Overexpression of MxbHLH18 Increased Iron and High Salinity Stress Tolerance in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
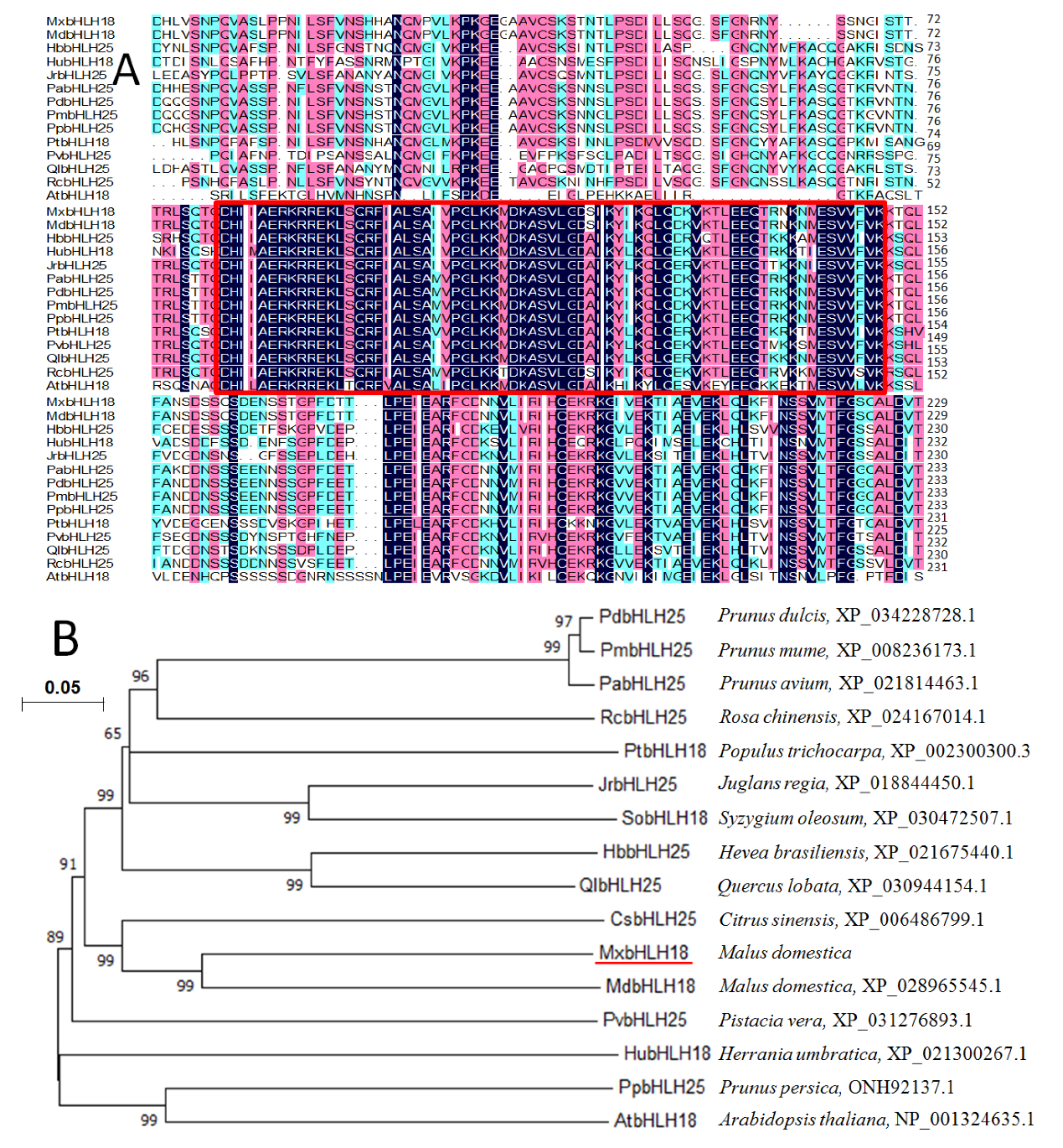
2.1. Sequence Analysis of MxbHLH18
2.2. Phylogenetic Analysis of MxbHLH18
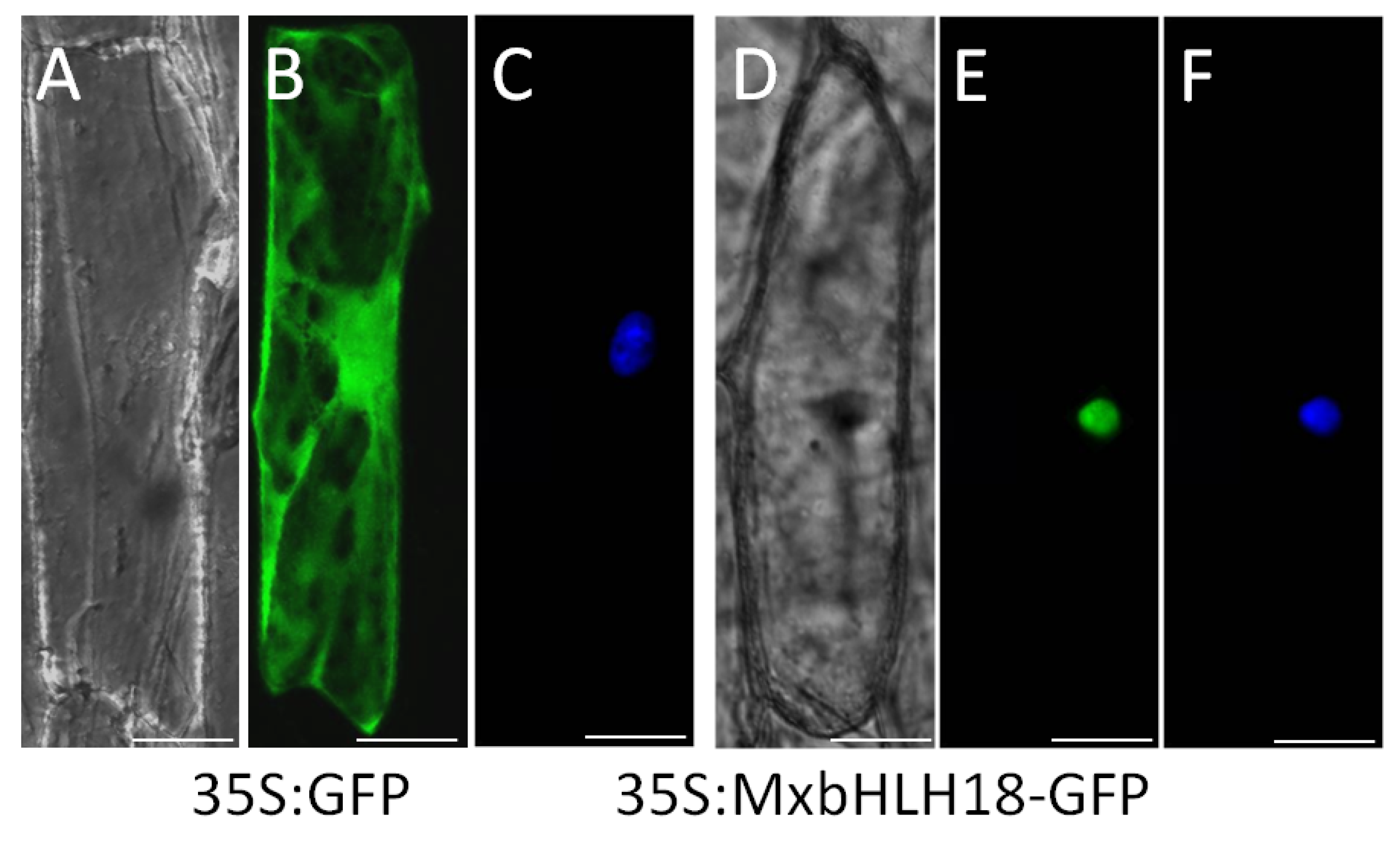
2.3. MxbHLH18 Was Localized in the Nucleus
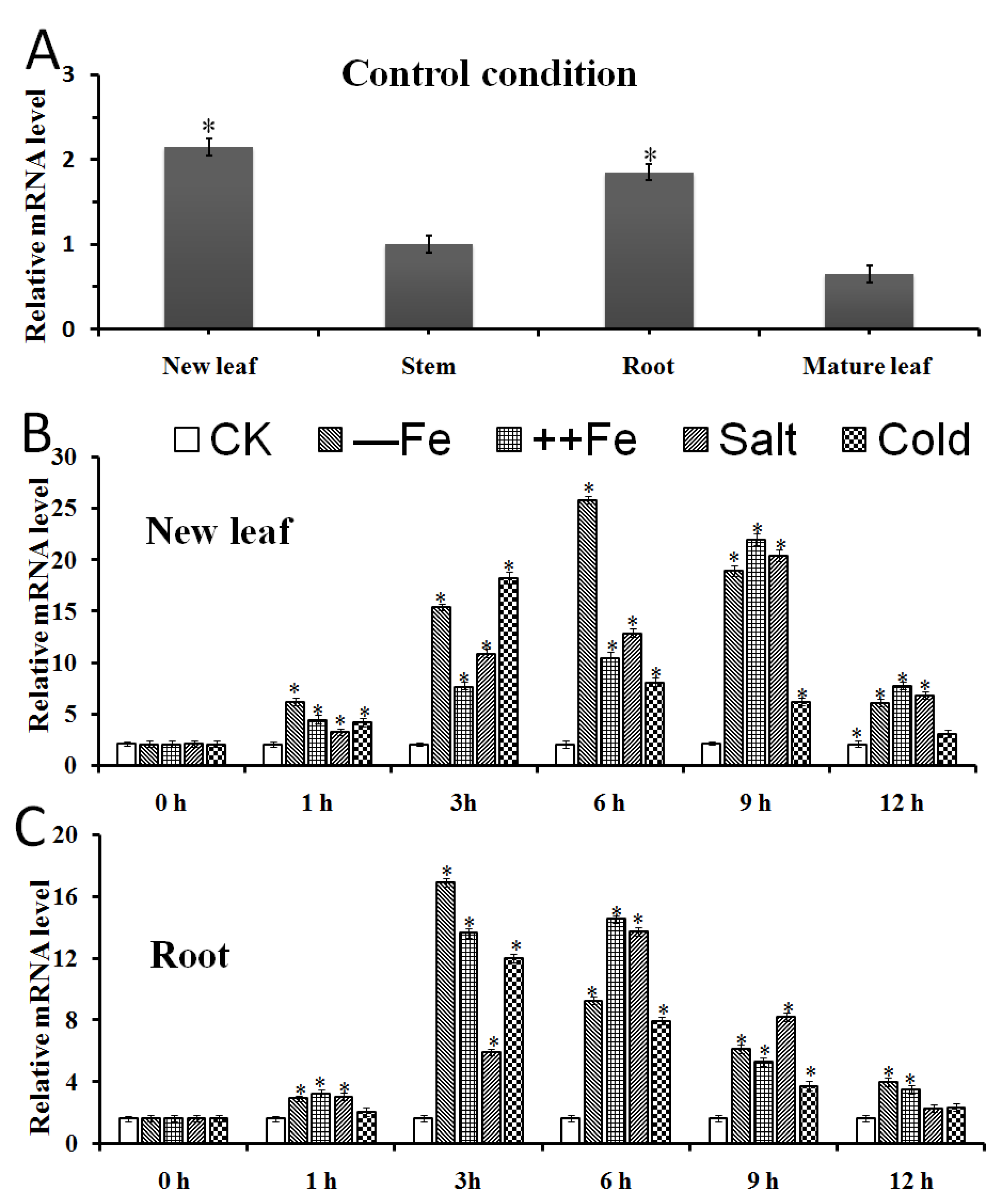
2.4. The Expression Patterns of MxbHLH18 in M. xiaojinensis
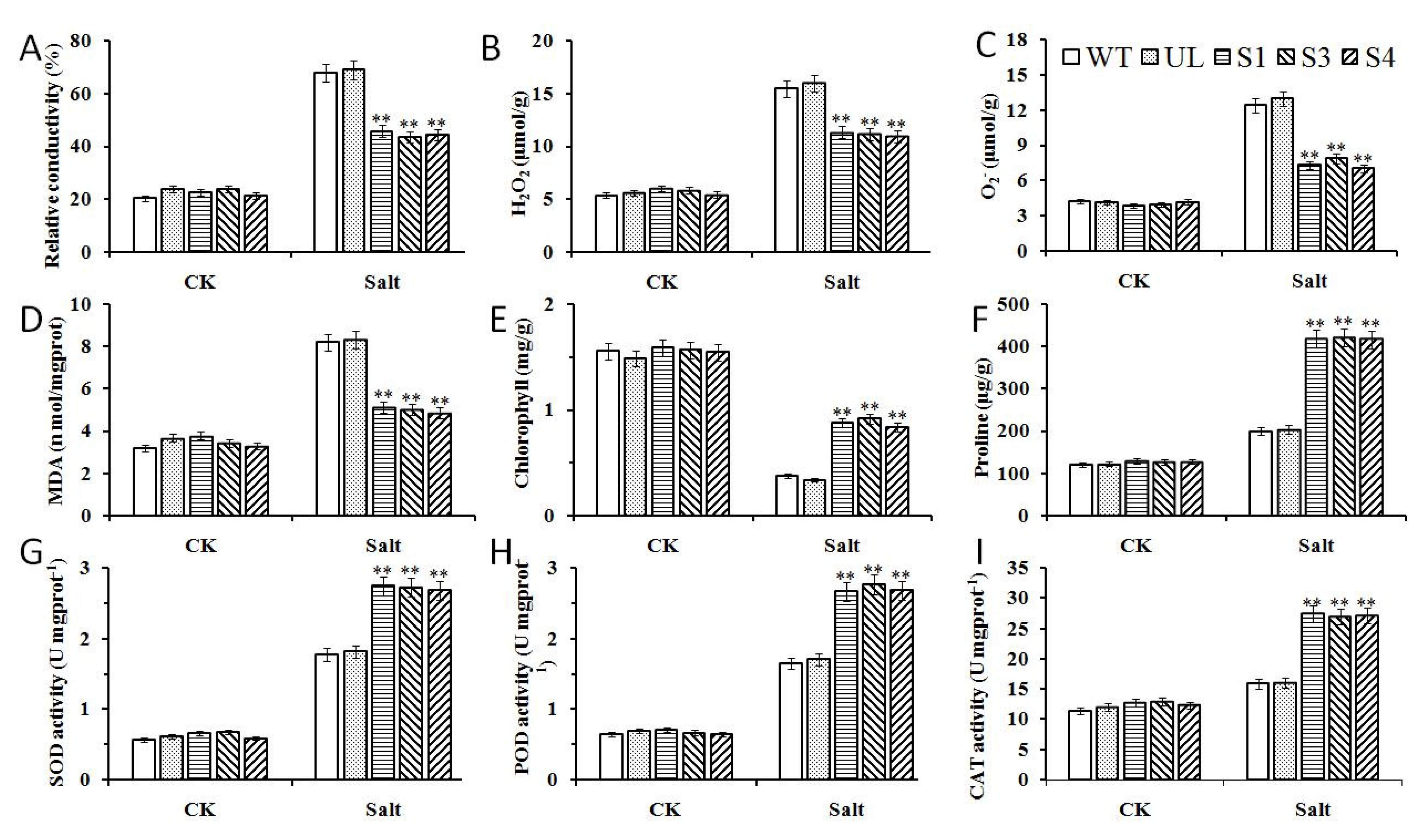
2.5. Overexpression of MxbHLH18 Contributed to Higher Salinity Tolerance in Transgenic A. thaliana
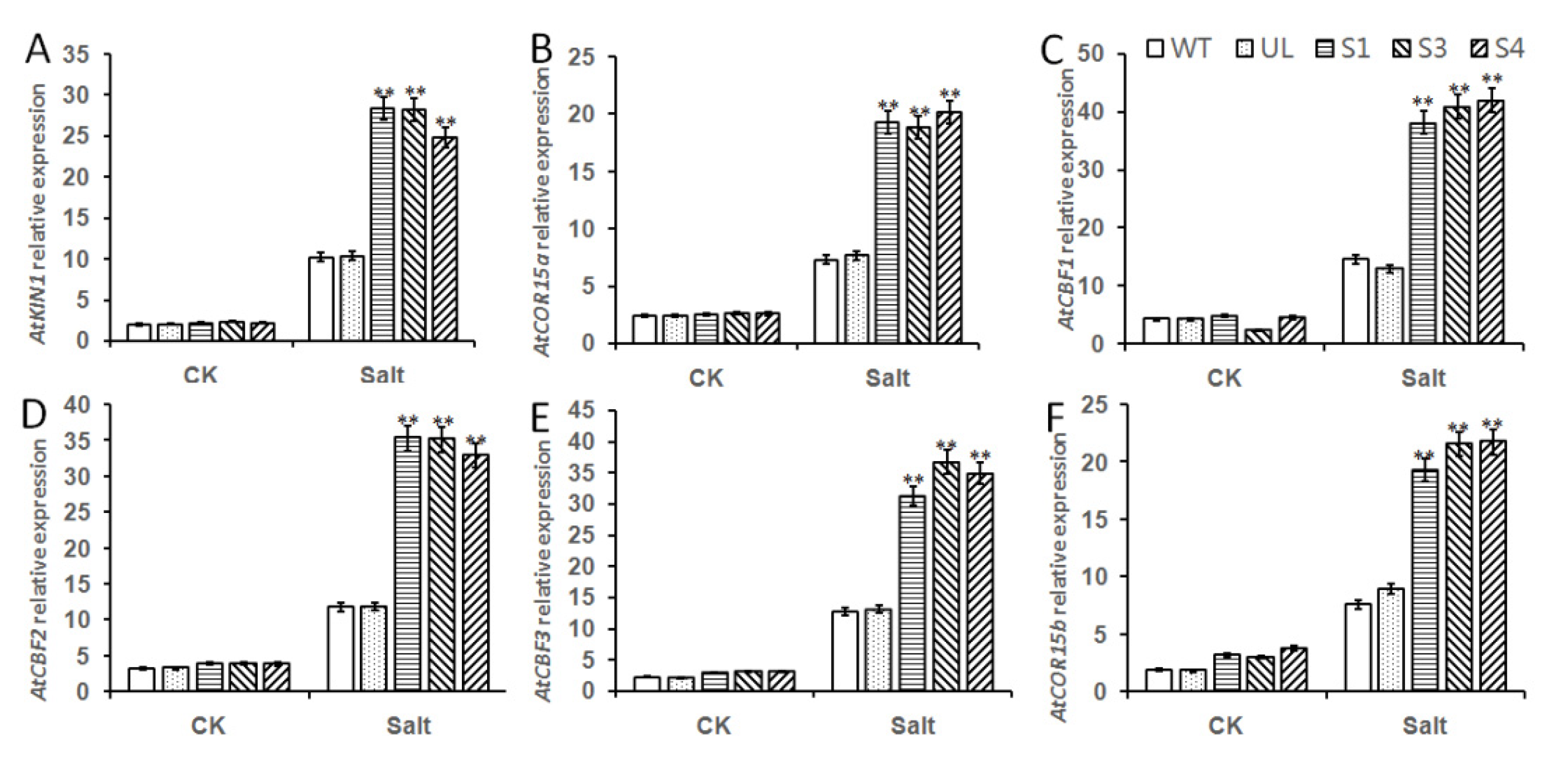
2.6. Overexpression of MbMYB108 Promotes the Expression of Salt Stress-Related Genes
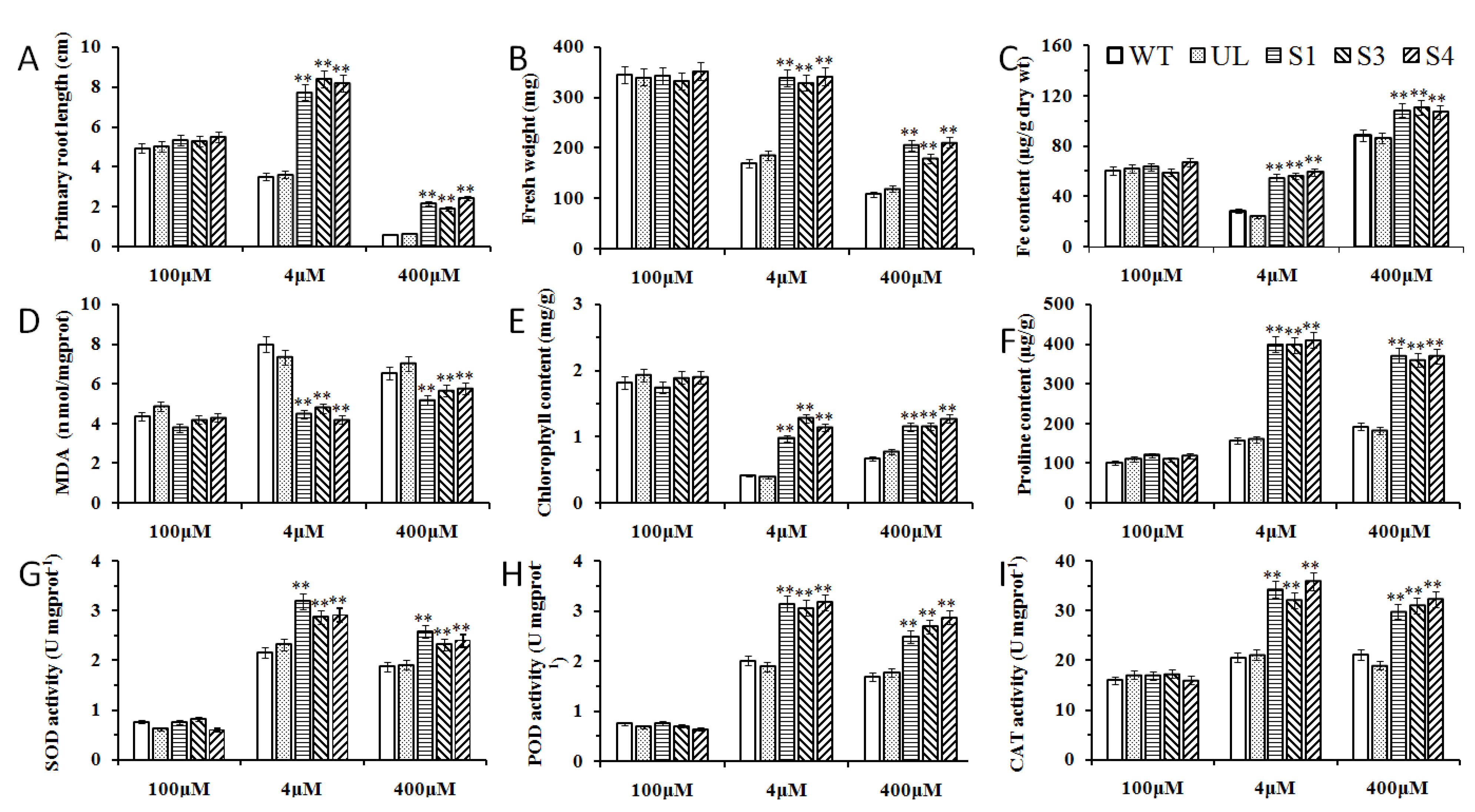
2.7. Overexpression of MxbHLH18 Enhanced High and/or Low Fe Stress Tolerance in Transgenic A. thaliana
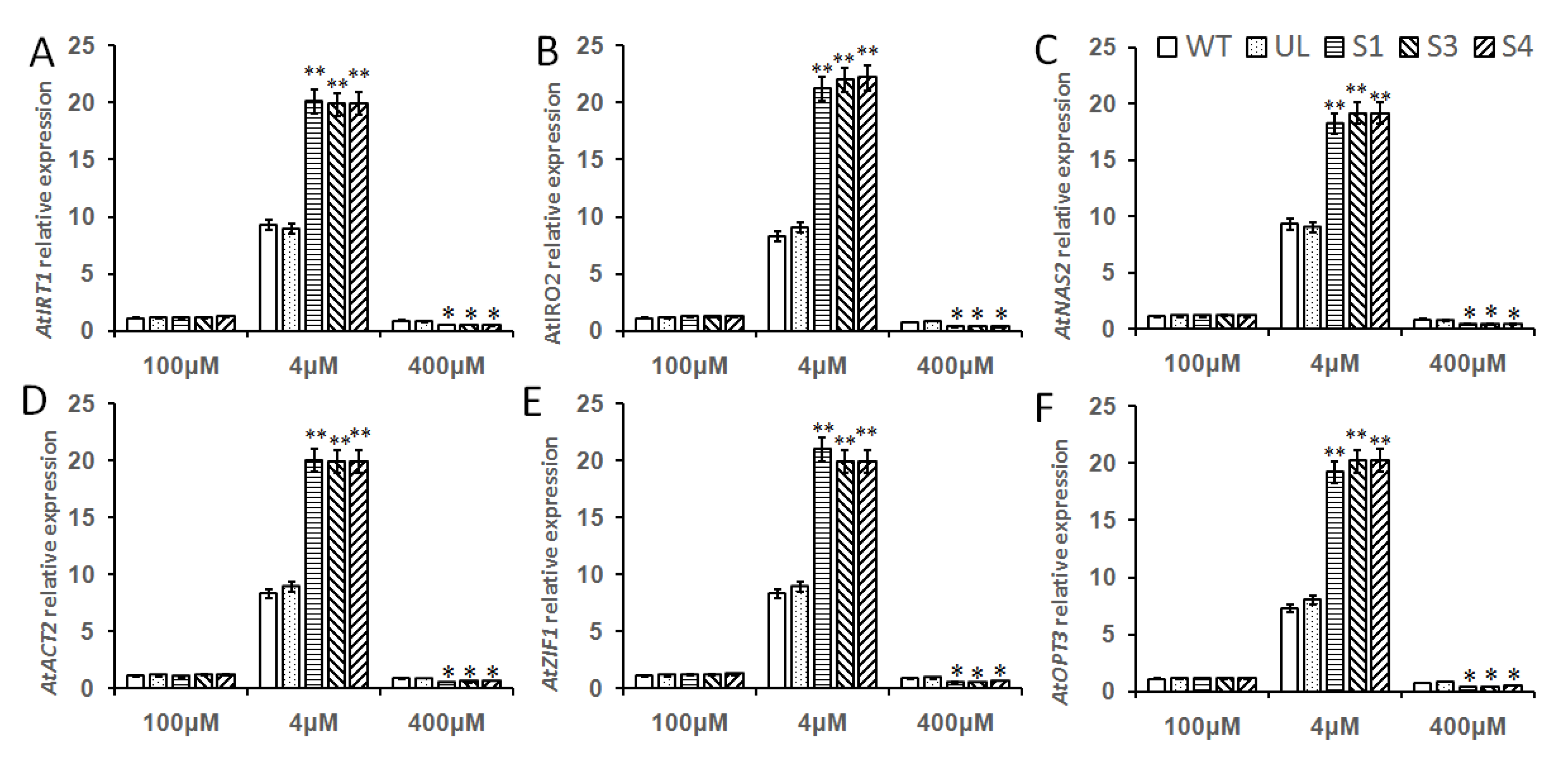
2.8. Overexpression of MbMYB108 Promotes the Expression of High and/or Low Fe Stress-Related Genes
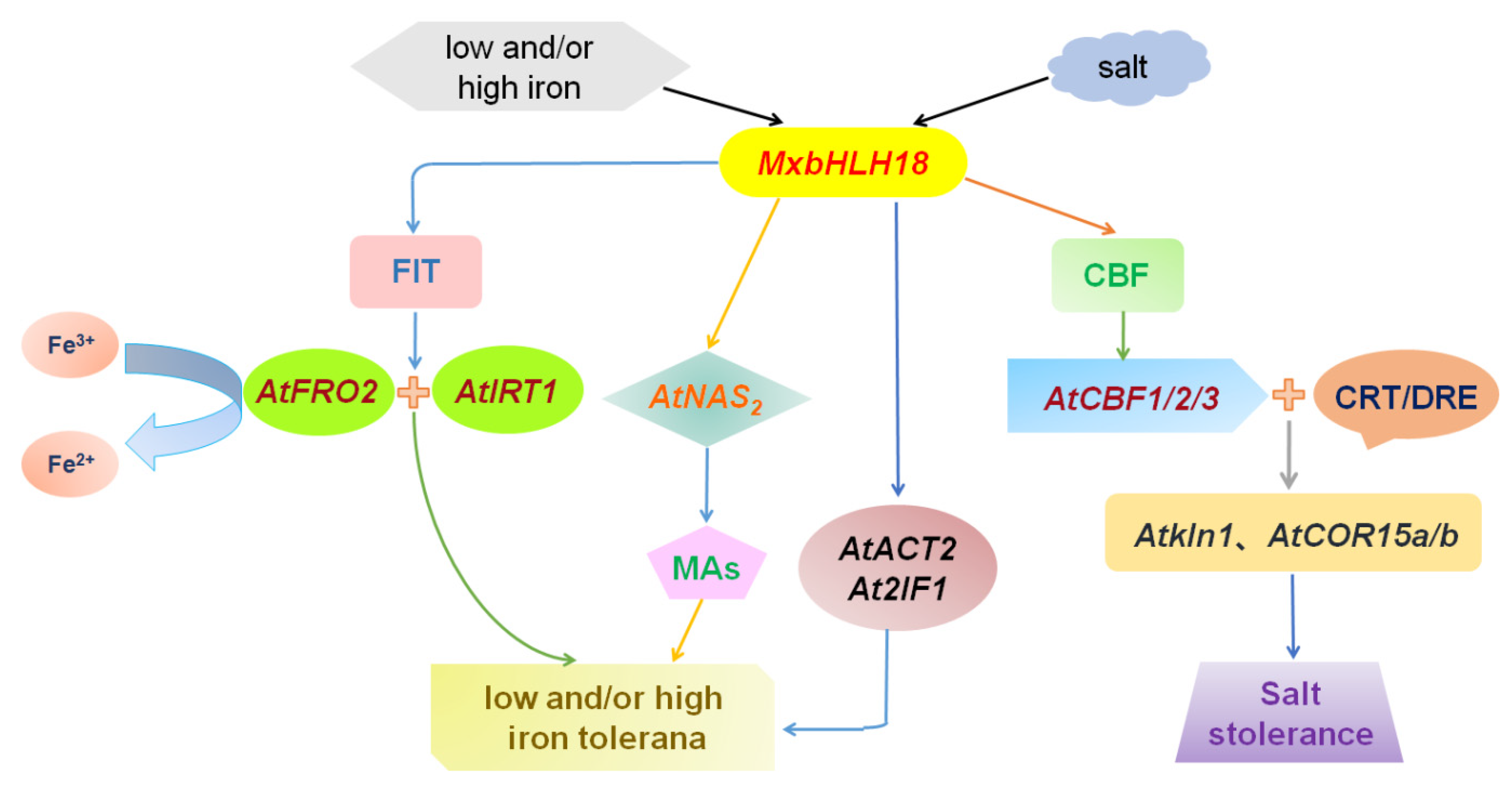
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Isolation and qPCR Analysis of MxbHLH18
4.3. Sequence Analysis of MxbHLH18
4.4. Subcellular Localization of the MxbHLH18 Protein
4.5. Generation of Transgenic A. thaliana Overexpressing MxbHLH18
4.6. Fe and Salt Stress Treatments to Transgenic A. thaliana
4.7. Determination of Relevant Physiological Indexes
4.8. Expression Analysis of MbMYB108 Downstream Genes
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2008, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Zhao, Y.; Wang, Y.; Lin, Y.; Peng, X.; Li, Q.; Chang, Y.; Jiang, H.; Xiang, Y.; Cheng, B. Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 119, 565–577. [Google Scholar] [CrossRef]
- Ding, W.; Fang, W.; Shi, S.; Zhao, Y.; Li, X.; Xiao, K. Wheat WRKY Type Transcription Factor Gene TaWRKY1 is Essential in Mediating Drought Tolerance Associated with an ABA-Dependent Pathway. Plant Mol. Biol. Rep. 2016, 34, 1111–1126. [Google Scholar] [CrossRef]
- Li, G.; Wang, Z.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.; Li, G.; Wu, Y.; Yang, J.; Ma, J.; et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice. New Phytol. 2018, 219, 149–162. [Google Scholar] [CrossRef] [Green Version]
