Transcriptome Analysis to Explore the Cause of the Formation of Different Inflorescences in Tomato
Abstract
:1. Introduction
2. Results
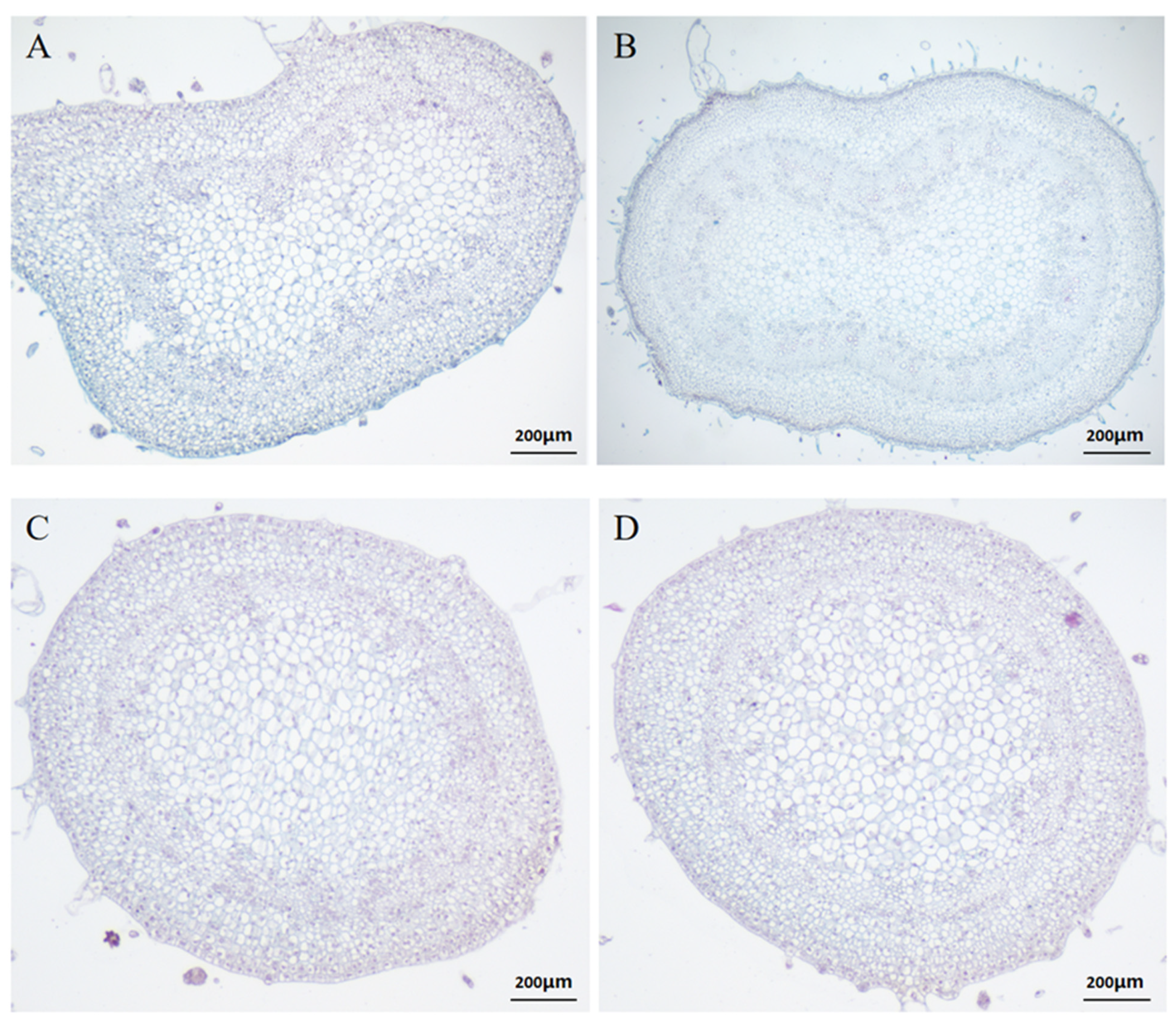
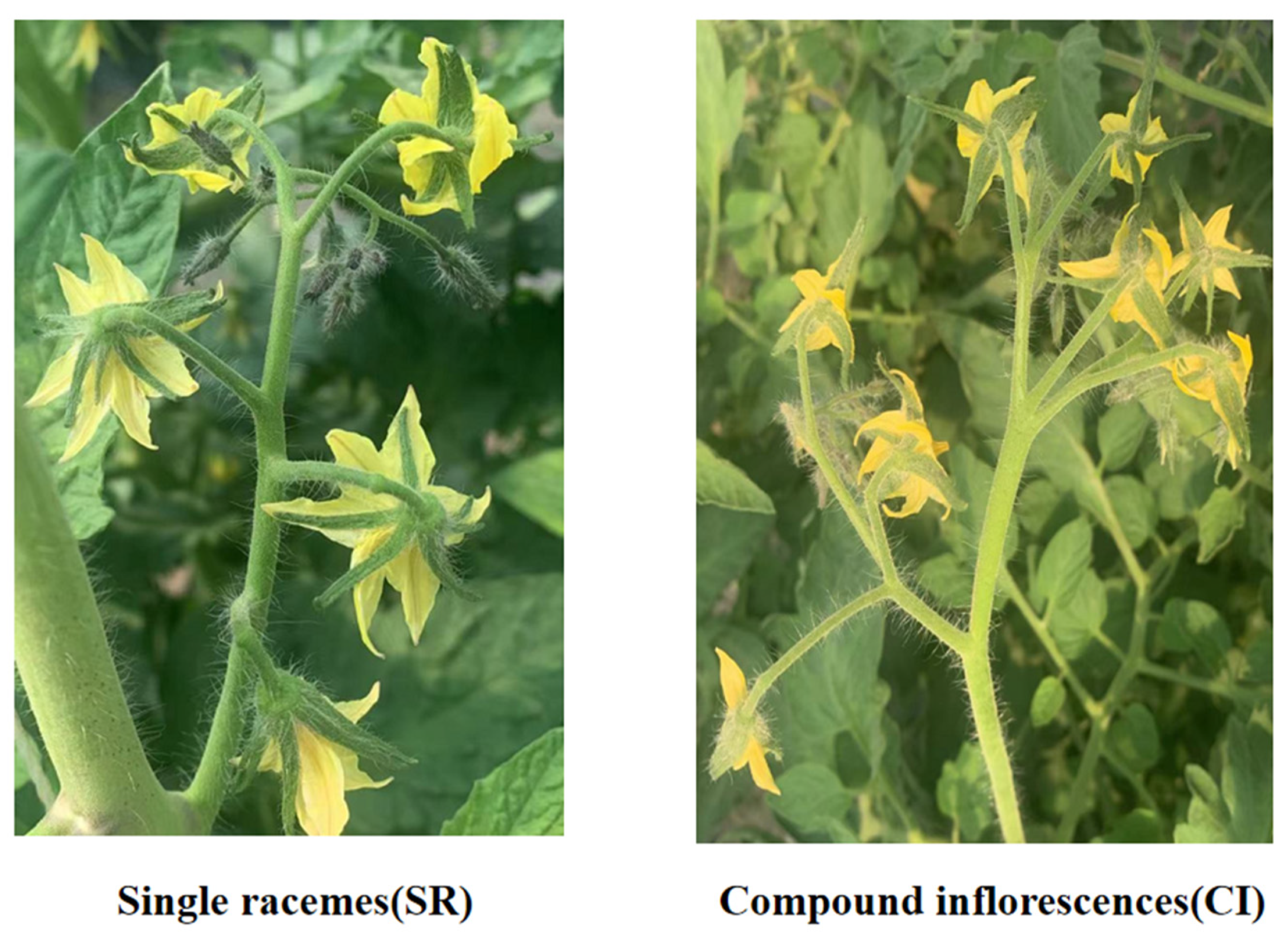
2.1. Paraffin Sectioning of Tomato Inflorescences
2.2. RNA Sequencing Data and Functional Analysis
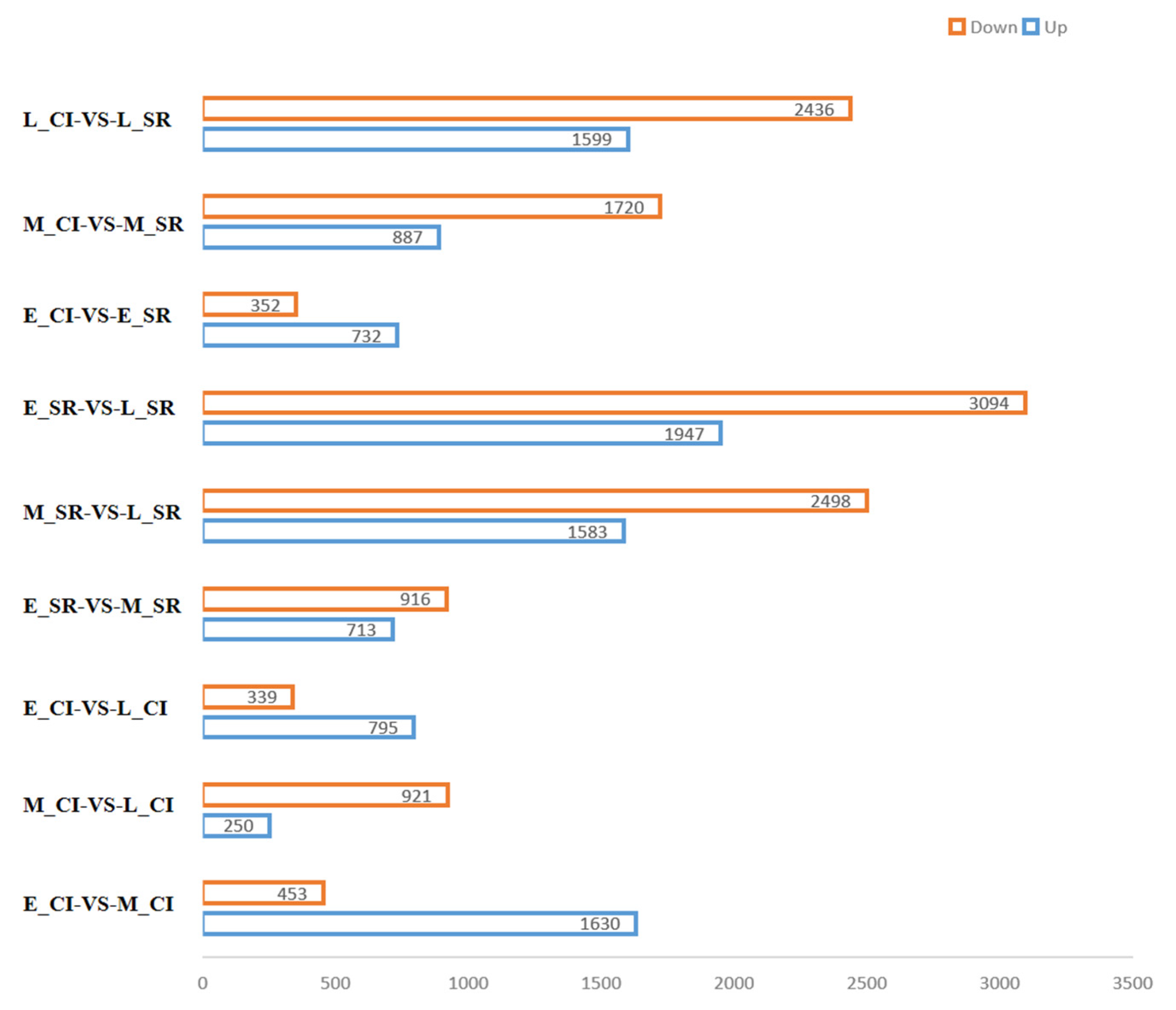
2.3. Expression and Analysis of Differentially Expressed Genes
2.4. GO Functional Classification Analysis of Differentially Expressed Genes
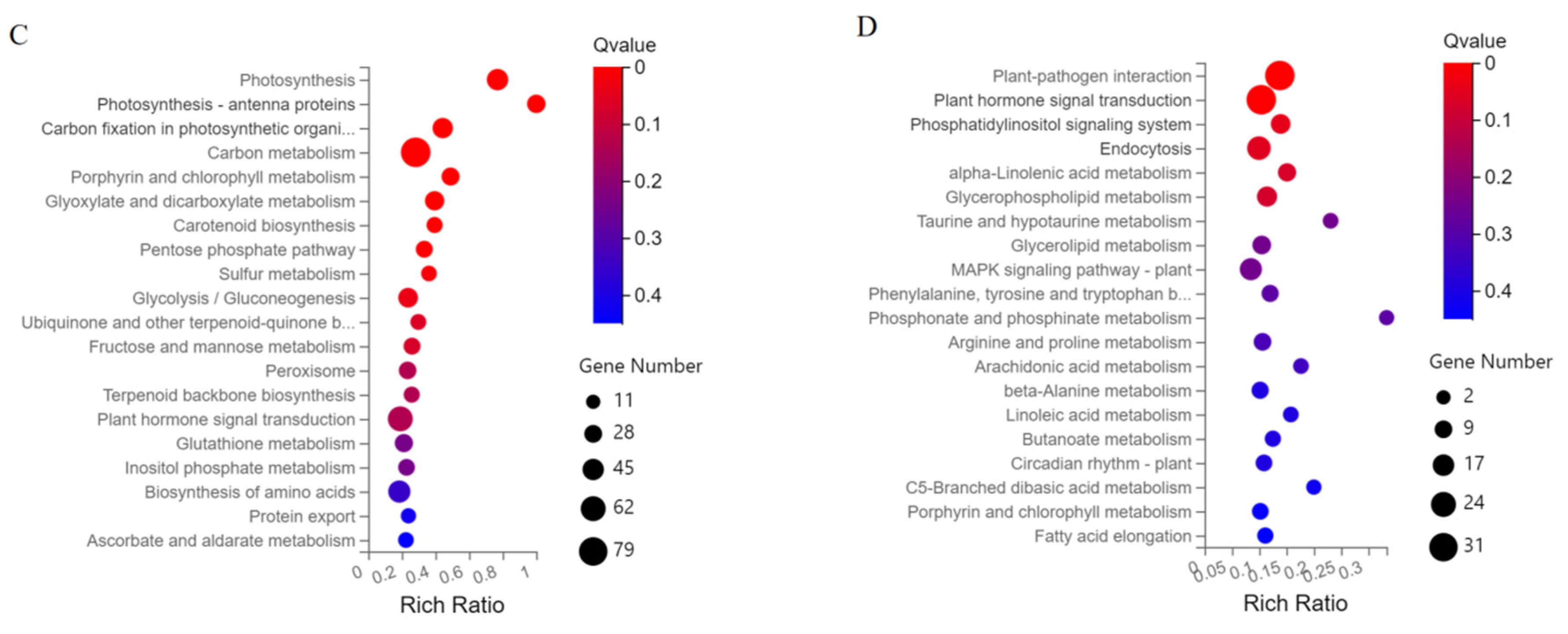
2.5. Classification of DEGs in KEGG Pathways
2.6. Screening of Genes Affecting Tomato Inflorescence Traits
2.7. Gene Co-Expression Network Analysis
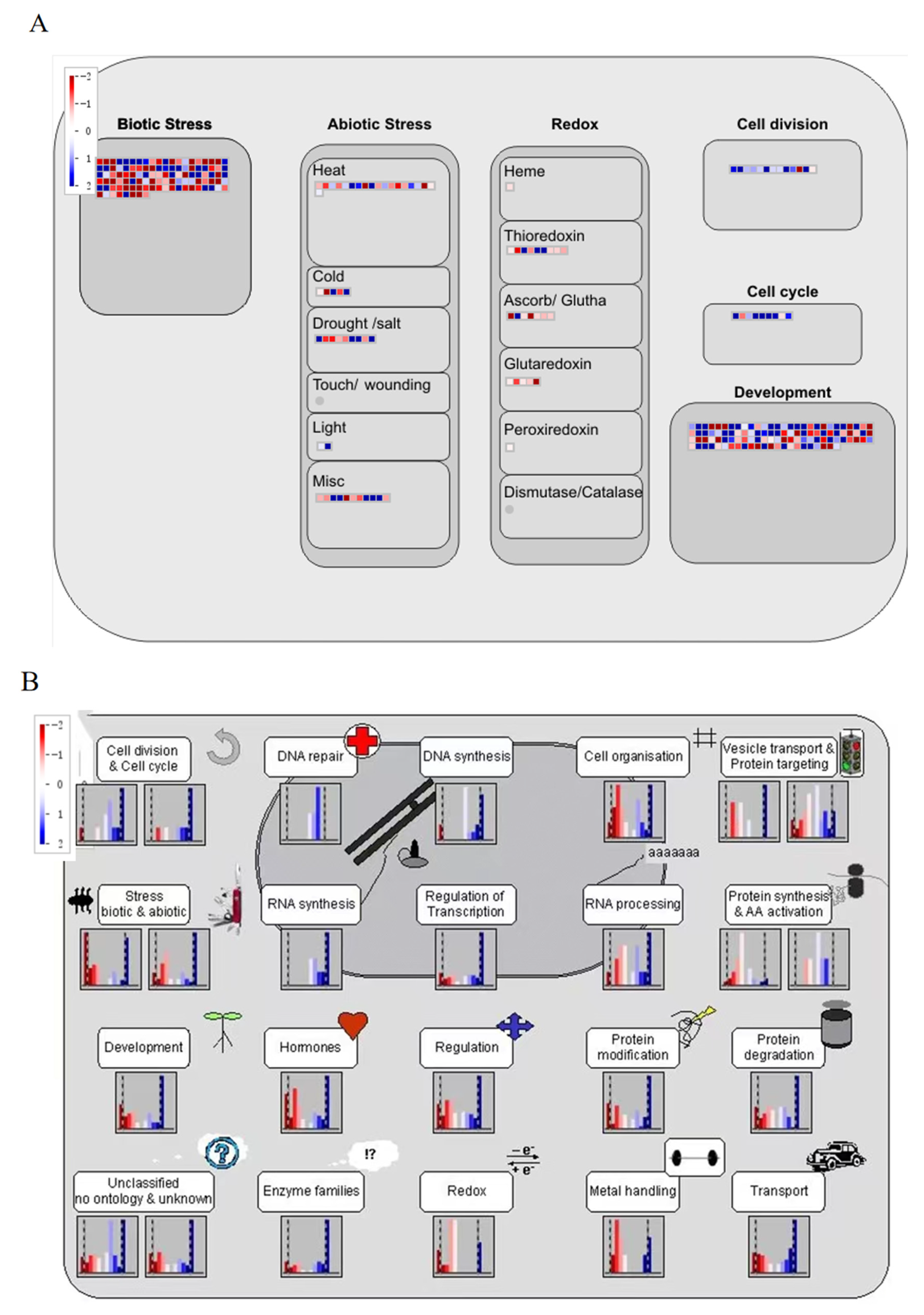
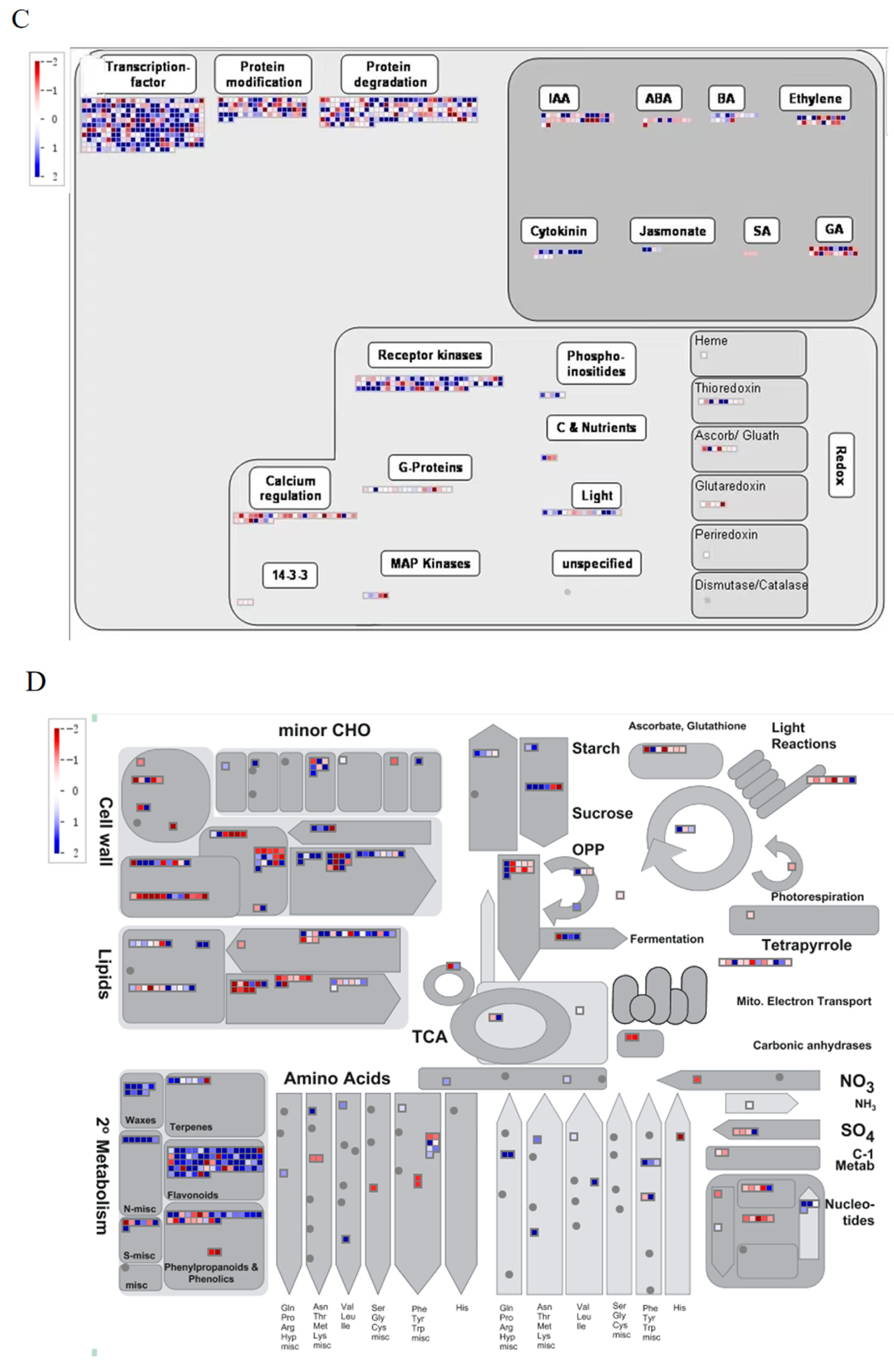
2.8. MapMan Software Analyzes DEGs
2.9. Validation of RNA-Seq Data via qRT–PCR
3. Discussion
4. Materials and Methods
4.1. Test Materials
4.2. Paraffin Sectioning
4.3. RNA Extraction and Illumina Sequencing
4.4. DEG Identification
4.5. KEGG Analysis of Differentially Expressed Genes
4.6. GO Enrichment Analysis of Differentially Expressed Genes
4.7. Weighted Coexpression Gene Network Analysis (WGCNA)
4.8. Related Genes Were Verified by qRT–PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Welty, N.; Radovich, C.; Meulia, T.; van der Knaap, E. Inflorescence development in two tomato species. Can. J. Bot. 2007, 85, 111–118. [Google Scholar] [CrossRef] [Green Version]
- Lippman, Z.B.; Cohen, O.; Alvarez, J.P.; Abu-Abied, M.; Pekker, I.; Paran, I.; Eshed, Y.; Zamir, D. The Making of a Compound Inflorescence in Tomato and Related Nightshades. PLoS Biol. 2008, 6, e288. [Google Scholar] [CrossRef]
- Teo, Z.W.N.; Song, S.; Wang, Y.Q.; Liu, J.; Yu, H. New insights into the regulation of inflorescence architecture. Trends Plant Sci. 2014, 19, 158–165. [Google Scholar] [CrossRef]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 1997, 89, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, J.C.; Brand, U.; Running, M.P.; Simon, R.; Meyerowitz, E.M. Signaling of Cell Fate Decisions by CLAVATA3 in Arabidopsis Shoot Meristems. Science 1999, 283, 1911–1914. [Google Scholar] [CrossRef]
