Alternatively Spliced Isoforms of the P2X7 Receptor: Structure, Function and Disease Associations
Abstract
1. Introduction
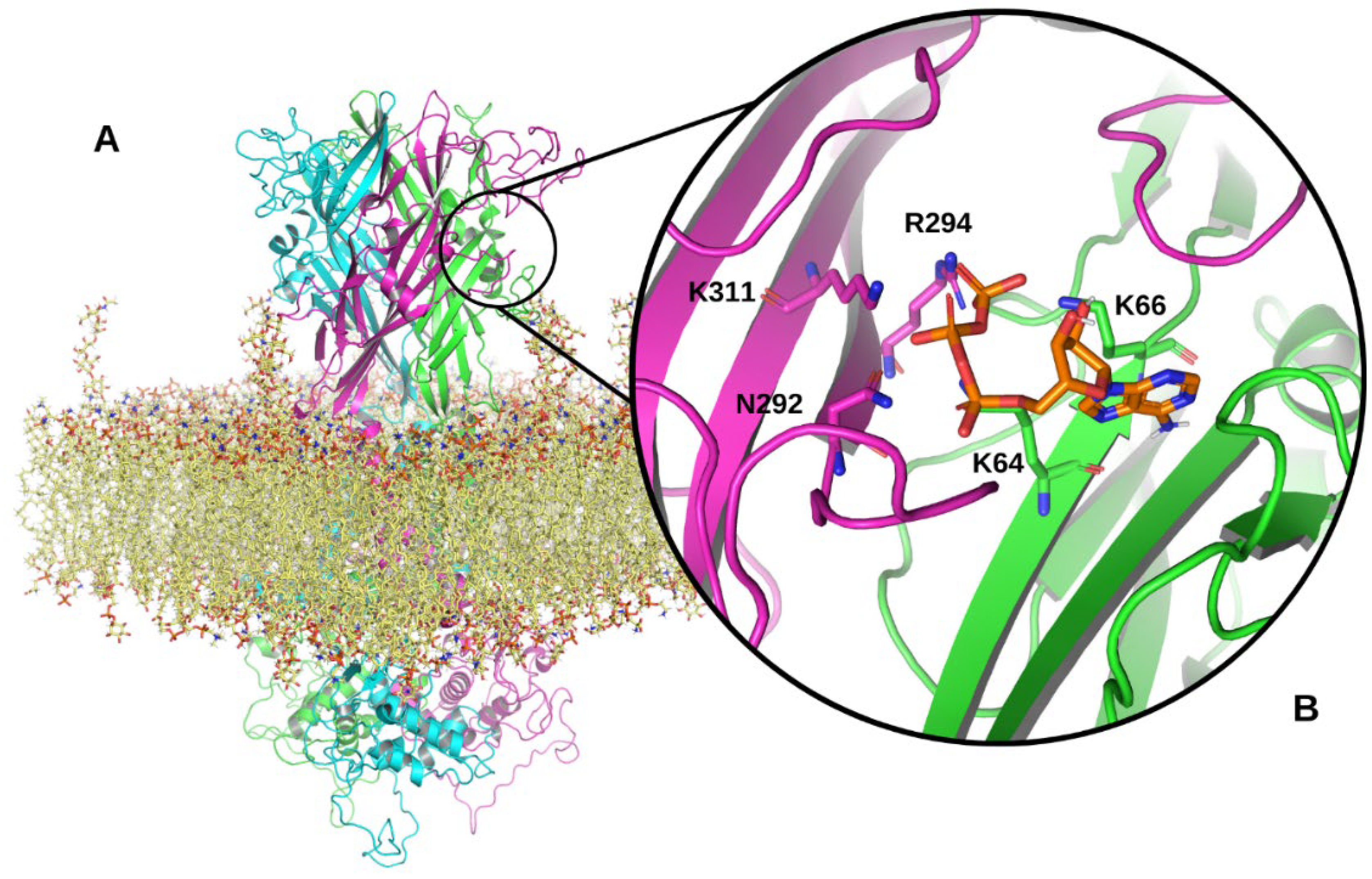
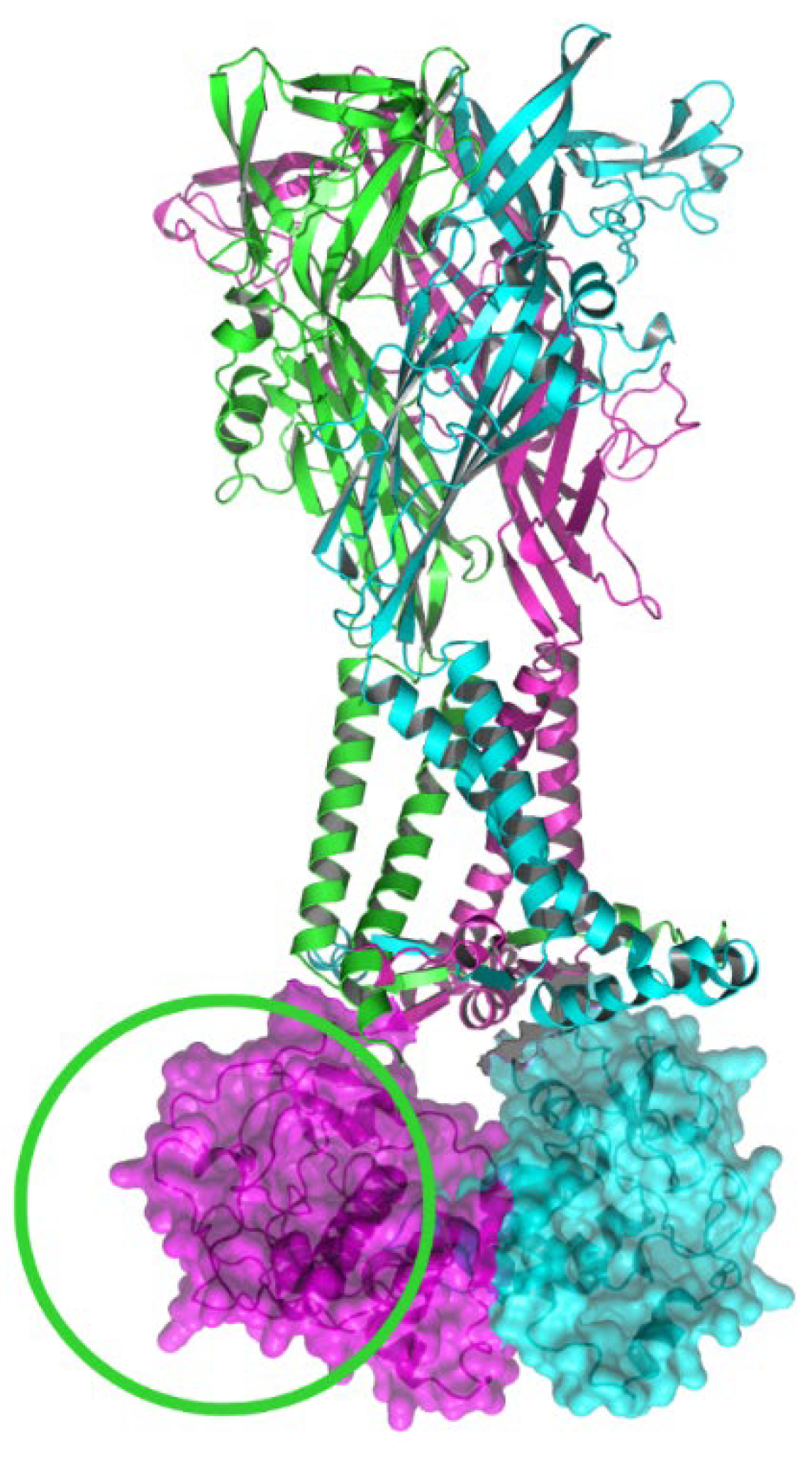
2. Structure of the P2X7R
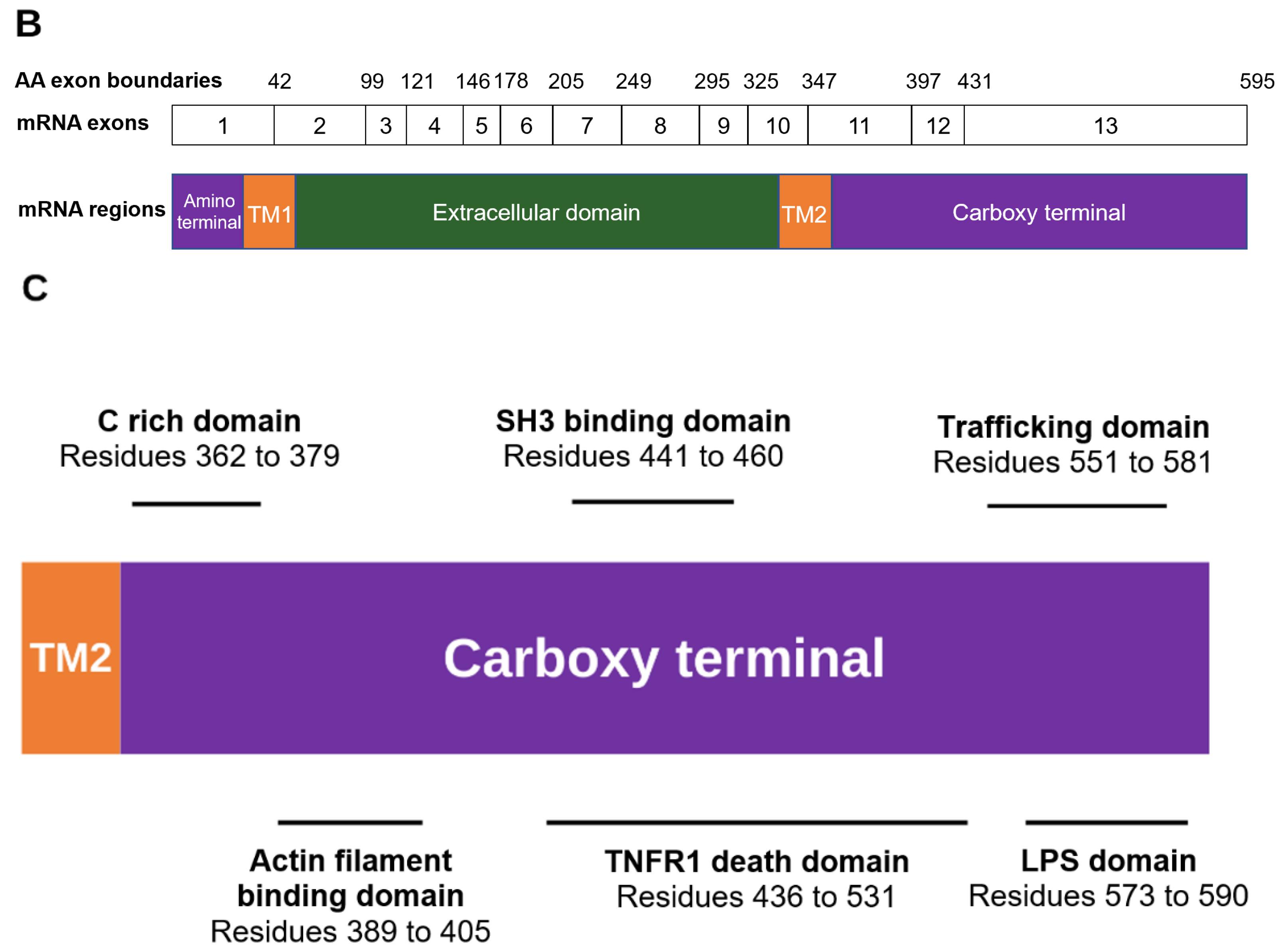
3. P2X7R Domains
3.1. Amino Terminal
3.2. Extracellular Domain
3.3. Carboxy Terminal
4. Alternative Splicing

5. Inherited Variation in P2RX7 mRNA Splice Sites
6. Role of P2X7R Isoforms in Cancer
6.1. Adenocarcinoma of the Lung
6.2. Neuroblastoma
6.3. Osteosarcoma
6.4. Cervical Cancer
6.5. Melanoma
6.6. Glioblastoma Multiforme
6.7. Leukaemia
7. Huntington’s Disease
8. Inflammation
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Chessell, I.P.; Simon, J.; Hibell, A.D.; Michel, A.D.; Barnard, E.A.; Humphrey, P.P. Cloning and functional characterisation of the mouse P2X7 receptor. FEBS Lett. 1998, 439, 26–30. [Google Scholar] [CrossRef]
- Agboh, K.C.; Webb, T.E.; Evans, R.J.; Ennion, S.J. Functional characterization of a P2X receptor from Schistosoma mansoni. J. Biol. Chem. 2004, 279, 41650–41657. [Google Scholar] [CrossRef] [PubMed]
- Bo, X.; Schoepfer, R.; Burnstock, G. Molecular cloning and characterization of a novel ATP P2X receptor subtype from embryonic chick skeletal muscle. J. Biol. Chem. 2000, 275, 14401–14407. [Google Scholar] [CrossRef] [PubMed]
- Boue-Grabot, E.; Akimenko, M.A.; Seguela, P. Unique functional properties of a sensory neuronal P2X ATP-gated channel from zebrafish. J. Neurochem. 2000, 75, 1600–1607. [Google Scholar] [CrossRef]
- Fountain, S.J.; Parkinson, K.; Young, M.T.; Cao, L.; Thompson, C.R.; North, R.A. An intracellular P2X receptor required for osmoregulation in Dictyostelium discoideum. Nature 2007, 448, 200–203. [Google Scholar] [CrossRef]
- Skarratt, K.K.; Gu, B.J.; Lovelace, M.D.; Milligan, C.J.; Stokes, L.; Glover, R.; Petrou, S.; Wiley, J.S.; Fuller, S.J. A P2RX7 single nucleotide polymorphism haplotype promotes exon 7 and 8 skipping and disrupts receptor function. FASEB J. 2020, 34, 3884–3901. [Google Scholar] [CrossRef]
- Burnstock, G.; Knight, G.E. Cellular distribution and functions of P2 receptor subtypes in different systems. Int. Rev. Cytol. 2004, 240, 31–304. [Google Scholar] [CrossRef]
- Solle, M.; Labasi, J.; Perregaux, D.G.; Stam, E.; Petrushova, N.; Koller, B.H.; Griffiths, R.J.; Gabel, C.A. Altered cytokine production in mice lacking P2X(7) receptors. J. Biol. Chem. 2001, 276, 125–132. [Google Scholar] [CrossRef]
- Chessell, I.P.; Hatcher, J.P.; Bountra, C.; Michel, A.D.; Hughes, J.P.; Green, P.; Egerton, J.; Murfin, M.; Richardson, J.; Peck, W.L.; et al. Disruption of the P2X7 purinoceptor gene abolishes chronic inflammatory and neuropathic pain. Pain 2005, 114, 386–396. [Google Scholar] [CrossRef]
- Basso, A.M.; Bratcher, N.A.; Harris, R.R.; Jarvis, M.F.; Decker, M.W.; Rueter, L.E. Behavioral profile of P2X7 receptor knockout mice in animal models of depression and anxiety: Relevance for neuropsychiatric disorders. Behav. Brain Res. 2009, 198, 83–90. [Google Scholar] [CrossRef]
- Ke, H.Z.; Qi, H.; Weidema, A.F.; Zhang, Q.; Panupinthu, N.; Crawford, D.T.; Grasser, W.A.; Paralkar, V.M.; Li, M.; Audoly, L.P.; et al. Deletion of the P2X7 nucleotide receptor reveals its regulatory roles in bone formation and resorption. Mol. Endocrinol. 2003, 17, 1356–1367. [Google Scholar] [CrossRef]
- Notomi, S.; Hisatomi, T.; Kanemaru, T.; Takeda, A.; Ikeda, Y.; Enaida, H.; Kroemer, G.; Ishibashi, T. Critical involvement of extracellular ATP acting on P2RX7 purinergic receptors in photoreceptor cell death. Am. J. Pathol. 2011, 179, 2798–2809. [Google Scholar] [CrossRef]
- Sluyter, R.; Stokes, L. Significance of P2X7 receptor variants to human health and disease. Recent Pat. Gene Seq. 2011, 5, 41–54. [Google Scholar] [CrossRef]
- Wiley, J.S.; Sluyter, R.; Gu, B.J.; Stokes, L.; Fuller, S.J. The human P2X7 receptor and its role in innate immunity. Tissue Antigens 2011, 78, 321–332. [Google Scholar] [CrossRef]
- Gu, B.J.; Wiley, J.S. P2X7 as a scavenger receptor for innate phagocytosis in the brain. Br. J. Pharm. 2018, 175, 4195–4208. [Google Scholar] [CrossRef]