- Luan, Q.; Chen, C.; Liu, M.; Li, Q.; Wang, L.; Ren, Z. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci. 2018, 279, 59–69. [Google Scholar] [CrossRef]
- Yao, C.; Li, X.; Li, Y.; Yang, G.; Liu, W.; Shao, B.; Zhong, J.; Huang, P.; Han, D. Overexpression of a Malus baccata MYB Transcription Factor Gene MbMYB4 Increases Cold and Drought Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 1794. [Google Scholar] [CrossRef]
- Yao, C.; Li, W.; Liang, X.; Ren, C.; Liu, W.; Yang, G.; Zhao, M.; Yang, T.; Li, X.; Han, D. Molecular cloning and characterization of MbMYB108, a Malus baccata MYB transcription factor gene, with functions in tolerance to cold and drought stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 4846. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1198. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.; Cho, Y.-G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Wu, K.-L.; Guo, Z.-J.; Wang, H.-H.; Li, J. The WRKY Family of Transcription Factors in Rice and Arabidopsis and Their Origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [Green Version]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 Transcription Factor Is a Modulator of Phosphate Acquisition and Root Development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.-L.; Xu, K.-D.; Pan, Y.-Z.; Jiang, B.-B.; Liu, G.-L.; Jia, Y.; Zhang, H.-Q. Functional Analysis of a Novel Chrysanthemum WRKY Transcription Factor Gene Involved in Salt Tolerance. Plant Mol. Biol. Rep. 2013, 32, 282–289. [Google Scholar] [CrossRef]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive Activation of Transcription Factor OsbZIP46 Improves Drought Tolerance in Rice. Plant Physiol. 2012, 158, 1755–1768. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; White, M.J.; Macrae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Singh, K.; Foley, R.C.; Oñatesánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant. Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Marschner, H.; Römheld, V. Strategies of plants for acquisition of iron. Plant Soil 1994, 165, 261–274. [Google Scholar] [CrossRef]