- Jeong, S.; Trotochaud, A.E.; Clark, S.E. The Arabidopsis CLAVATA2 Gene Encodes a Receptor-Like Protein Required for the Stability of the CLAVATA1 Receptor-Like Kinase. Plant Cell 1999, 11, 1925. [Google Scholar] [CrossRef] [Green Version]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The Stem Cell Population of Arabidopsis Shoot Meristems Is Maintained by a Regulatory Loop between the CLAVATA and WUSCHEL Genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Ohyama, K.; Shinohara, H.; Ogawa-Ohnishi, M.; Matsubayashi, Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 2009, 5, 578–580. [Google Scholar] [CrossRef]
- Stahl, Y.; Simon, R. Plant primary meristems: Shared functions and regulatory mechanisms. Curr. Opin. Plant Biol. 2010, 13, 53–58. [Google Scholar] [CrossRef]
- Pautler, M.; Tanaka, W.; Hirano, H.Y.; Jackson, D. Grass Meristems I: Shoot Apical Meristem Maintenance, Axillary Meristem Determinacy and the Floral Transition. Plant Cell Physiol. 2013, 54, 302–312. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.-H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef]
- Rodriguez-Leal, D.; Xu, C.; Kwon, C.-T.; Soyars, C.; Demesa-Arevalo, E.; Man, J.; Liu, L.; Lemmon, Z.H.; Jones, D.S.; Van Eck, J.; et al. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 2019, 51, 786–792. [Google Scholar] [CrossRef]
- Molinero-Rosales, N.; Latorre, A.; Jamilena, M.; Lozano, R. SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 2004, 218, 427–434. [Google Scholar] [CrossRef]
- Lifschitz, E.; Eviatar, T.; Rozman, A.; Shalit, A.; Goldshmidt, A.; Amsellem, Z.; Alvarez, J.P.; Eshed, Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 2006, 103, 6398–6403. [Google Scholar] [CrossRef] [Green Version]
- Quinet, M.; Dubois, C.; Goffin, M.C.; Chao, J.; Dielen, V.; Batoko, H.; Boutry, M.; Kinet, J.M. Characterization of tomato (Solanum Lycopersicum L.) mutants affected in their flowering time and in the morphogenesis of their reproductive structure. J. Exp. Bot. 2006, 57, 1381–1390. [Google Scholar] [CrossRef]
- Szymkowiak, E.J.; Irish, E.E. Interactions between jointless and wild-type tomato tissues during development of the pedicel abscission zone and the inflorescence meristem. Plant Cell 1999, 11, 159–175. [Google Scholar] [CrossRef] [Green Version]