- Jiang, L.H.; Caseley, E.A.; Muench, S.P.; Roger, S. Structural basis for the functional properties of the P2X7 receptor for extracellular ATP. Purinergic Signal. 2021, 17, 331–344. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Mazziotta, C.; Lanzillotti, C.; Stefani, C.; Badiale, G.; Campione, G.; Martini, F.; Tognon, M. The Role of Purinergic P2X7 Receptor in Inflammation and Cancer: Novel Molecular Insights and Clinical Applications. Cancers 2022, 14, 1116. [Google Scholar] [CrossRef]
- Kasuya, G.; Yamaura, T.; Ma, X.B.; Nakamura, R.; Takemoto, M.; Nagumo, H.; Tanaka, E.; Dohmae, N.; Nakane, T.; Yu, Y.; et al. Structural insights into the competitive inhibition of the ATP-gated P2X receptor channel. Nat. Commun. 2017, 8, 876. [Google Scholar] [CrossRef]
- Karasawa, A.; Kawate, T. Structural basis for subtype-specific inhibition of the P2X7 receptor. eLife 2016, 5, e22153. [Google Scholar] [CrossRef]
- Hattori, M.; Gouaux, E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature 2012, 485, 207–212. [Google Scholar] [CrossRef]
- Kawate, T.; Michel, J.C.; Birdsong, W.T.; Gouaux, E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature 2009, 460, 592–598. [Google Scholar] [CrossRef]
- McCarthy, A.E.; Yoshioka, C.; Mansoor, S.E. Full-Length P2X7 Structures Reveal How Palmitoylation Prevents Channel Desensitization. Cell 2019, 179, 659–670.e613. [Google Scholar] [CrossRef]
- Schrödinger, L.; DeLano, W. PyMOL. 2020. Available online: http://www.pymol.org/pymol (accessed on 22 June 2022).
- Buell, G.N.; Talabot, F.; Gos, A.; Lorenz, J.; Lai, E.; Morris, M.A.; Antonarakis, S.E. Gene structure and chromosomal localization of the human P2X7 receptor. Recept. Channels 1998, 5, 347–354. [Google Scholar]
- Newbolt, A.; Stoop, R.; Virginio, C.; Surprenant, A.; North, R.A.; Buell, G.; Rassendren, F. Membrane topology of an ATP-gated ion channel (P2X receptor). J. Biol. Chem. 1998, 273, 15177–15182. [Google Scholar] [CrossRef]
- Torres, G.E.; Egan, T.M.; Voigt, M.M. Topological analysis of the ATP-gated ionotropic [correction of ionotrophic] P2X2 receptor subunit. FEBS Lett. 1998, 425, 19–23. [Google Scholar] [CrossRef][Green Version]
- Rassendren, F.; Buell, G.; Newbolt, A.; North, R.A.; Surprenant, A. Identification of amino acid residues contributing to the pore of a P2X receptor. EMBO J. 1997, 16, 3446–3454. [Google Scholar] [CrossRef]
- Egan, T.M.; Haines, W.R.; Voigt, M.M. A domain contributing to the ion channel of ATP-gated P2X2 receptors identified by the substituted cysteine accessibility method. J. Neurosci. 1998, 18, 2350–2359. [Google Scholar] [CrossRef]
- Khakh, B.S.; Bao, X.R.; Labarca, C.; Lester, H.A. Neuronal P2X transmitter-gated cation channels change their ion selectivity in seconds. Nat. Neurosci. 1999, 2, 322–330. [Google Scholar] [CrossRef]
- Nicke, A.; Baumert, H.G.; Rettinger, J.; Eichele, A.; Lambrecht, G.; Mutschler, E.; Schmalzing, G. P2X1 and P2X3 receptors form stable trimers: A novel structural motif of ligand-gated ion channels. Embo J. 1998, 17, 3016–3028. [Google Scholar] [CrossRef]
- Rassendren, F.; Buell, G.N.; Virginio, C.; Collo, G.; North, R.A.; Surprenant, A. The permeabilizing ATP receptor, P2X7. Cloning and expression of a human cDNA. J. Biol. Chem. 1997, 272, 5482–5486. [Google Scholar] [CrossRef]
- Steinberg, T.H.; Newman, A.S.; Swanson, J.A.; Silverstein, S.C. ATP4- permeabilizes the plasma membrane of mouse macrophages to fluorescent dyes. J. Biol. Chem. 1987, 262, 8884–8888. [Google Scholar] [CrossRef]
- Virginio, C.; MacKenzie, A.; North, R.A.; Surprenant, A. Kinetics of cell lysis, dye uptake and permeability changes in cells expressing the rat P2X7 receptor. J. Physiol. 1999, 519 Pt 2, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Riedel, T.; Schmalzing, G.; Markwardt, F. Influence of extracellular monovalent cations on pore and gating properties of P2X7 receptor-operated single-channel currents. Biophys. J. 2007, 93, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Toombes, G.E.; Silberberg, S.D.; Swartz, K.J. Physical basis of apparent pore dilation of ATP-activated P2X receptor channels. Nat. Neurosci. 2015, 18, 1577–1583. [Google Scholar] [CrossRef]
- Pippel, A.; Stolz, M.; Woltersdorf, R.; Kless, A.; Schmalzing, G.; Markwardt, F. Localization of the gate and selectivity filter of the full-length P2X7 receptor. Proc. Natl. Acad. Sci. USA 2017, 114, E2156–E2165. [Google Scholar] [CrossRef]
- Browne, L.E.; Compan, V.; Bragg, L.; North, R.A. P2X7 receptor channels allow direct permeation of nanometer-sized dyes. J. Neurosci. 2013, 33, 3557–3566. [Google Scholar] [CrossRef]
