 2008. Field Crops Res. 2011, 122, 225–233. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [Green Version]
- Eckardt, N.A. Cytoplasmic Male Sterility and Fertility Restoration. Plant Cell 2006, 18, 515–517. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Chatrath, R.; Mishra, B. Perspective of hybrid wheat research: A review. Indian J. Agric. Sci. 2010, 80, 1013–1027. [Google Scholar]
- Sun, H.; Zhang, J.; Wang, Y.; Zhao, L. A review of utilization of heterosis in three legume crops of pigeonpea, Alfalfa and Soybean. Sci. Agric. Sin. 2009, 42, 1528–1539. [Google Scholar]
- Gan, Y.; Gong, D.H.; Yang, Z.L.; Jia, X.Y.; Xiang, G.L.; Huang, Z.H.; Pan, J.H. Breeding of Jianyou 388, a New Hybrid Rice Variety. Guizhou Agric. Sci. 2012, 40, 12–14. [Google Scholar]
- Fu, D.; Xiao, M.; Hayward, A.; Fu, Y.; Liu, G.; Jiang, G.; Zhang, H. Utilization of crop heterosis. Euphytica 2014, 197, 161–173. [Google Scholar] [CrossRef]
- Zhang, L.L.; Wang, C.J.; Yu, Y.G.; Zhang, Y.M.; Song, Y.L.; Li, Z.; Wang, S.P.; Zhang, Y.F.; Guo, X.F.; Liu, D.; et al. Cyclin-Dependent Kinase Inhibitor Gene TaICK1 acts as a Potential Contributor to Wheat Male Sterility induced by a Chemical Hybridizing Agent. Int. J. Mol. Sci. 2020, 21, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.F.; Diao, X.M. Progress in Identification of Plant Male Sterility Related Nuclear Genes. J. Plant Genet. Resour. 2013, 13, 1108–1117. [Google Scholar]
- Ku, S.; Yoon, H.; Suh, H.S.; Chung, Y.-Y. Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 2003, 217, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hu, S.W.; Li, W.; Yu, C.Y. Breeding of RGMS lines and investigation on their cytology in Brassica napus L. J. Northwest A&F Univ. Nat. Sci. Ed. 2010, 38, 111–118+124. [Google Scholar]
- Su, Y.W.; Zhao, S.Y.; Zhang, Y.J.; Shi, H.L.; He, Q.W. Study on Cytomorphology of Microsporogenesis in Male Sterile Line of Radish (Raphanus sativus L.). Acta Hortic. Sin. 1995, 4, 348–352. [Google Scholar]
- Zhao, H.F.; Huang, W.; Ahmed, S.S.; Gong, Z.H.; Zhao, L.M. The pollen wall and tapetum are altered in the cytoplasmic male sterile line RC(7) of Chinese cabbage (Brassica campestris ssp. pekinensis). Genet. Mol. Res. 2012, 11, 4145–4156. [Google Scholar] [CrossRef]
- Yao, Y.Q.; Zhang, G.S.; Liu, H.W.; Wang, J.W. Studies on correlation between intine and K-type cytoplasmic male sterility in wheat. Acta Bot. Boreali-Occident. Sin. 2002, 2, 125–129+249. [Google Scholar]
- Han, X.B.; Li, R.Q.; Xu, N.Y.; Wang, J.B.; Xu, Z.Y. A Comparative Cytological Study on Pollen Abortion of Different Cytoplasmic Male Sterile Lines of Wheat. Acta Agron. Sin. 1996, 6, 646–651+769–770. [Google Scholar]
- Osthoff, K.S.; Wiermann, R. Phenols as Integrated Compounds of Sporopollenin from Pinus Pollen. J. Plant Physiol. 1987, 131, 5–15. [Google Scholar] [CrossRef]
- Ma, H. Molecular Genetic Analyses of Microsporogenesis and Microgametogenesis in Flowering Plants. Monash Bioeth. Rev. 2005, 56, 393–434. [Google Scholar] [CrossRef] [PubMed]
- Paxson-Sowders, D.M.; Dodrill, C.H.; Owen, H.A.; Makaroff, C.A. DEX1, a Novel Plant Protein, Is Required for Exine Pattern Formation during Pollen Development in Arabidopsis. Plant Physiol. 2001, 127, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Morant, M.; Jorgensen, K.; Schaller, H.; Pinot, F.; Moller, B.L.; Werck-Reichhart, D.; Bak, S. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 2007, 19, 1473–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piffanelli, P.; Ross, J.H.E.; Murphy, D.J. Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 1998, 11, 65–80. [Google Scholar] [CrossRef]
- Bubert, H.; Lambert, J.; Steuernagel, S.; Ahlers, F.; Wiermann, R. Continuous Decomposition of Sporopollenin from Pollen of Typha angustifolia L. by Acidic Methanolysis. Z. Nat. C J. Biosci. 2002, 57, 1035–1041. [Google Scholar] [CrossRef]
- Schnurr, J.; Shockey, J.; Browse, J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 2004, 16, 629–642. [Google Scholar] [CrossRef] [Green Version]