- Thouet, J.; Quinet, M.; Lutts, S.; Kinet, J.M.; Perilleux, C. Repression of Floral Meristem Fate Is Crucial in Shaping Tomato Inflorescence. PLoS ONE 2012, 7, e31096. [Google Scholar] [CrossRef] [Green Version]
- Molinero-Rosales, N.; Jamilena, M.; Zurita, S.; Gómez, P.; Capel, J.; Lozano, R. FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. Cell Mol. Biol. 1999, 20, 685–693. [Google Scholar] [CrossRef]
- Zheng, H.; Kawabata, S. Identification and Validation of New Alleles of FALSIFLORA and COMPOUND INFLORESCENCE Genes Controlling the Number of Branches in Tomato Inflorescence. Int. J. Mol. Sci. 2017, 18, 1572. [Google Scholar] [CrossRef] [Green Version]
- MacAlister, C.A.; Park, S.J.; Jiang, K.; Marcel, F.; Bendahmane, A.; Izkovich, Y.; Eshed, Y.; Lippman, Z.B. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat. Genet. 2012, 44, 1393–1398. [Google Scholar] [CrossRef]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Jiang, K.; Schatz, M.C.; Lippman, Z.B. Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 2012, 109, 639–644. [Google Scholar] [CrossRef] [Green Version]
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E.; et al. Bypassing Negative Epistasis on Yield in Tomato Imposed by a Domestication Gene. Cell 2017, 169, 1142–1155.e12. [Google Scholar] [CrossRef] [Green Version]
- Soyk, S.; Lemmon, Z.H.; Sedlazeck, F.J.; Jiménez-Gómez, J.M.; Alonge, M.; Hutton, S.F.; Van Eck, J.; Schatz, M.C.; Lippman, Z.B. Duplication of a domestication locus neutralized a cryptic variant that caused a breeding barrier in tomato. Nat. Plants 2019, 5, 471–479.e23. [Google Scholar] [CrossRef]
- Alonge, M.; Wang, X.; Benoit, M.; Soyk, S.; Pereira, L.; Zhang, L.; Suresh, H.; Ramakrishnan, S.; Maumus, F.; Ciren, D.; et al. Major Impacts of Widespread Structural Variation on Gene Expression and Crop Improvement in Tomato. Cell 2020, 182, 145–161. [Google Scholar] [CrossRef]
- Bisbis, B.; Delmas, F.; Joubès, J.; Sicard, A.; Hernould, M.; Inzé, D.; Mouras, A.; Chevalier, C. Cyclin-dependent kinase (CDK) inhibitors regulate the CDK-cyclin complex activities in endoreduplicating cells of developing tomato fruit. J. Biol. Chem. 2006, 281, 7374–7383. [Google Scholar] [CrossRef] [Green Version]