- Chaumont, S.; Khakh, B.S. Patch-clamp coordinated spectroscopy shows P2X2 receptor permeability dynamics require cytosolic domain rearrangements but not Panx-1 channels. Proc. Natl. Acad. Sci. USA 2008, 105, 12063–12068. [Google Scholar] [CrossRef]
- Pelegrin, P.; Surprenant, A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 2006, 25, 5071–5082. [Google Scholar] [CrossRef]
- Mariathasan, S.; Weiss, D.S.; Newton, K.; McBride, J.; O’Rourke, K.; Roose-Girma, M.; Lee, W.P.; Weinrauch, Y.; Monack, D.M.; Dixit, V.M. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 2006, 440, 228–232. [Google Scholar] [CrossRef]
- Garbers, C.; Janner, N.; Chalaris, A.; Moss, M.L.; Floss, D.M.; Meyer, D.; Koch-Nolte, F.; Rose-John, S.; Scheller, J. Species specificity of ADAM10 and ADAM17 proteins in interleukin-6 (IL-6) trans-signaling and novel role of ADAM10 in inducible IL-6 receptor shedding. J. Biol. Chem. 2011, 286, 14804–14811. [Google Scholar] [CrossRef]
- Gu, B.J.; Wiley, J.S. Rapid ATP-induced release of matrix metalloproteinase 9 is mediated by the P2X7 receptor. Blood 2006, 107, 4946–4953. [Google Scholar] [CrossRef]
- Gu, B.J.; Saunders, B.M.; Jursik, C.; Wiley, J.S. The P2X7-nonmuscle myosin membrane complex regulates phagocytosis of nonopsonized particles and bacteria by a pathway attenuated by extracellular ATP. Blood 2010, 115, 1621–1631. [Google Scholar] [CrossRef]
- Boué-Grabot, É.; Archambault, V.; Séguéla, P. A Protein Kinase C Site Highly Conserved in P2X Subunits Controls the Desensitization Kinetics of P2X2 ATP-gated Channels*. J. Biol. Chem. 2000, 275, 10190–10195. [Google Scholar] [CrossRef]
- Allsopp, R.C.; Evans, R.J. The Intracellular Amino Terminus Plays a Dominant Role in Desensitization of ATP-gated P2X Receptor Ion Channels*. J. Biol. Chem. 2011, 286, 44691–44701. [Google Scholar] [CrossRef]
- Habermacher, C.; Dunning, K.; Chataigneau, T.; Grutter, T. Molecular structure and function of P2X receptors. Neuropharmacology 2016, 104, 18–30. [Google Scholar] [CrossRef]
- North, R.A. Molecular physiology of P2X receptors. Physiol. Rev. 2002, 82, 1013–1067. [Google Scholar] [CrossRef]
- Worthington, R.A.; Smart, M.L.; Gu, B.J.; Williams, D.A.; Petrou, S.; Wiley, J.S.; Barden, J.A. Point mutations confer loss of ATP-induced human P2X(7) receptor function. FEBS Lett. 2002, 512, 43–46. [Google Scholar] [CrossRef]
- Bin Dayel, A.; Evans, R.J.; Schmid, R. Mapping the Site of Action of Human P2X7 Receptor Antagonists AZ11645373, Brilliant Blue G, KN-62, Calmidazolium, and ZINC58368839 to the Intersubunit Allosteric Pocket. Mol. Pharmacol. 2019, 96, 355–363. [Google Scholar] [CrossRef]
- Allsopp, R.C.; Dayl, S.; Bin Dayel, A.; Schmid, R.; Evans, R.J. Mapping the Allosteric Action of Antagonists A740003 and A438079 Reveals a Role for the Left Flipper in Ligand Sensitivity at P2X7 Receptors. Mol. Pharmacol. 2018, 93, 553–562. [Google Scholar] [CrossRef]
- Surprenant, A.; Rassendren, F.; Kawashima, E.; North, R.A.; Buell, G. The cytolytic P2Z receptor for extracellular ATP identified as a P2X receptor (P2X7). Science 1996, 272, 735–738. [Google Scholar] [CrossRef]
- Adinolfi, E.; Cirillo, M.; Woltersdorf, R.; Falzoni, S.; Chiozzi, P.; Pellegatti, P.; Callegari, M.G.; Sandona, D.; Markwardt, F.; Schmalzing, G.; et al. Trophic activity of a naturally occurring truncated isoform of the P2X7 receptor. FASEB J. 2010, 24, 3393–3404. [Google Scholar] [CrossRef]
- Gu, B.J.; Zhang, W.; Worthington, R.A.; Sluyter, R.; Dao-Ung, P.; Petrou, S.; Barden, J.A.; Wiley, J.S. A Glu-496 to Ala polymorphism leads to loss of function of the human P2X7 receptor. J. Biol. Chem. 2001, 276, 11135–11142. [Google Scholar] [CrossRef]
- El-Moatassim, C.; Dubyak, G.R. A novel pathway for the activation of phospholipase D by P2z purinergic receptors in BAC1.2F5 macrophages. J. Biol. Chem. 1992, 267, 23664–23673. [Google Scholar] [CrossRef]
- Denlinger, L.C.; Fisette, P.L.; Sommer, J.A.; Watters, J.J.; Prabhu, U.; Dubyak, G.R.; Proctor, R.A.; Bertics, P.J. Cutting edge: The nucleotide receptor P2X7 contains multiple protein- and lipid-interaction motifs including a potential binding site for bacterial lipopolysaccharide. J. Immunol. 2001, 167, 1871–1876. [Google Scholar] [CrossRef]
- Gibson, T.J. Cell regulation: Determined to signal discrete cooperation. Trends Biochem. Sci. 2009, 34, 471–482. [Google Scholar] [CrossRef]