- Kienow, L.; Schneider, K.; Bartsch, M.; Stuible, H.P.; Weng, H.; Miersch, O.; Wasternack, C.; Kombrink, E. Jasmonates meet fatty acids: Functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J. Exp. Bot. 2008, 59, 403–419. [Google Scholar] [CrossRef]
- de Azevedo Souza, C.; Kim, S.S.; Koch, S.; Kienow, L.; Schneider, K.; McKim, S.M.; Haughn, G.W.; Kombrink, E.; Douglas, C.J. A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 2009, 21, 507–525. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y.; et al. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef] [Green Version]
- Aarts, M.; Hodge, R.; Kalantidis, K.; Florack, D.; Wilson, Z.; Mulligan, B.; Stiekema, W.; Scott, R.; Pereira, A. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 1997, 12, 616–623. [Google Scholar] [CrossRef]
- Doan, T.T.; Carlsson, A.S.; Hamberg, M.; Bulow, L.; Stymne, S.; Olsson, P. Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J. Plant Physiol. 2009, 166, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yu, X.-H.; Zhang, K.; Shi, J.; Oliveira, S.D.; Schreiber, L.; Shanklin, J.; Zhang, D. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 2011, 157, 842–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Tan, H.; Yu, X.H.; Liu, Y.; Liang, W.; Ranathunge, K.; Franke, R.B.; Schreiber, L.; Wang, Y.; Kai, G.; et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 2011, 23, 2225–2246. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.M.; Xia, Q.; Xie, W.S.; Dumonceaux, T.; Zou, J.; Datla, R.; Selvaraj, G. Male gametophyte development in bread wheat (Triticum aestivum L.): Molecular, cellular, and biochemical analyses of a sporophytic contribution to pollen wall ontogeny. Plant J. 2002, 30, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Yang, C.Y.; Yuan, Z.; Zhang, D.S.; Gondwe, M.Y.; Ding, Z.W.; Liang, W.Q.; Zhang, D.B.; Wilson, Z.A. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 2010, 22, 91–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, N.N.; Liang, W.Q.; Yang, X.J.; Jin, W.L.; Wilson, Z.A.; Hu, J.P.; Zhang, D.B. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Shi, J.X.; Zhao, G.C.; Zhang, D.B.; Liang, W.Q. Post-meiotic deficient anther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. J. Plant Biol. 2013, 56, 59–68. [Google Scholar] [CrossRef]
- Huysmans, S.; El-Ghazaly, G.; Smets, E. Orbicules in angiosperms: Morphology, function, distribution, and relation with tapetum types. Bot. Rev. 1998, 64, 240–272. [Google Scholar] [CrossRef]
- Xiong, S.X.; Lu, J.Y.; Lou, Y.; Teng, X.D.; Gu, J.N.; Zhang, C.; Shi, Q.S.; Yang, Z.N.; Zhu, J. The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana. Plant J. 2016, 88, 936–946. [Google Scholar] [CrossRef]
- Wang, K.; Guo, Z.L.; Zhou, W.T.; Zhang, C.; Zhang, Z.Y.; Lou, Y.; Xiong, S.X.; Yao, X.Z.; Fan, J.J.; Zhu, J.; et al. The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol. 2018, 178, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Zhang, D.S.; Liu, H.S.; Yin, C.S.; Li, X.X.; Liang, W.Q.; Yuan, Z.; Xu, B.; Chu, H.W.; Wang, J.; et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 2006, 18, 2999–3014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, W.L.; Zhang, G.S.; Liu, H.W.; Wang, J.W.; Wang, X.L.; Zhao, Z.J. Studies on fertility restoration performance of male sterile lines of non-1BL/1RS wheat with Ae.kotschyi, Ae. variabilis and Ae. ventricosa cytoplasms. Acta Bot. Boreali-Occident. Sin. 2003, 1, 69–74. [Google Scholar]
- Li, H.X.; Zhang, L.Y.; Zhang, G.S.; Niu, N.; Zhu, Z.W. Construction on cDNA library from fertility-related genes of male sterile wheat with aegilops kotschyi cytoplasm by SSH. Acta Agron. Sin. 2008, 6, 965–971. [Google Scholar]
- Niu, N.; Ma, S.C.; Wang, J.W.; Song, Y.L.; Zhang, G.S. Construction of nucleo-cytoplasmic male sterility-restorer near-isogenic lines of wheat with Ae. kotschyi cytoplasm and detection of their genetic background. J. South. Agric. 2016, 47, 1247–1250. [Google Scholar]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef]