- Aoki, K.; Yano, K.; Suzuki, A.; Kawamura, S.; Sakurai, N.; Suda, K.; Kurabayashi, A.; Suzuki, T.; Tsugane, T.; Watanabe, T.; et al. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genom. 2010, 11, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemhauser, J.L.; Zambryski, P.C.; Roe, J.L. Auxin signaling in Arabidopsis flower development. Curr. Opin. Plant Biol. 1998, 1, 531–535. [Google Scholar] [CrossRef]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhardt, D.; Mandel, T.; Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 2000, 12, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.; Jones, M.L.; Banowetz, G.M.; Clark, D.G. Overproduction of cytokinins in petunia flowers transforms with PSAG12-IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol. 2003, 132, 2174–2183. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Cerny, R.E.; Qi, Y.; Bhat, D.; Aydt, C.M.; Hanson, D.D.; Malloy, K.P.; Ness, L.A. Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol. 2003, 131, 1270–1282. [Google Scholar] [CrossRef] [Green Version]
- Young, T.E.; Giesler-Lee, J.; Gallie, D.R. Senescence-induced expression of cytokinin reverses pistil abortion during maize flower development. Plant J. 2004, 38, 910–922. [Google Scholar] [CrossRef]
- Dong, B.; Deng, Y.; Wang, H.; Gao, R.; Stephen, G.K.; Chen, S.; Jiang, J.; Chen, F. Gibberellic Acid Signaling Is Required to Induce Flowering of Chrysanthemums Grown under Both Short and Long Days. Int. J. Mol. Sci. 2017, 18, 1259. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Chen, G.; Tang, B.; Zhang, J.; Zhou, S.; Hu, Z. The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 2018, 267, 65–73. [Google Scholar] [CrossRef]
- Chersicola, M.; Kladnik, A.; Znidaric, M.T.; Mrak, T.; Gruden, K. 1-Aminocyclopropane-1-Carboxylate Oxidase Induction in Tomato Flower Pedicel Phloem and Abscission Related Processes Are Differentially Sensitive to Ethylene. Front. Plant Sci. 2017, 8, 464. [Google Scholar] [CrossRef] [Green Version]
- Jianhong, H.; Alon, I.; Naomi, O.; Tai-Ping, S. The Interaction between DELLA and ARF/IAA Mediates Crosstalk between Gibberellin and Auxin Signaling to Control Fruit Initiation in Tomato. Plant Cell 2018, 30, 1710–1728. [Google Scholar]