- Adinolfi, E.; Raffaghello, L.; Giuliani, A.L.; Cavazzini, L.; Capece, M.; Chiozzi, P.; Bianchi, G.; Kroemer, G.; Pistoia, V.; Di Virgilio, F. Expression of P2X7 receptor increases in vivo tumor growth. Cancer Res. 2012, 72, 2957–2969. [Google Scholar] [CrossRef]
- Pfeiffer, Z.A.; Aga, M.; Prabhu, U.; Watters, J.J.; Hall, D.J.; Bertics, P.J. The nucleotide receptor P2X7 mediates actin reorganization and membrane blebbing in RAW 264.7 macrophages via p38 MAP kinase and Rho. J. Leukoc. Biol. 2004, 75, 1173–1182. [Google Scholar] [CrossRef]
- Hsu, H.; Xiong, J.; Goeddel, D.V. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell 1995, 81, 495–504. [Google Scholar] [CrossRef]
- Park, Y.H.; Jeong, M.S.; Jang, S.B. Death domain complex of the TNFR-1, TRADD, and RIP1 proteins for death-inducing signaling. Biochem. Biophys. Res. Commun. 2014, 443, 1155–1161. [Google Scholar] [CrossRef]
- Dhanasekaran, D.N.; Reddy, E.P. JNK signaling in apoptosis. Oncogene 2008, 27, 6245–6251. [Google Scholar] [CrossRef]
- Hsu, H.; Huang, J.; Shu, H.-B.; Baichwal, V.; Goeddel, D.V. TNF-Dependent Recruitment of the Protein Kinase RIP to the TNF Receptor-1 Signaling Complex. Immunity 1996, 4, 387–396. [Google Scholar] [CrossRef]
- Cunha, S.R.; Mohler, P.J. Ankyrin protein networks in membrane formation and stabilization. J. Cell. Mol. Med. 2009, 13, 4364–4376. [Google Scholar] [CrossRef] [PubMed]
- Smart, M.L.; Gu, B.; Panchal, R.G.; Wiley, J.; Cromer, B.; Williams, D.A.; Petrou, S. P2X7 receptor cell surface expression and cytolytic pore formation are regulated by a distal C-terminal region. J. Biol. Chem. 2003, 278, 8853–8860. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, I.; Schönefeld, M.; Aichele, D.; Groer, G.; Gessner, A.; Schnare, M. Murine Bactericidal/Permeability-Increasing Protein Inhibits the Endotoxic Activity of Lipopolysaccharide and Gram-Negative Bacteria. J. Immunol. 2008, 180, 7546–7552. [Google Scholar] [CrossRef]
- Gonnord, P.; Delarasse, C.; Auger, R.; Benihoud, K.; Prigent, M.; Cuif, M.H.; Lamaze, C.; Kanellopoulos, J.M. Palmitoylation of the P2X7 receptor, an ATP-gated channel, controls its expression and association with lipid rafts. FASEB J. 2009, 23, 795–805. [Google Scholar] [CrossRef]
- Karasawa, A.; Michalski, K.; Mikhelzon, P.; Kawate, T. The P2X7 receptor forms a dye-permeable pore independent of its intracellular domain but dependent on membrane lipid composition. eLife 2017, 6, e31186. [Google Scholar] [CrossRef]
- Robinson, L.E.; Shridar, M.; Smith, P.; Murrell-Lagnado, R.D. Plasma membrane cholesterol as a regulator of human and rodent P2X7 receptor activation and sensitization. J. Biol. Chem. 2014, 289, 31983–31994. [Google Scholar] [CrossRef]
- Cheewatrakoolpong, B.; Gilchrest, H.; Anthes, J.C.; Greenfeder, S. Identification and characterization of splice variants of the human P2X7 ATP channel. Biochem. Biophys. Res. Commun. 2005, 332, 17–27. [Google Scholar] [CrossRef]
- Costa-Junior, H.M.; Sarmento Vieira, F.; Coutinho-Silva, R. C terminus of the P2X7 receptor: Treasure hunting. Purinergic Signal. 2011, 7, 7–19. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Furlanis, E.; Scheiffele, P. Regulation of Neuronal Differentiation, Function, and Plasticity by Alternative Splicing. Annu. Rev. Cell Dev. Biol. 2018, 34, 451–469. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Hsiao, Y.-H.E.; Bahn, J.H.; Lin, X.; Chan, T.-M.; Wang, R.; Xiao, X. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Res. 2016, 26, 440–450. [Google Scholar] [CrossRef]
- Fuller, S.J.; Stokes, L.; Skarratt, K.K.; Gu, B.J.; Wiley, J.S. Genetics of the P2X7 receptor and human disease. Purinergic Signal. 2009, 5, 257–262. [Google Scholar] [CrossRef]
- Stokes, L.; Fuller, S.J.; Sluyter, R.; Skarratt, K.K.; Gu, B.J.; Wiley, J.S. Two haplotypes of the P2X(7) receptor containing the Ala-348 to Thr polymorphism exhibit a gain-of-function effect and enhanced interleukin-1beta secretion. FASEB J. 2010, 24, 2916–2927. [Google Scholar] [CrossRef]
- Sun, C.; Chu, J.; Singh, S.; Salter, R.D. Identification and characterization of a novel variant of the human P2X(7) receptor resulting in gain of function. Purinergic Signal. 2010, 6, 31–45. [Google Scholar] [CrossRef]
