Transcriptomic Profiling of Human Limbus-Derived Stromal/Mesenchymal Stem Cells—Novel Mechanistic Insights into the Pathways Involved in Corneal Wound Healing
Abstract
:1. Introduction
2. Results
2.1. Expansion of Limbal Stem Cells in Culture
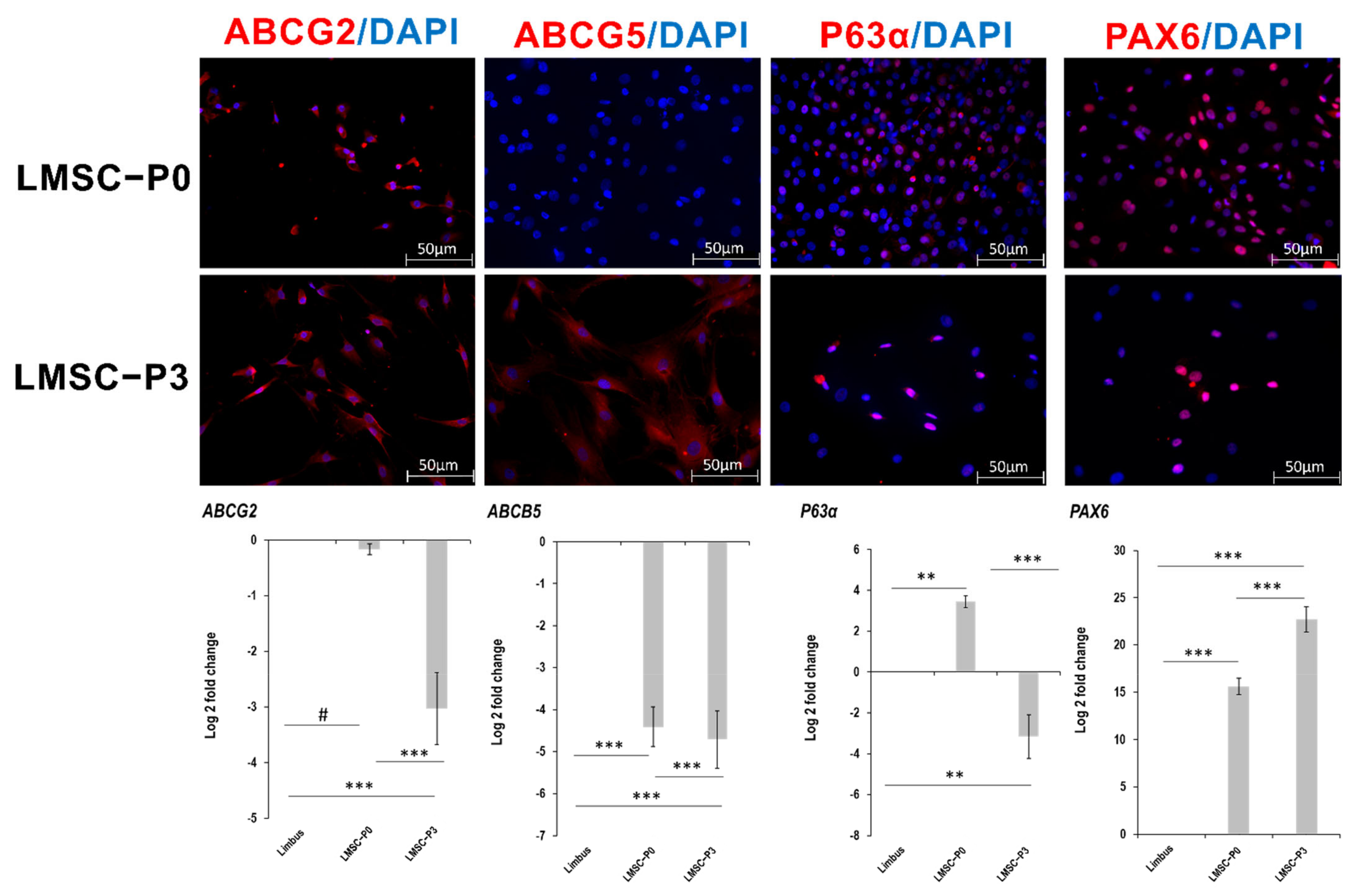
2.2. Cell Type Biomarker Changes during Culture Passages
2.2.1. Stem Cell and Ocular Biomarkers
2.2.2. Mesenchymal Stem Cell Markers
2.3. Genome Wide Transcriptomics Analysis Using RNA-Seq
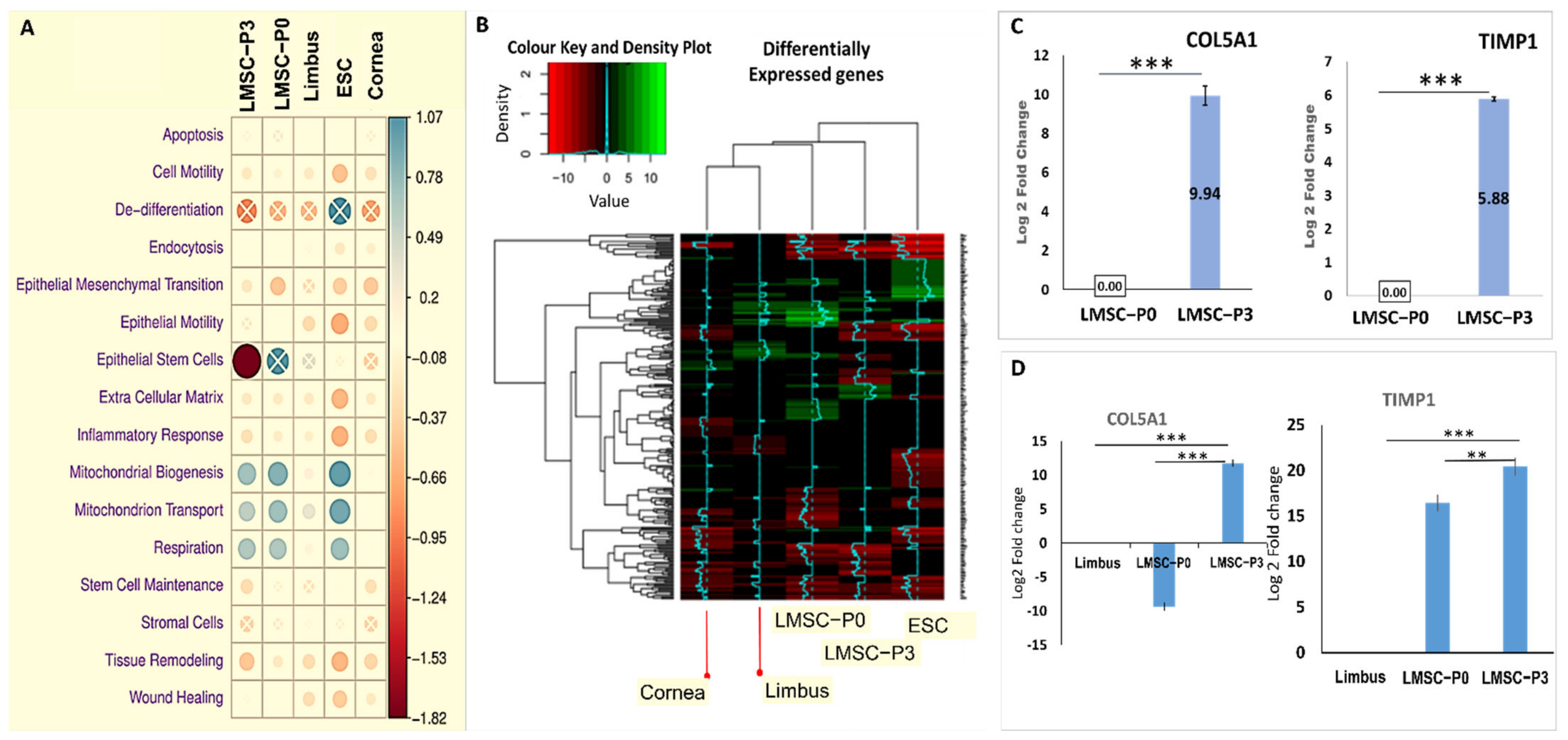
2.3.1. Transcriptome Overview Using Principal Components Analysis Plot
2.3.2. Visualizing the Asymmetry in Gene Expression of Various Tissues
2.4. Tissue-Specific Differential Expression and Pathway Enrichment Analysis
2.4.1. Interpretations from Gene Ontology Enrichment Analysis
2.4.2. GO Pathway Level Gene Expression Changes with Respect to Whole Transcriptome
2.4.3. Genes of Multiple Cell Signaling Pathways
Genes Involved in Wound Healing Pathway
Other Signaling Pathways
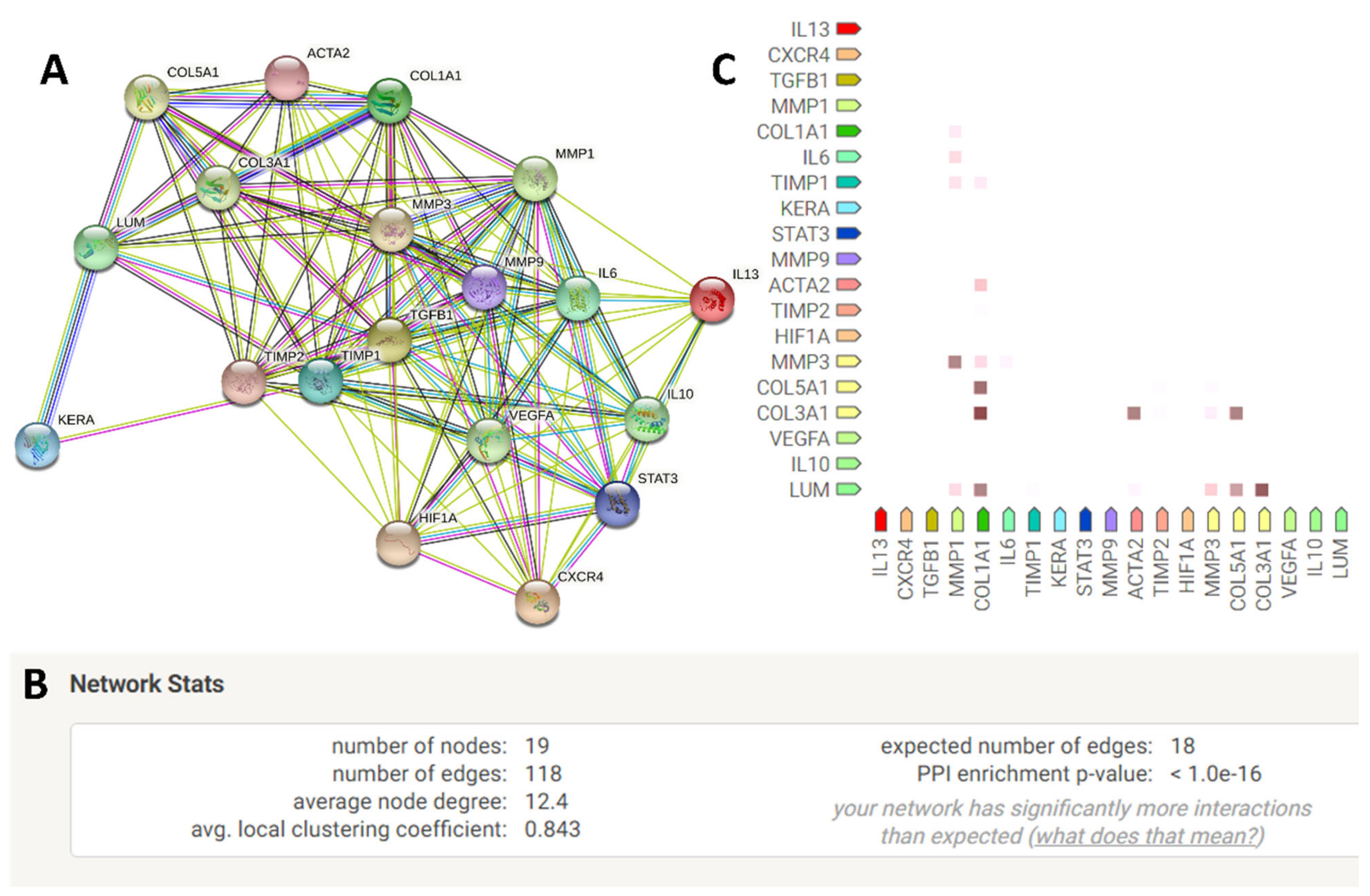
2.5. Quantification of Genes Interacting among the Exclusively Up-Regulated Genes in LMSC−P3
3. Discussion
4. Materials and Methods
4.1. Ethics Approval and Tissue Collection
4.2. Establishment of Limbal Stem Cell Culture
4.3. Immunofluorescence Assay
4.4. RNA Isolation
4.5. Next Generation RNA Sequencing (RNA-Seq) and Library Preparation
4.5.1. Pre-Processing of the RNA-Seq Data for Data Analysis
4.5.2. Differential Expression Analysis
4.5.3. Delineating Cell-Specific Gene Expression Patterns and Testing for Pathway Enrichment
4.5.4. Gene Ontology Pathway-Specific Gene Expression Changes
4.6. Reverse Transcriptase PCR