- Serrani, J.C.; Sanjuán, R.; Ruiz-Rivero, O.; Fos, M.; García-Martínez, J.L. Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 2007, 145, 246–257. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yan, F.; Tang, Y.; Yuan, Y.; Deng, W.; Li, Z. Auxin Response Gene SlARF3 Plays Multiple Roles in Tomato Development and is Involved in the Formation of Epidermal Cells and Trichomes. Plant Cell Physiol. 2015, 56, 2110–2124. [Google Scholar]
- Li, Z.; Vickrey, T.L.; McNally, M.G.; Sato, S.J.; Clemente, T.E.; Mower, J.P. Assessing Anthocyanin Biosynthesis in Solanaceae as a Model Pathway for Secondary Metabolism. Genes 2019, 10, 559. [Google Scholar] [CrossRef] [Green Version]
- Krieger, U.; Lippman, Z.B.; Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010, 42, 459–463. [Google Scholar] [CrossRef]
- Lifschitz, E.; Eshed, Y. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 2006, 57, 3405–3414. [Google Scholar] [CrossRef]
- Pnueli, L.; Carmel-Goren, L.; Hareven, D.; Gutfinger, T.; Alvarez, J.; Ganal, M.; Zamir, D.; Lifschitz, E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 1998, 125, 1979–1989. [Google Scholar] [CrossRef]
- Pnueli, L.; Gutfinger, T.; Hareven, D.; Ben-Naim, O.; Ron, N.; Adir, N.; Lifschitz, E. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 2001, 13, 2687–2702. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, G.; Tillmann, E.; Carriero, F.; Fiore, C.; Cellini, F.; Theres, K. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 2002, 99, 1064–1069. [Google Scholar] [CrossRef] [Green Version]
- Souer, E.; Rebocho, A.B.; Bliek, M.; Kusters, E.; de Bruin, R.A.; Koes, R. Patterning of Inflorescences and Flowers by the F-Box Protein DOUBLE TOP and the LEAFY Homolog ABERRANT LEAF AND FLOWER of Petunia(W). Plant Cell 2008, 20, 2033–2048. [Google Scholar] [CrossRef] [Green Version]
- Olimpieri, I.; Mazzucato, A. Phenotypic and genetic characterization of the pistillate mutation in tomato. Theor. Appl. Genetics. Theor. Angew. Genet. 2008, 118, 151–163. [Google Scholar] [CrossRef]
- Lee, T.H.; Kim, Y.J.; Rou, W.S.; Eun, H.S. Fabrication of formalin-fixed, paraffin-embedded (FFPE) circulating tumor cell (CTC) block using a hydrogel core-mediated method. Micromachines 2021, 12, 1128. [Google Scholar] [CrossRef]