- Feng, Y.H.; Li, X.; Wang, L.; Zhou, L.; Gorodeski, G.I. A truncated P2X7 receptor variant (P2X7-j) endogenously expressed in cervical cancer cells antagonizes the full-length P2X7 receptor through hetero-oligomerization. J. Biol. Chem. 2006, 281, 17228–17237. [Google Scholar] [CrossRef]
- Skarratt, K.K.; Fuller, S.J.; Sluyter, R.; Dao-Ung, L.P.; Gu, B.J.; Wiley, J.S. A 5′ intronic splice site polymorphism leads to a null allele of the P2X7 gene in 1-2% of the Caucasian population. FEBS Lett. 2005, 579, 2675–2678. [Google Scholar] [CrossRef]
- Nicke, A.; Kuan, Y.H.; Masin, M.; Rettinger, J.; Marquez-Klaka, B.; Bender, O.; Gorecki, D.C.; Murrell-Lagnado, R.D.; Soto, F. A functional P2X7 splice variant with an alternative transmembrane domain 1 escapes gene inactivation in P2X7 knock-out mice. J. Biol. Chem. 2009, 284, 25813–25822. [Google Scholar] [CrossRef]
- Pan, H.; Ni, H.; Zhang, L.; Xing, Y.; Fan, J.; Li, P.; Li, T.; Jia, R.; Ge, S.; Zhang, H.; et al. P2RX7-V3 is a novel oncogene that promotes tumorigenesis in uveal melanoma. Tumour Biol. 2016, 37, 13533–13543. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Hu, X.; Feng, Y.; Zhang, D.; Zhao, S.D.; Hu, Z.; Greshock, J.; Zhang, Y.; Yang, L.; Zhong, X.; Wang, L.P.; et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 2014, 26, 344–357. [Google Scholar] [CrossRef]
- Masin, M.; Young, C.; Lim, K.; Barnes, S.J.; Xu, X.J.; Marschall, V.; Brutkowski, W.; Mooney, E.R.; Gorecki, D.C.; Murrell-Lagnado, R. Expression, assembly and function of novel C-terminal truncated variants of the mouse P2X7 receptor: Re-evaluation of P2X7 knockouts. Br. J. Pharmacol. 2012, 165, 978–993. [Google Scholar] [CrossRef]
- Xu, X.J.; Boumechache, M.; Robinson, L.E.; Marschall, V.; Gorecki, D.C.; Masin, M.; Murrell-Lagnado, R.D. Splice variants of the P2X7 receptor reveal differential agonist dependence and functional coupling with pannexin-1. J. Cell Sci. 2012, 125, 3776–3789. [Google Scholar] [CrossRef]
- Truty, R.; Ouyang, K.; Rojahn, S.; Garcia, S.; Colavin, A.; Hamlington, B.; Freivogel, M.; Nussbaum, R.L.; Nykamp, K.; Aradhya, S. Spectrum of splicing variants in disease genes and the ability of RNA analysis to reduce uncertainty in clinical interpretation. Am. J. Hum. Genet. 2021, 108, 696–708. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Misquitta, C.; Zhang, W.; Saltzman, A.L.; Mohammad, N.; Babak, T.; Siu, H.; Hughes, T.R.; Morris, Q.D.; et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell 2004, 16, 929–941. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Královicová, J.; Lei, H.; Vorechovský, I. Phenotypic consequences of branch point substitutions. Hum. Mutat. 2006, 27, 803–813. [Google Scholar] [CrossRef]
- Cartegni, L.; Chew, S.L.; Krainer, A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002, 3, 285–298. [Google Scholar] [CrossRef]
- Nembaware, V.; Lupindo, B.; Schouest, K.; Spillane, C.; Scheffler, K.; Seoighe, C. Genome-wide survey of allele-specific splicing in humans. BMC Genom. 2008, 9, 265. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nembaware, V.; Wolfe, K.H.; Bettoni, F.; Kelso, J.; Seoighe, C. Allele-specific transcript isoforms in human. FEBS Lett. 2004, 577, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.G.; Muniz-Medina, V.; Shahlavi, T.; Baker, C.C.; Inui, H.; Ueda, T.; Emmert, S.; Schneider, T.D.; Kraemer, K.H. The human XPC DNA repair gene: Arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res. 2002, 30, 3624–3631. [Google Scholar] [CrossRef] [PubMed]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 2009, 106, 9362–9367. [Google Scholar] [CrossRef]
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef]
- Pellegatti, P.; Raffaghello, L.; Bianchi, G.; Piccardi, F.; Pistoia, V.; Di Virgilio, F. Increased level of extracellular ATP at tumor sites: In vivo imaging with plasma membrane luciferase. PLoS ONE 2008, 3, e2599. [Google Scholar] [CrossRef]
- Rodriguez-Canales, J.; Parra-Cuentas, E.; Wistuba, I.I. Diagnosis and Molecular Classification of Lung Cancer. Cancer Treat. Res. 2016, 170, 25–46. [Google Scholar] [CrossRef]
- Boldrini, L.; Giordano, M.; Alì, G.; Servadio, A.; Pelliccioni, S.; Niccoli, C.; Mussi, A.; Fontanini, G. P2X7 protein expression and polymorphism in non-small cell lung cancer (NSCLC). J. Negat. Results Biomed. 2014, 13, 16. [Google Scholar] [CrossRef]