4.7. qRT-PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meek, K.M.; Knupp, C. Corneal structure and transparency. Prog. Retin. Eye Res. 2015, 49, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DelMonte, D.W.; Kim, T. Anatomy and physiology of the cornea. J. Cataract Refract. Surg. 2011, 37, 588–598. [Google Scholar] [CrossRef]
- Eghrari, A.O.; Riazuddin, S.A.; Gottsch, J.D. Overview of the Cornea: Structure, Function, and Development. Prog. Mol. Biol. Transl. Sci. 2015, 134, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Yun, H.; Funderburgh, M.L.; Du, Y. Regenerative therapy for the Cornea. Prog. Retin. Eye Res. 2021, 87, 101011. [Google Scholar] [CrossRef] [PubMed]
- Barrientez, B.; Nicholas, S.E.; Whelchel, A.; Sharif, R.; Hjortdal, J.; Karamichos, D. Corneal injury: Clinical and molecular aspects. Exp. Eye Res. 2019, 186, 107709. [Google Scholar] [CrossRef] [PubMed]
- Van Buskirk, E.M. The anatomy of the limbus. Eye 1989, 3, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitragotri, N.; Damala, M.; Singh, V.; Basu, S. Limbal Stromal Stem Cells in Corneal Wound Healing: Current Perspectives and Future Applications. In Corneal Regeneration; Alió, J.L., Alió del Barrio, J.L., Arnalich-Montiel, F., Eds.; Essentials in Ophthalmology; Springer International Publishing: Cham, Switzerland, 2019; pp. 387–402. ISBN 978-3-030-01303-5. [Google Scholar]
- Seyed-Safi, A.G.; Daniels, J.T. The limbus: Structure and function. Exp. Eye Res. 2020, 197, 108074. [Google Scholar] [CrossRef]
- Dua, H.S.; Shanmuganathan, V.A.; Powell-Richards, A.O.; Tighe, P.J.; Joseph, A. Limbal epithelial crypts: A novel anatomical structure and a putative limbal stem cell niche. Br. J. Ophthalmol. 2005, 89, 529–532. [Google Scholar] [CrossRef] [Green Version]