- Xia, Y.; Li, Z.; Wang, J.W.; Li, Y.H.; Ren, Y.; Du, J.J.; Song, Q.L.; Ma, S.C.; Song, Y.L.; Zhao, H.Y.; et al. Isolation and Identification of a TaTDR-Like Wheat Gene Encoding a bHLH Domain Protein, Which Negatively Regulates Chlorophyll Biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.S.; Yang, T.Z. A preliminary on the male sterile lines of wheat with Ae. ventricosa and Ae. kotschyi and Ae. variabilis cytoplasms. Acta Agron. Sin. 1989, 1, 1–10. [Google Scholar]
- Wei, Y.M.; Zheng, Y.L.; Zhou, Y.H.; Liu, D.C.; Lan, X.J.; Zhou, R.H.; Jia, J.Z. The effects of chromosome 1RS/1BL translocation on the agronomic characters of common wheat revealed by RFLP markers. Southwest China J. Agric. Sci. 1999, S2, 105–110. [Google Scholar]
- Zhou, Y.; He, Z.H.; Zhang, G.S.; Xia, L.Q.; Yu, G. Utilization of 1BL/1RS translocation in wheat breeding in China. Acta Agron. Sin. 2004, 30, 531–535. [Google Scholar]
- Fu, X.D.; Liu, Q.; Li, Z.S.; Zhang, A.M.; Ling, H.Q.; Tong, Y.P.; Liu, Z.Y. Research achievement and prospect development on wheat genome. Bull. Chin. Acad. Sci. 2018, 33, 909–914. [Google Scholar]
- Dobritsa, A.A.; Shrestha, J.; Morant, M.; Pinot, F.; Matsuno, M.; Swanson, R.; Moller, B.L.; Preuss, D. CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 2009, 151, 574–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quilichini, T.D.; Friedmann, M.C.; Samuels, A.L.; Douglas, C.J. ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 2010, 154, 678–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, X.Y.; Yang, K.Z.; Zhang, Y.; Wang, W.; Liu, X.L.; Chen, L.Q.; Zhang, X.Q.; Ye, D. WBC27, an adenosine tri-phosphate-binding cassette protein, controls pollen wall formation and patterning in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 74–88. [Google Scholar] [CrossRef]
- Mondol, P.C.; Xu, D.W.; Duan, L.D.; Shi, J.X.; Wang, C.H.; Chen, X.F.; Chen, M.J.; Hu, J.P.; Liang, W.Q.; Zhang, D.B. Defective Pollen Wall 3 (DPW3), a novel alpha integrin-like protein, is required for pollen wall formation in rice. New Phytol. 2020, 225, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Zhang, G.S.; Li, Y.X.; Zhang, L.Y.; Wang, S.P.; Zhao, X.L.; Wang, L.M.; Song, Y.L. The relationship on anther tapetum, sporopollenin and expression of RAFTIN1 in physiological male sterile wheat. Sci. Agric. Sin. 2011, 44, 3937–3944. [Google Scholar]
- Xu, J.; Ding, Z.W.; Vizcay-Barrena, G.; Shi, J.X.; Liang, W.Q.; Yuan, Z.; Werck-Reichhart, D.; Schreiber, L.; Wilson, Z.A.; Zhang, D.B. ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 2014, 26, 1544–1556. [Google Scholar] [CrossRef] [Green Version]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [Green Version]
- Preston, J.; Wheeler, J.; Heazlewood, J.; Li, S.F.; Parish, R.W. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2004, 40, 979–995. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, Y.J.; Timofejeva, L.; Chen, C.B.; Grossniklaus, U.; Ma, H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 2006, 133, 3085–3095. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Li, H.; Shi, Q.L.; Jiang, H.; Chen, H.; Zhong, X.L.; Gao, J.F.; Cui, Y.L.; Yang, Z.N. The Arabidopsis bHLH transcription factor DYTI is essential for anther development by regulating callose dissolution. J. Shanghai Norm. Univ. 2009, 34, 2. [Google Scholar]
- Ji, C.H.; Li, H.Y.; Chen, L.B.; Xie, M.; Wang, F.P.; Chen, Y.L.; Liu, Y.G. A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol. Plant 2013, 6, 1715–1718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.F.; Sun, L.P.; Zhang, P.P.; Zhang, Y.X.; Yu, P.; Liu, L.; Abbas, A.; Xiang, X.J.; Wu, W.X.; Zhan, X.D.; et al. TDR INTERACTING PROTEIN 3, encoding a PHD-finger transcription factor, regulates Ubisch bodies and pollen wall formation in rice. Plant J. 2019, 99, 844–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.S.; Liang, W.Q.; Yuan, Z.; Li, N.; Shi, J.; Wang, J.; Liu, Y.M.; Yu, W.J.; Zhang, D.B. Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 2008, 1, 599–610. [Google Scholar] [CrossRef]