- Benzaquen, J.; Dit Hreich, S.J.; Heeke, S.; Juhel, T.; Lalvee, S.; Bauwens, S.; Saccani, S.; Lenormand, P.; Hofman, V.; Butori, M.; et al. P2RX7B is a new theranostic marker for lung adenocarcinoma patients. Theranostics 2020, 10, 10849–10860. [Google Scholar] [CrossRef]
- Irwin, M.S.; Naranjo, A.; Zhang, F.F.; Cohn, S.L.; London, W.B.; Gastier-Foster, J.M.; Ramirez, N.C.; Pfau, R.; Reshmi, S.; Wagner, E.; et al. Revised Neuroblastoma Risk Classification System: A Report from the Children’s Oncology Group. J. Clin. Oncol. 2021, 39, 3229–3241. [Google Scholar] [CrossRef]
- Ulrich, H.; Ratajczak, M.Z.; Schneider, G.; Adinolfi, E.; Orioli, E.; Ferrazoli, E.G.; Glaser, T.; Corrêa-Velloso, J.; Martins, P.C.M.; Coutinho, F.; et al. Kinin and Purine Signaling Contributes to Neuroblastoma Metastasis. Front. Pharmacol. 2018, 9, 500. [Google Scholar] [CrossRef]
- Jørgensen, N.R.; Henriksen, Z.; Sørensen, O.H.; Eriksen, E.F.; Civitelli, R.; Steinberg, T.H. Intercellular calcium signaling occurs between human osteoblasts and osteoclasts and requires activation of osteoclast P2X7 receptors. J. Biol. Chem. 2002, 277, 7574–7580. [Google Scholar] [CrossRef]
- Gartland, A.; Hipskind, R.A.; Gallagher, J.A.; Bowler, W.B. Expression of a P2X7 receptor by a subpopulation of human osteoblasts. J. Bone Miner. Res. 2001, 16, 846–856. [Google Scholar] [CrossRef]
- Genetos, D.C.; Kephart, C.J.; Zhang, Y.; Yellowley, C.E.; Donahue, H.J. Oscillating fluid flow activation of gap junction hemichannels induces ATP release from MLO-Y4 osteocytes. J. Cell. Physiol. 2007, 212, 207–214. [Google Scholar] [CrossRef]
- Worthley, D.L.; Churchill, M.; Compton, J.T.; Tailor, Y.; Rao, M.; Si, Y.; Levin, D.; Schwartz, M.G.; Uygur, A.; Hayakawa, Y.; et al. Gremlin 1 Identifies a Skeletal Stem Cell with Bone, Cartilage, and Reticular Stromal Potential. Cell 2015, 160, 269–284. [Google Scholar] [CrossRef]
- Jafari, F.; Javdansirat, S.; Sanaie, S.; Naseri, A.; Shamekh, A.; Rostamzadeh, D.; Dolati, S. Osteosarcoma: A comprehensive review of management and treatment strategies. Ann. Diagn. Pathol. 2020, 49, 151654. [Google Scholar] [CrossRef]
- Tattersall, L.; Shah, K.M.; Lath, D.L.; Singh, A.; Down, J.M.; De Marchi, E.; Williamson, A.; Di Virgilio, F.; Heymann, D.; Adinolfi, E.; et al. The P2RX7B splice variant modulates osteosarcoma cell behaviour and metastatic properties. J. Bone Oncol. 2021, 31, 100398. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, H.; Li, W.; Wu, H.; Yang, Y. Highly-expressed P2X7 receptor promotes growth and metastasis of human HOS/MNNG osteosarcoma cells via PI3K/Akt/GSK3β/β-catenin and mTOR/HIF1α/VEGF signaling. Int. J. Cancer 2019, 145, 1068–1082. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef]
- Okunade, K.S. Human papillomavirus and cervical cancer. J. Obstet. Gynaecol. 2020, 40, 602–608. [Google Scholar] [CrossRef]
- Francies, F.Z.; Bassa, S.; Chatziioannou, A.; Andreas Martin, K.; Dlamini, Z. Splicing Genomics Events in Cervical Cancer: Insights for Phenotypic Stratification and Biomarker Potency. Genes 2021, 12, 130. [Google Scholar] [CrossRef] [PubMed]
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Barsouk, A. Epidemiology of Melanoma. Med. Sci. 2021, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- White, N.; Butler, P.E.; Burnstock, G. Human melanomas express functional P2 X(7) receptors. Cell Tissue Res. 2005, 321, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014, 20, 1531–1541. [Google Scholar] [CrossRef]
- Fan, J.; Xing, Y.; Wen, X.; Jia, R.; Ni, H.; He, J.; Ding, X.; Pan, H.; Qian, G.; Ge, S.; et al. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015, 16, 139. [Google Scholar] [CrossRef]
- Silantyev, A.S.; Falzone, L.; Libra, M.; Gurina, O.I.; Kardashova, K.S.; Nikolouzakis, T.K.; Nosyrev, A.E.; Sutton, C.W.; Mitsias, P.D.; Tsatsakis, A. Current and Future Trends on Diagnosis and Prognosis of Glioblastoma: From Molecular Biology to Proteomics. Cells 2019, 8, 863. [Google Scholar] [CrossRef]
- Khaddour, K.; Johanns, T.M.; Ansstas, G. The Landscape of Novel Therapeutics and Challenges in Glioblastoma Multiforme: Contemporary State and Future Directions. Pharmaceuticals 2020, 13, 389. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncol. 2018, 20, iv1–iv86. [Google Scholar] [CrossRef]
- Monif, M.; O’Brien, T.J.; Drummond, K.J.; Reid, C.A.; Liubinas, S.V.; Williams, D.A. P2X7 receptors are a potential novel target for anti-glioma therapies. J. Inflamm. 2014, 11, 25. [Google Scholar] [CrossRef]