- Dziasko, M.A.; Armer, H.E.; Levis, H.J.; Shortt, A.J.; Tuft, S.; Daniels, J.T. Localisation of epithelial cells capable of holoclone formation in vitro and direct interaction with stromal cells in the native human limbal crypt. PLoS ONE 2014, 9, e94283. [Google Scholar] [CrossRef]
- Higa, K.; Shimmura, S.; Miyashita, H.; Shimazaki, J.; Tsubota, K. Melanocytes in the corneal limbus interact with K19-positive basal epithelial cells. Exp. Eye Res. 2005, 81, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wang, B.; Wan, P.; Liang, X.; Wang, X.; Liu, Y.; Zhou, Q.; Wang, Z. Roles of limbal microvascular net and limbal stroma in regulating maintenance of limbal epithelial stem cells. Cell Tissue Res. 2015, 359, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Gage, P.J.; Rhoades, W.; Prucka, S.K.; Hjalt, T. Fate maps of neural crest and mesoderm in the mammalian eye. Investig. Ophthalmol. Vis. Sci. 2005, 46, 4200–4208. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Funderburgh, M.L.; Mann, M.M.; SundarRaj, N.; Funderburgh, J.L. Multipotent stem cells in human corneal stroma. Stem Cells Dayt. Ohio 2005, 23, 1266–1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polisetty, N.; Fatima, A.; Madhira, S.L.; Sangwan, V.S.; Vemuganti, G.K. Mesenchymal cells from limbal stroma of human eye. Mol. Vis. 2008, 14, 431–442. [Google Scholar] [PubMed]
- Li, G.-G.; Zhu, Y.-T.; Xie, H.-T.; Chen, S.-Y.; Tseng, S.C.G. Mesenchymal stem cells derived from human limbal niche cells. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5686–5697. [Google Scholar] [CrossRef] [Green Version]