- Xue, D.-Q.; Chen, X.-L.; Zhang, H.; Chai, X.-F.; Jiang, J.-B.; Xu, X.-Y.; Li, J.-F. Transcriptome Analysis of the Cf-12-Mediated Resistance Response to Cladosporium fulvum in Tomato. Front. Plant Sci. 2017, 7, 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Song, W.; Zhang, Z.Q.; Wang, H.D.; Yang, M.M.; Guo, R.J.; Li, M.L. Transcriptome Analysis of Dastarcus helophoroides (Coleoptera: Bothrideridae) Using Illumina HiSeq Sequencing. PLoS ONE 2014, 9, e100673. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewey, C.N.; Bo, L. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 106. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Di Pietro, A.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Feng, Z.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Thimm, O.; Blaesing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Yang, M.; Wu, X.-Q.; Ding, C.-B.; Zhang, G.-F.; Li, M.; Lv, L.-N.; Li, Y.-H.; Sun, D.-W.; Zhao, J.-J. Weighted gene co-expression network analysis identifies specific modules and hub genes related to Parkinson’s disease. Neuroreport 2021, 32, 1073–1081. [Google Scholar] [CrossRef]
- Rotenberg, D.; Thompson, T.S.; German, T.L.; Willis, D.K. Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods 2006, 138, 49–59. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]





| Sample | Total Mapping (%) | Uniquely Mapping (%) | Total Raw Reads (M) | Total Clean Reads (M) | Total Clean Bases (Gb) | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Clean Reads Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| E_CI_1 | 92.54 | 91.01 | 47.33 | 45.03 | 6.76 | 96.58 | 91.56 | 95.15 |
| E_CI_2 | 92.9 | 91.47 | 47.33 | 45.18 | 6.78 | 96.62 | 91.70 | 95.47 |
| E_CI_3 | 93.23 | 91.85 | 45.57 | 43.92 | 6.59 | 96.71 | 91.86 | 96.37 |
| E_SR_1 | 92.72 | 91.29 | 47.33 | 45.40 | 6.81 | 96.81 | 92.13 | 95.92 |
| E_SR_2 | 92.92 | 91.44 | 47.33 | 45.33 | 6.80 | 96.59 | 91.55 | 95.77 |
| E_SR_3 | 92.24 | 90.81 | 47.33 | 44.78 | 6.72 | 96.64 | 91.67 | 94.63 |
| M_CI_1 | 93.10 | 91.68 | 45.57 | 44.16 | 6.62 | 96.66 | 91.72 | 96.89 |
| M_CI_2 | 93.11 | 91.66 | 45.57 | 44.12 | 6.62 | 96.58 | 91.56 | 96.81 |
| M_CI_3 | 92.85 | 91.38 | 47.33 | 45.39 | 6.81 | 96.86 | 92.20 | 95.91 |