- Ryu, J.K.; Jantaratnotai, N.; Serrano-Perez, M.C.; McGeer, P.L.; McLarnon, J.G. Block of purinergic P2X7R inhibits tumor growth in a C6 glioma brain tumor animal model. J. Neuropathol. Exp. Neurol. 2011, 70, 13–22. [Google Scholar] [CrossRef]
- Tamajusuku, A.S.; Villodre, E.S.; Paulus, R.; Coutinho-Silva, R.; Battasstini, A.M.; Wink, M.R.; Lenz, G. Characterization of ATP-induced cell death in the GL261 mouse glioma. J. Cell. Biochem. 2010, 109, 983–991. [Google Scholar] [CrossRef]
- Fang, J.; Chen, X.; Zhang, L.; Chen, J.; Liang, Y.; Li, X.; Xiang, J.; Wang, L.; Guo, G.; Zhang, B.; et al. P2X7R suppression promotes glioma growth through epidermal growth factor receptor signal pathway. Int. J. Biochem. Cell Biol. 2013, 45, 1109–1120. [Google Scholar] [CrossRef]
- Kan, L.K.; Seneviratne, S.; Drummond, K.J.; Williams, D.A.; O’Brien, T.J.; Monif, M. P2X7 receptor antagonism inhibits tumour growth in human high-grade gliomas. Purinergic Signal. 2020, 16, 327–336. [Google Scholar] [CrossRef]
- Allsopp, R.C.; Dayl, S.; Schmid, R.; Evans, R.J. Unique residues in the ATP gated human P2X7 receptor define a novel allosteric binding pocket for the selective antagonist AZ10606120. Sci. Rep. 2017, 7, 725. [Google Scholar] [CrossRef]
- Jiang, L.H.; Mackenzie, A.B.; North, R.A.; Surprenant, A. Brilliant blue G selectively blocks ATP-gated rat P2X(7) receptors. Mol. Pharm. 2000, 58, 82–88. [Google Scholar] [CrossRef]
- Di Virgilio, F. Novel data point to a broader mechanism of action of oxidized ATP: The P2X7 receptor is not the only target. Br. J. Pharm. 2003, 140, 441–443. [Google Scholar] [CrossRef]
- Zanoni, M.; Sarti, A.C.; Zamagni, A.; Cortesi, M.; Pignatta, S.; Arienti, C.; Tebaldi, M.; Sarnelli, A.; Romeo, A.; Bartolini, D.; et al. Irradiation causes senescence, ATP release, and P2X7 receptor isoform switch in glioblastoma. Cell Death Dis. 2022, 13, 80. [Google Scholar] [CrossRef]
- Zhang, X.J.; Zheng, G.G.; Ma, X.T.; Yang, Y.H.; Li, G.; Rao, Q.; Nie, K.; Wu, K.F. Expression of P2X7 in human hematopoietic cell lines and leukemia patients. Leuk. Res. 2004, 28, 1313–1322. [Google Scholar] [CrossRef]
- Pegoraro, A.; Orioli, E.; De Marchi, E.; Salvestrini, V.; Milani, A.; Di Virgilio, F.; Curti, A.; Adinolfi, E. Differential sensitivity of acute myeloid leukemia cells to daunorubicin depends on P2X7A versus P2X7B receptor expression. Cell Death Dis. 2020, 11, 876. [Google Scholar] [CrossRef]
- Lecciso, M.; Ocadlikova, D.; Sangaletti, S.; Trabanelli, S.; De Marchi, E.; Orioli, E.; Pegoraro, A.; Portararo, P.; Jandus, C.; Bontadini, A.; et al. ATP Release from Chemotherapy-Treated Dying Leukemia Cells Elicits an Immune Suppressive Effect by Increasing Regulatory T Cells and Tolerogenic Dendritic Cells. Front. Immunol. 2017, 8, 1918. [Google Scholar] [CrossRef]
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Olla, I.; Santos-Galindo, M.; Elorza, A.; Lucas, J.J. P2X7 Receptor Upregulation in Huntington’s Disease Brains. Front. Mol. Neurosci. 2020, 13, 567430. [Google Scholar] [CrossRef]
- Díaz-Hernández, M.; Díez-Zaera, M.; Sánchez-Nogueiro, J.; Gómez-Villafuertes, R.; Canals, J.M.; Alberch, J.; Miras-Portugal, M.T.; Lucas, J.J. Altered P2X7-receptor level and function in mouse models of Huntington’s disease and therapeutic efficacy of antagonist administration. FASEB J. 2009, 23, 1893–1906. [Google Scholar] [CrossRef]
- Mehta, V.B.; Hart, J.; Wewers, M.D. ATP-stimulated release of interleukin (IL)-1 beta and IL-18 requires priming by lipopolysaccharide and is independent of caspase-1 cleavage. J. Biol. Chem. 2001, 276, 3820–3826. [Google Scholar] [CrossRef]
- Hung, S.C.; Choi, C.H.; Said-Sadier, N.; Johnson, L.; Atanasova, K.R.; Sellami, H.; Yilmaz, Ö.; Ojcius, D.M. P2X4 assembles with P2X7 and pannexin-1 in gingival epithelial cells and modulates ATP-induced reactive oxygen species production and inflammasome activation. PLoS ONE 2013, 8, e70210. [Google Scholar] [CrossRef]
- Orioli, E.; De Marchi, E.; Giuliani, A.L.; Adinolfi, E. P2X7 Receptor Orchestrates Multiple Signalling Pathways Triggering Inflammation, Autophagy and Metabolic/Trophic Responses. Curr. Med. Chem. 2017, 24, 2261–2275. [Google Scholar] [CrossRef]
- Biswas, S.K.; Chittezhath, M.; Shalova, I.N.; Lim, J.Y. Macrophage polarization and plasticity in health and disease. Immunol. Res. 2012, 53, 11–24. [Google Scholar] [CrossRef]