- Funderburgh, J.L.; Funderburgh, M.L.; Du, Y. Stem Cells in the Limbal Stroma. Ocul. Surf. 2016, 14, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Pinnamaneni, N.; Funderburgh, J.L. Concise review: Stem cells in the corneal stroma. Stem Cells Dayt. Ohio 2012, 30, 1059–1063. [Google Scholar] [CrossRef] [Green Version]
- Sidney, L.E.; Hopkinson, A. Corneal keratocyte transition to mesenchymal stem cell phenotype and reversal using serum-free medium supplemented with fibroblast growth factor-2, transforming growth factor-β3 and retinoic acid. J. Tissue Eng. Regen. Med. 2018, 12, e203–e215. [Google Scholar] [CrossRef]
- Park, S.H.; Kim, K.W.; Chun, Y.S.; Kim, J.C. Human mesenchymal stem cells differentiate into keratocyte-like cells in keratocyte-conditioned medium. Exp. Eye Res. 2012, 101, 16–26. [Google Scholar] [CrossRef]
- Basu, S.; Hertsenberg, A.J.; Funderburgh, M.L.; Burrow, M.K.; Mann, M.M.; Du, Y.; Lathrop, K.L.; Syed-Picard, F.N.; Adams, S.M.; Birk, D.E.; et al. Human limbal biopsy-derived stromal stem cells prevent corneal scarring. Sci. Transl. Med. 2014, 6, 266ra172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, A.; Balayan, A.; Funderburgh, M.L.; Ngo, J.; Funderburgh, J.L.; Deng, S.X. Differentiation Capacity of Human Mesenchymal Stem Cells into Keratocyte Lineage. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3013–3023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashmani, K.; Branch, M.J.; Sidney, L.E.; Dhillon, P.S.; Verma, M.; McIntosh, O.D.; Hopkinson, A.; Dua, H.S. Characterization of corneal stromal stem cells with the potential for epithelial transdifferentiation. Stem Cell Res. Ther. 2013, 4, 75. [Google Scholar] [CrossRef] [Green Version]