- López-Millán, A.F.; Morales, F.; Gogorcena, Y.; Abadía, A.; Abadía, J. Metabolic responses in iron deficient tomato plants. J. Plant Physiol. 2009, 166, 375–384. [Google Scholar] [CrossRef]
- Wang, M.; Gong, J.; Bhullar, N.K. Iron deficiency triggered transcriptome changes in bread wheat. Comput. Struct. Biotechnol. J. 2020, 18, 2709–2722. [Google Scholar] [CrossRef]
- Yang, G.; Li, J.; Liu, W.; Yu, Z.; Shi, Y.; Lv, B.; Wang, B.; Han, D. Molecular cloning and characterization of MxNAS2, a gene encoding nicotianamine synthase in Malus xiaojinensis, with functions in tolerance to iron stress and misshapen flower in transgenic tobacco. Sci. Hortic. 2015, 183, 77–86. [Google Scholar] [CrossRef]
- Wang, J.; Hu, Z.; Zhao, T.; Yang, Y.; Chen, T.; Yang, M.; Yu, W.; Zhang, B. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genom. 2015, 16, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledent, V.; Vervoort, M. The Basic Helix-Loop-Helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, Y.; Liu, Z. Basic Helix-Loop-Helix (bHLH) transcription factor family in Yellow horn (Xanthoceras sorbifolia Bunge): Genome-wide characterization, chromosome location, phylogeny, structures and expression patterns. Int. J. Biol. Macromol. 2020, 160, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Roychoudhury, A. Regulation of physiological aspects in plants by hydrogen sulfide and nitric oxide under challenging environment. Physiol. Plant. 2019, 168, 374–393. [Google Scholar] [CrossRef] [Green Version]
- Jin, R.; Kim, H.S.; Yu, T.; Zhang, A.; Yang, Y.; Liu, M.; Yu, W.; Zhao, P.; Zhang, Q.; Cao, Q.; et al. Identification and function analysis of bHLH genes in response to cold stress in sweetpotato. Plant Physiol. Biochem. 2021, 169, 224–235. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Villanova, I.R.; Martinez-Garcia, J.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [Green Version]
- Shen, T.; Wen, X.; Wen, Z.; Qiu, Z.; Hou, Q.; Li, Z.; Mei, L.; Yu, H.; Qiao, G. Genome-wide identification and expression analysis of bHLH transcription factor family in response to cold stress in sweet cherry (Prunus avium L.). Sci. Hortic. 2021, 279, 109905. [Google Scholar] [CrossRef]