- Lavin, Y.; Mortha, A.; Rahman, A.; Merad, M. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 2015, 15, 731–744. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, B.P.; Danhorn, T.; De Arras, L.; Flatley, B.R.; Marcus, R.A.; Farias-Hesson, E.; Leach, S.M.; Alper, S. Regulation of toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS Genet. 2015, 11, e1004932. [Google Scholar] [CrossRef] [PubMed]
- Alasoo, K.; Martinez, F.O.; Hale, C.; Gordon, S.; Powrie, F.; Dougan, G.; Mukhopadhyay, S.; Gaffney, D.J. Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription. Sci. Rep. 2015, 5, 12524. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef]
- Pelegrin, P.; Surprenant, A. Dynamics of macrophage polarization reveal new mechanism to inhibit IL-1beta release through pyrophosphates. EMBO J. 2009, 28, 2114–2127. [Google Scholar] [CrossRef]

| NCBI | ENSEMBL | UniPRotKB | Possible P2X7-Like Proteins with at Least 1 TMD | |||||
|---|---|---|---|---|---|---|---|---|
| Common mRNA Isoform Name | Accession Number | Other Transcripts with Corresponding Exonic Structure | Transcript Name | Accession Number | Accession Number | Number of Amino Acids | Predicted Molecular Weight (kDa) | |
| Transcript Name | Accession Number | |||||||
| A | P2X7 receptor | Y09561 | P2X7-202 | ENST00000328963.10 | Q99572-1 | 595 | 68.6 | |
| GQ1801221 | Variant 1 | NM_002562.5 a | ||||||
| B | AY847298.1 | Variant 5 | NR_033951.2 | P2X7-203 | ENST00000535250.5 | Q99572-2 | 364 | 41.8 |
| C | AY847299.1 | Variant 6 | NR_033952.2 | P2X7-212 | ENST00000541716.5 | Q99572-3 | 128 | 14.7 |
| 113 | 13.4 | |||||||
| D | AY847300.1 | Variant 8 | NR_033954.2 b | P2X7-201 | ENST0000261826.10 | J3KN30 | 149 | 17.1 |
| Q99572-4 | 425 | 49.2 | ||||||
| E | AY847301.1 | P2X7-204 | ENST00000535600.2 | Q99572-5 | 275 | 31.3 | ||
| F | AY847302.1 | Variant 10 | NR_033956.2 | P2X7-210 | ENST00000541022.5 | 128 c | 14.7 | |
| Q99572-6 | 306 | 35.5 | ||||||
| G | AY847303.1 | Variant 2 | NR_033948.2 | P2X7-208 | ENST00000539606.5 | F5H2X6 | 127 | 14.7 |
| Q99572-7 | 274 | 31.4 | ||||||
| H | AY847304.1 | Variant 3 | NR_033949.2 | P2X7-207 | ENST00000538011.5 | F5H2X6 | 127 | 14.7 |
| Q99572-8 | 505 | 58.2 | ||||||
| J | DQ399293.1 | Variant 9 | NR_033955.2 | P2X7-211 | ENST00000541564.5 | F5H8E7 | 258 | 29.3 |
| 306 d | 35.5 | |||||||
| L | MK465687.1 | Predicted variant X1 | XM_047428912.1 | 506 | 58.0 | |||
| Predicted variant X2 | XM_011538419.4 | 491 | 56.7 | |||||
| ∆E2 e | - | - | ||||||
| N | MK465688.1 | 207 | 23.5 | |||||
| 344 f | 40.1 | |||||||
| O | MK465689.1 | 213 | 24.3 | |||||
| 272 | 31.9 | |||||||
| P | MK465690.1 | 258 | 29.3 | |||||
| Q | MK465691.1 | 128 c | 14.7 | |||||
| 306 d | 35.6 | |||||||
| Variant 4 | NR_033950.2 g | P2X7-209 | ENST00000539695.5 | 148 | 17 | |||
| Variant 7 | NR_033953.2 h | P2X7-206 | ENST00000537312.5 | F5H237 | 47 i | 5.5 | ||
| 260 | 29.9 | |||||||
| P2X7-213 j | ENST00000545434.5 | F5H237 | 47 i | 5.5 | ||||
| P2X7-205 k | ENST00000535928.5 | F5H237 | 47 i | 5.5 | ||||
| Predicted Variant X3 | XM_017019367.3 | 47 i | 5.5 | |||||
| 344 f | 40.2 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Salis, S.K.F.; Li, L.; Chen, Z.; Lam, K.W.; Skarratt, K.K.; Balle, T.; Fuller, S.J. Alternatively Spliced Isoforms of the P2X7 Receptor: Structure, Function and Disease Associations. Int. J. Mol. Sci. 2022, 23, 8174. https://doi.org/10.3390/ijms23158174
De Salis SKF, Li L, Chen Z, Lam KW, Skarratt KK, Balle T, Fuller SJ. Alternatively Spliced Isoforms of the P2X7 Receptor: Structure, Function and Disease Associations. International Journal of Molecular Sciences. 2022; 23(15):8174. https://doi.org/10.3390/ijms23158174
Chicago/Turabian StyleDe Salis, Sophie K. F., Lanxin Li, Zheng Chen, Kam Wa Lam, Kristen K. Skarratt, Thomas Balle, and Stephen J. Fuller. 2022. "Alternatively Spliced Isoforms of the P2X7 Receptor: Structure, Function and Disease Associations" International Journal of Molecular Sciences 23, no. 15: 8174. https://doi.org/10.3390/ijms23158174
APA StyleDe Salis, S. K. F., Li, L., Chen, Z., Lam, K. W., Skarratt, K. K., Balle, T., & Fuller, S. J. (2022). Alternatively Spliced Isoforms of the P2X7 Receptor: Structure, Function and Disease Associations. International Journal of Molecular Sciences, 23(15), 8174. https://doi.org/10.3390/ijms23158174