- Katikireddy, K.R.; Dana, R.; Jurkunas, U.V. Differentiation potential of limbal fibroblasts and bone marrow mesenchymal stem cells to corneal epithelial cells. Stem Cells Dayt. Ohio 2014, 32, 717–729. [Google Scholar] [CrossRef]
- Espana, E.M.; Kawakita, T.; Romano, A.; Di Pascuale, M.; Smiddy, R.; Liu, C.; Tseng, S.C.G. Stromal niche controls the plasticity of limbal and corneal epithelial differentiation in a rabbit model of recombined tissue. Investig. Ophthalmol. Vis. Sci. 2003, 44, 5130–5135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziasko, M.A.; Tuft, S.J.; Daniels, J.T. Limbal melanocytes support limbal epithelial stem cells in 2D and 3D microenvironments. Exp. Eye Res. 2015, 138, 70–79. [Google Scholar] [CrossRef]
- Kureshi, A.K.; Dziasko, M.; Funderburgh, J.L.; Daniels, J.T. Human corneal stromal stem cells support limbal epithelial cells cultured on RAFT tissue equivalents. Sci. Rep. 2015, 5, 16186. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Zenkel, M.; Menzel-Severing, J.; Kruse, F.E.; Schlötzer-Schrehardt, U. Cell Adhesion Molecules and Stem Cell-Niche-Interactions in the Limbal Stem Cell Niche. Stem Cells Dayt. Ohio 2016, 34, 203–219. [Google Scholar] [CrossRef]
- Yazdanpanah, G.; Jabbehdari, S.; Djalilian, A.R. Limbal and corneal epithelial homeostasis. Curr. Opin. Ophthalmol. 2017, 28, 348–354. [Google Scholar] [CrossRef]
- Polisetti, N.; Gießl, A.; Zenkel, M.; Heger, L.; Dudziak, D.; Naschberger, E.; Stich, L.; Steinkasserer, A.; Kruse, F.E.; Schlötzer-Schrehardt, U. Melanocytes as emerging key players in niche regulation of limbal epithelial stem cells. Ocul. Surf. 2021, 22, 172–189. [Google Scholar] [CrossRef]