- Wang, P.; Su, L.; Gao, H.; Jiang, X.; Wu, X.; Li, Y.; Zhang, Q.; Wang, Y.; Ren, F. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018, 9, 64. [Google Scholar] [CrossRef] [Green Version]
- Gong, X.-Q.; Liu, J.-H. Genetic transformation and genes for resistance to abiotic and biotic stresses in Citrus and its related genera. Plant Cell Tissue Organ Cult. (PCTOC) 2012, 113, 137–147. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Li, G.X.; Zheng, S.J. WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 2013, 76, 825–835. [Google Scholar] [CrossRef]
- Lee, H.; Changhyun, C.; Choi, C.; Choi, N.; Ji, H.-S.; Park, S.R.; Lee, S.; Hwang, D.-J. Rice WRKY11 Plays a Role in Pathogen Defense and Drought Tolerance. Rice 2018, 11, 1–12. [Google Scholar] [CrossRef]
- Li, L.; Cai, Z.; Ma, Q.; Xia, Z.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 enhances drought and salt tolerance through an ABA-mediated pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [Google Scholar]
- He, L.; Gao, C.; Wang, Y.; Wu, Y.; Liu, Z. A Basic Helix-Loop-Helix Gene from Poplar is Regulated by a Basic Leucine-Zipper Protein and is Involved in the ABA-Dependent Signaling Pathway. Plant Mol. Biol. Rep. 2012, 31, 344–351. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, H.; Kong, W.; Peng, R.; Liu, Q.; Yao, Q. The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis. Planta 2016, 244, 59–73. [Google Scholar] [CrossRef]
- Han, Z.; Shen, T.; Korcak, R.F.; Baligar, V.C. Iron absorption by iron-efficient and inefficient species of apples. J. Plant. Nutr. 1998, 21, 181–190. [Google Scholar] [CrossRef]
- Ricachenevsky, F.K.; Sperotto, R.A.; Menguer, P.K.; Fett, J.P. Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol. Biol. Rep. 2010, 37, 3735–3745. [Google Scholar] [CrossRef]
- Yan, J.Y.; Li, C.X.; Sun, L.; Ren, J.Y.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY Transcription Factor Regulates Fe Translocation under Fe Deficiency. Plant Physiol. 2016, 171, 2017–2027. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Wang, Y.; Zhang, Z.; Pu, Q.; Ding, H.; Han, J.; Fan, T.; Bai, X.; Yang, G. Isolation and functional analysis of MxCS3: A gene encoding a citrate synthase in Malus xiaojinensis, with functions in tolerance to iron stress and abnormal flower in transgenic Arabidopsis thaliana. Plant Growth Regul. 2017, 82, 479–489. [Google Scholar] [CrossRef]