- Yang, Z.F.; Liu, L.; Sun, L.P.; Yu, P.; Zhang, P.P.; Abbas, A.; Xiang, X.J.; Wu, W.X.; Zhang, Y.X.; Cao, L.Y.; et al. OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 2019, 99, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Fang, Z.J.; Zhu, J.; Gao, J.F.; Yang, Z.N. OsMYB103 is required for rice anther development by regulating tapetum development and exine formation. Chin. Sci. Bull. 2010, 55, 3288–3297. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene 2011, 484, 61–68. [Google Scholar] [CrossRef]
- Qi, T.C.; Huang, H.; Song, S.S.; Xie, D.X. Regulation of Jasmonate-Mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef] [Green Version]
- Lou, Y.; Zhou, H.S.; Han, Y.; Zeng, Q.Y.; Zhu, J.; Yang, Z.N. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana. New Phytol. 2018, 217, 378–391. [Google Scholar] [CrossRef] [Green Version]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.A.; Iacuone, S.; Li, S.F.; Parish, R.W. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 2011, 23, 2209–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Walter, M.; Chaban, C.; Schutze, K.; Batistic, O.; Weckermann, K.; Nake, C.; Blazevic, D.; Grefen, C.; Schumacher, K.; Oecking, C.; et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004, 40, 428–438. [Google Scholar] [CrossRef]
- Lu, X.D.; Zhou, C.M.; Xu, P.B.; Luo, Q.; Lian, H.L.; Yang, H.Q. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 2015, 8, 467–478. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.M.; Zhu, Q.G.; Deng, C.L.; Luo, Z.R.; Sun, N.J.; Grierson, D.; Yin, X.R.; Chen, K.S. Hypoxia-responsive ERFs involved in postdeastringency softening of persimmon fruit. Plant Biotechnol. J. 2017, 15, 1409–1419. [Google Scholar] [CrossRef] [Green Version]
- Zeng, J.K.; Li, X.; Xu, Q.; Chen, J.Y.; Yin, X.R.; Ferguson, I.B.; Chen, K.S. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 2015, 13, 1325–1334. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Song, Q.L.; Zhang, L.L.; Li, Z.; Wang, C.S.; Zhang, G.S. Comparative proteomic analysis of developmental changes in P-type cytoplasmic nale sterile and maintainer anthers in wheat. Int. J. Mol. Sci. 2021, 22, 2012. [Google Scholar] [CrossRef] [PubMed]
- Johnson-Brousseau, S.A.; McCormick, S. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 2004, 39, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.J.; Li, H.F.; Su, Y.L.; Li, W.Q.; Shi, C.H. G1/ELE functions in the development of rice lemmas in addition to determining identities of empty glumes. Front. Plant Sci. 2016, 7, 1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, B.; Xia, Y.; Zhang, G.; Wang, J.; Ma, S.; Song, Y.; Yang, Z.; Dennis, E.S.; Niu, N. The Transcription Factors TaTDRL and TaMYB103 Synergistically Activate the Expression of TAA1a in Wheat, Which Positively Regulates the Development of Microspore in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 7996. https://doi.org/10.3390/ijms23147996
Wu B, Xia Y, Zhang G, Wang J, Ma S, Song Y, Yang Z, Dennis ES, Niu N. The Transcription Factors TaTDRL and TaMYB103 Synergistically Activate the Expression of TAA1a in Wheat, Which Positively Regulates the Development of Microspore in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(14):7996. https://doi.org/10.3390/ijms23147996
Chicago/Turabian StyleWu, Baolin, Yu Xia, Gaisheng Zhang, Junwei Wang, Shoucai Ma, Yulong Song, Zhiquan Yang, Elizabeth S. Dennis, and Na Niu. 2022. "The Transcription Factors TaTDRL and TaMYB103 Synergistically Activate the Expression of TAA1a in Wheat, Which Positively Regulates the Development of Microspore in Arabidopsis" International Journal of Molecular Sciences 23, no. 14: 7996. https://doi.org/10.3390/ijms23147996
APA StyleWu, B., Xia, Y., Zhang, G., Wang, J., Ma, S., Song, Y., Yang, Z., Dennis, E. S., & Niu, N. (2022). The Transcription Factors TaTDRL and TaMYB103 Synergistically Activate the Expression of TAA1a in Wheat, Which Positively Regulates the Development of Microspore in Arabidopsis. International Journal of Molecular Sciences, 23(14), 7996. https://doi.org/10.3390/ijms23147996