| M_SR_1 | 93.04 | 91.56 | 45.57 | 43.84 | 6.58 | 96.66 | 91.68 | 96.20 |
| M_SR_2 | 92.70 | 91.28 | 47.33 | 45.08 | 6.76 | 96.76 | 91.93 | 95.24 |
| M_SR_3 | 92.98 | 91.55 | 47.33 | 45.46 | 6.82 | 96.59 | 91.55 | 96.06 |
| L_CI_1 | 92.64 | 91.19 | 47.33 | 45.43 | 6.81 | 96.67 | 91.79 | 95.99 |
| L_CI_2 | 92.49 | 91.10 | 47.33 | 45.01 | 6.75 | 96.72 | 91.92 | 95.11 |
| L_CI_3 | 93.06 | 91.59 | 47.33 | 45.42 | 6.81 | 96.73 | 91.88 | 95.98 |
| L_SR_1 | 93.37 | 91.92 | 45.57 | 43.86 | 6.58 | 96.69 | 91.73 | 96.25 |
| L_SR_2 | 92.91 | 91.44 | 47.33 | 45.44 | 6.82 | 96.48 | 91.31 | 96.01 |
| L_SR_3 | 93.44 | 91.96 | 45.57 | 43.99 | 6.60 | 96.65 | 91.69 | 96.52 |
| Gene ID | Symbol | Log2 Fold-Chang | Pathway | |
|---|---|---|---|---|
| E116-vs-E115 | L116-vs-L115 | |||
| 101252689 | LOC101252689 | 0.98 | 1.16 | Plant hormone signal transduction |
| 101267446 | LOC101267446 | 0.96 | 1.12 | Plant hormone signal transduction |
| 778363 | ARF3 | 0.5 | 0.64 | Plant hormone signal transduction |
| 101262709 | LOC101262709 | 1.38 | 1.85 | Plant hormone signal transduction |
| 778241 | ARF4 | 0.51 | 1.11 | Plant hormone signal transduction |
| 101248511 | LOC101248511 | −1 | −0.93 | Plant hormone signal transduction |
| 100134907 | GA2ox1 | −2.59 | −3.24 | Plant hormone signal transduction |
| 101258926 | LOC101258926 | −3.04 | −2.2 | Plant hormone signal transduction |
| 100736482 | F3H | 1.01 | 1.63 | Flavonoid biosynthesis |
| 101249699 | LOC101249699 | 1.32 | 1.44 | Flavonoid biosynthesis |
| 101266223 | LOC101266223 | 0.95 | 1.24 | Flavonoid biosynthesis |
| 104648079 | LOC104648079 | 0.93 | 1.24 | Zeatin biosynthesis |
| 101256765 | LOC101256765 | −4.5 | −4.19 | Zeatin biosynthesis |
| 101243791 | LOC101243791 | 10.07 | 9.9 | Carbon metabolism |
| 100529127 | LOC100529127 | 4.85 | 7.92 | Cell cycle |
| 543785 | krp2 | 0.66 | 1.93 | Cell cycle |
| 101254782 | LOC101254782 | 1.58 | 1.14 | Chloroplastic |
| 101255133 | LOC101255133 | 1.47 | 1.57 | Chloroplastic |
| 101255274 | LOC101255274 | 8.92 | 8.65 | Chloroplastic |
| 101258716 | LOC101258716 | 2.11 | 1.91 | Chloroplastic |
| 101261442 | LOC101261442 | 1.12 | 1.16 | Chloroplastic |
| 101266861 | LOC101266861 | 1.08 | 1.56 | Chloroplastic |