- Zhou, J.; Li, F.; Wang, J.-L.; Ma, Y.; Chong, K.; Xu, Y.-Y. Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J. Plant Physiol. 2009, 166, 1296–1306. [Google Scholar] [CrossRef]
- Cui, J.; You, C.; Zhu, E.; Huang, Q.; Ma, H.; Chang, F. Feedback Regulation of DYT1 by Interactions with Downstream bHLH Factors Promotes DYT1 Nuclear Localization and Anther Development. Plant Cell 2016, 28, 1078–1093. [Google Scholar] [CrossRef] [Green Version]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The Basic Helix-Loop-Helix Transcription Factor Family in Plants: A Genome-Wide Study of Protein Structure and Functional Diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W. bHLH122 is important for drought and osmotic stress resistancein Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Mao, K.; Dong, Q.; Li, C.; Liu, C.; Ma, F. Genome Wide Identification and Characterization of Apple bHLH Transcription Factors and Expression Analysis in Response to Drought and Salt Stress. Front. Plant Sci. 2017, 8, 480. [Google Scholar] [CrossRef] [Green Version]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB)f unction as transcriptional activators in abscisic acid signaling. Plant Cell. 2003, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Abe, H.; Urao, T. Role of Arabidopsis MYC and MYB homologs in drought and ab-scisic acid regulated gene expression. Plant Cell. 1997, 9, 1859–1868. [Google Scholar]
- Feng, X.; Zhao, Q.; Zhao, L.; Qiao, Y.; Xie, X.; Feng, H..; Yao, Y.; You, C.; Hao, Y. The cold-induced basic helix-loop-helixtranscription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 2012, 12, 22. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kim, H. Unctional analysis of a calcium-binding transcription factor involved in plant salt stress signaling. FEBS Lett. 2006, 580, 5251–5256. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Jiang, H.; Mao, Z.; Liu, W.; Jiang, S.; Xu, H.; Su, M.; Zhang, J.; Wang, N.; Zhang, Z.; et al. Ethylene increases the cold tolerance of apple via the MdERF1B-MdCIbHLH1 regulatory module. Plant J. 2021, 106, 379–393. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Sui, X.-Y.; Yang, J.-S.; Xiang, X.-H.; Li, Z.-Q.; Wang, Y.-Y.; Zhou, Z.-C.; Hu, R.-S.; Liu, D. A novel bHLH transcription factor, NtbHLH1, modulates iron homeostasis in tobacco (Nicotiana tabacum L.). Biochem. Biophys. Res. Commun. 2019, 522, 233–239. [Google Scholar] [CrossRef]
- Han, D.; Hou, Y.; Wang, Y.; Ni, B.; Li, Z.; Yang, G. Overexpression of a Malus baccata WRKY transcription factor gene (MbWRKY5) increases drought and salt tolerance in transgenic tobacco. Can. J. Plant Sci. 2019, 99, 173–183. [Google Scholar] [CrossRef]
- Zhou, S.; Zheng, W.-J.; Liu, B.-H.; Zheng, J.-C.; Dong, F.-S.; Liu, Z.-F.; Wen, Z.-Y.; Yang, F.; Wang, H.-B.; Xu, Z.-S.; et al. Characterizing the Role of TaWRKY13 in Salt Tolerance. Int. J. Mol. Sci. 2019, 20, 5712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Hou, L.; Xiao, P.; Guo, Y.; Deyholos, M.K.; Liu, X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2018, 280, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Xiu, H.; Nuruzzaman, M.; Guo, X.; Cao, H.; Huang, J.; Chen, X.; Wu, K.; Zhang, R.; Huang, Y.; Luo, J.; et al. Molecular Cloning and Expression Analysis of Eight PgWRKY Genes in Panax ginseng Responsive to Salt and Hormones. Int. J. Mol. Sci. 2016, 17, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parvaiz, A.; Satyawati, S. Salt stress and phyto-biochemical responses of plants—A review. Plant Soil Environ. 2008, 54, 89–99. [Google Scholar] [CrossRef]
- Han, D.; Han, J.; Yang, G.; Wang, S.; Xu, T.; Li, W. An ERF Transcription Factor Gene from Malus baccata (L.) Borkh, MbERF11, Affects Cold and Salt Stress Tolerance in Arabidopsis. Forests 2020, 11, 514. [Google Scholar] [CrossRef]
- Matysik, J.; Alia, A.; Bhalu, B.; Mohanty, P. Molecular mechanism of quenching of reactive oxygen species by proline under stress in plants. Curr. Sci. 2002, 82, 525–532. [Google Scholar]