| 104649068 | LOC104649068 | 1.26 | 1.3 | F-box |
| 112940016 | LOC112940016 | 6.79 | 6.53 | F-box |
| 101250115 | LOC101250115 | 12.31 | 12.13 | F-box |
| 101244792 | LOC101244792 | 9.56 | 9.4 | F-box family protein |
| 101261682 | LOC101261682 | 3.65 | 5.77 | F-box protein CPR1 |
| 778295 | CHS2 | 2.73 | 2.18 | Circadian rhythm |
| 101246029 | SFT | −1.3 | −4.15 | Circadian rhythm |
| 100301925 | AN | 1.59 | 1.12 | Making of a compound inflorescence |
| 543630 | FA | 0.94 | 2.61 | Floral meristem identity |
| 544038 | SP | 1.86 | 1.8 | Regulates vegetative to reproductive switching |
| 543703 | blind | 0.29 | 2.33 | Control the formation of lateral meristems |
| 101259221 | LOC101259221 | 0.79 | 1.11 | Starch and sucrose metabolism |
| 101266123 | LOC101266123 | 0.44 | 1.31 | Glycine, serine and threonine metabolism |
| 101268249 | LOC101268249 | 0.52 | 0.54 | Purine metabolism |
| 101268552 | LOC101268552 | 5.39 | 5.13 | Sulfur metabolism |
| 112940464 | LOC112940464 | 8.95 | 8.75 | Fructose and mannose metabolism |
| 101245153 | CYP736A1 | 1.35 | 1.83 | Tissue specific promoters |
| 101251053 | dxs2 | 0.8 | 2.15 | Terpenoid backbone biosynthesis |
| 101261618 | Cwp | 1.9 | 3.77 | Cuticular water permeability protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Transcriptome Analysis to Explore the Cause of the Formation of Different Inflorescences in Tomato. Int. J. Mol. Sci. 2022, 23, 8216. https://doi.org/10.3390/ijms23158216
Yang Y, Zhao T, Xu X, Jiang J, Li J. Transcriptome Analysis to Explore the Cause of the Formation of Different Inflorescences in Tomato. International Journal of Molecular Sciences. 2022; 23(15):8216. https://doi.org/10.3390/ijms23158216
Chicago/Turabian StyleYang, Yahui, Tingting Zhao, Xiangyang Xu, Jingbin Jiang, and Jingfu Li. 2022. "Transcriptome Analysis to Explore the Cause of the Formation of Different Inflorescences in Tomato" International Journal of Molecular Sciences 23, no. 15: 8216. https://doi.org/10.3390/ijms23158216
APA StyleYang, Y., Zhao, T., Xu, X., Jiang, J., & Li, J. (2022). Transcriptome Analysis to Explore the Cause of the Formation of Different Inflorescences in Tomato. International Journal of Molecular Sciences, 23(15), 8216. https://doi.org/10.3390/ijms23158216