- Han, D.; Zhang, Z.; Ding, H.; Chai, L.; Liu, W.; Li, H.; Yang, G. Isolation and characterization of MbWRKY2 gene involved in enhanced drought tolerance in transgenic tobacco. J. Plant Interact. 2018, 13, 163–172. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Zhang, X.; Wang, W.; Wang, Y.; Ming, F. The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS ONE 2013, 10, 1932–6203. [Google Scholar] [CrossRef]
- Han, D.; Zhang, Z.; Ding, H.; Wang, Y.; Liu, W.; Li, H.; Yang, G. Molecular cloning and functional analysis of MbWRKY3 involved in improved drought tolerance in transformed tobacco. J. Plant Interact. 2018, 13, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Zhang, Z.; Ni, B.; Ding, H.; Liu, W.; Li, W.; Chai, L.; Yang, G. Isolation and functional analysis of MxNAS3 involved in enhanced iron stress tolerance and abnormal flower in transgenic Arabidopsis. J. Plant Interact. 2018, 13, 433–441. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Han, D.; Shi, Y.; Yu, Z.; Liu, W.; Lv, B.; Wang, B.; Yang, G. Isolation and functional analysis of MdCS1: A gene encoding a citrate synthase in Malus domestica (L.) Borkh. Plant Growth Regul. 2014, 75, 209–218. [Google Scholar] [CrossRef]
- An, G.; Watson, B.D.; Chang, C.C. Transformation of tobacco, tomato, potato, and Arabidopsis thaliana using a binary Ti vector system. Plant Physiol. 1986, 81, 301–305. [Google Scholar] [CrossRef] [Green Version]
- Niu, C.-F.; Wei, W.; Zhou, Q.-Y.; Tian, A.; Hao, Y.-J.; Zhang, W.-K.; Ma, B.; Lin, Q.; Zhang, Z.-B.; Zhang, J.-S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef]
- Jaffar, M.A.; Song, A.; Faheem, M.; Chen, S.; Jiang, J.; Liu, C.; Fan, Q.; Chen, F. Involvement of CmWRKY10 in Drought Tolerance of Chrysanthemum through the ABA-Signaling Pathway. Int. J. Mol. Sci. 2016, 17, 693. [Google Scholar] [CrossRef]
- Jia, H.; Wang, C.; Wang, F.; Liu, S.; Li, G.; Guo, X. GhWRKY68 Reduces Resistance to Salt and Drought in Transgenic Nicotiana benthamiana. PLoS ONE 2015, 10, e0120646. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Xu, T.; Han, J.; Liu, W.; Wang, Y.; Li, X.; Sun, X.; Wang, X.; Li, T.; Yang, G. Overexpression of MxWRKY53 increased iron and high salinity stress tolerance in Arabidopsis thaliana. In Vitro Cell. Dev. Biol. 2021, 58, 266–278. [Google Scholar] [CrossRef]
- Liu, W.; Wu, T.; Li, Q.; Zhang, X.; Xu, X.; Li, T.; Han, Z.; Wang, Y. An ethylene response factor (MxERF4) functions as a repressor of Fe acquisition in Malus xiaojinensis. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Chen, M.; Li, L.; Ma, Y. Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef]
- Pan, Y.; Wu, L.J.; Yu, Z.L. Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul. 2006, 49, 157–165. [Google Scholar] [CrossRef]
- Ranieri, A.; Petacco, F.; Castagna, A.; Soldatini, G.F. Redox state and peroxidase system in sunflower plants exposed to ozone. Plant Sci. 2000, 159, 159–167. [Google Scholar] [CrossRef]
- Zhang, L.; Xi, D.; Luo, L.; Meng, F.; Li, Y.; Wu, C.-A.; Guo, X. Cotton GhMPK2 is involved in multiple signaling pathways and mediates defense responses to pathogen infection and oxidative stress. FEBS J. 2011, 278, 1367–1378. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 TF functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar]
- Liu, Y. Determination of hydrogen peroxide content in white fungus by spectrophotometry. Chem. Eng. Res. Des. 2019, 2, 139–140. [Google Scholar]
- Wang, C.; Deng, P.; Chen, L.; Wang, X.; Ma, H.; Hu, W.; Yao, N.; Feng, Y.; Chai, R.; Yang, G.; et al. A Wheat WRKY Transcription Factor TaWRKY10 Confers Tolerance to Multiple Abiotic Stresses in Transgenic Tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, X.; Li, Y.; Yao, A.; Liu, W.; Yang, T.; Zhao, M.; Zhang, B.; Han, D. Overexpression of MxbHLH18 Increased Iron and High Salinity Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 8007. https://doi.org/10.3390/ijms23148007
Liang X, Li Y, Yao A, Liu W, Yang T, Zhao M, Zhang B, Han D. Overexpression of MxbHLH18 Increased Iron and High Salinity Stress Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(14):8007. https://doi.org/10.3390/ijms23148007
Chicago/Turabian StyleLiang, Xiaoqi, Yingmei Li, Anqi Yao, Wanda Liu, Tianyu Yang, Mengfei Zhao, Bingxiu Zhang, and Deguo Han. 2022. "Overexpression of MxbHLH18 Increased Iron and High Salinity Stress Tolerance in Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 14: 8007. https://doi.org/10.3390/ijms23148007