- Zhao, J.; Mo, V.; Nagasaki, T. Distribution of label-retaining cells in the limbal epithelium of a mouse eye. J. Histochem. Cytochem. 2009, 57, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ksander, B.R.; Kolovou, P.E.; Wilson, B.J.; Saab, K.R.; Guo, Q.; Ma, J.; McGuire, S.P.; Gregory, M.S.; Vincent, W.J.B.; Perez, V.L.; et al. ABCB5 is a limbal stem cell gene required for corneal development and repair. Nature 2014, 511, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Amitai-Lange, A.; Altshuler, A.; Bubley, J.; Dbayat, N.; Tiosano, B.; Shalom-Feuerstein, R. Lineage tracing of stem and progenitor cells of the murine corneal epithelium. Stem Cells Dayt. Ohio 2015, 33, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Reinshagen, H.; Auw-Haedrich, C.; Sorg, R.V.; Boehringer, D.; Eberwein, P.; Schwartzkopff, J.; Sundmacher, R.; Reinhard, T. Corneal surface reconstruction using adult mesenchymal stem cells in experimental limbal stem cell deficiency in rabbits. Acta Ophthalmol. 2011, 89, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Samaeekia, R.; Rabiee, B.; Putra, I.; Shen, X.; Park, Y.J.; Hematti, P.; Eslani, M.; Djalilian, A.R. Effect of Human Corneal Mesenchymal Stromal Cell-derived Exosomes on Corneal Epithelial Wound Healing. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5194–5200. [Google Scholar] [CrossRef] [Green Version]
- Basu, S.; Damala, M.; Tavakkoli, F.; Mitragotri, N.; Singh, V. Human Limbus-derived Mesenchymal/Stromal Stem Cell Therapy for Superficial Corneal Pathologies: Two-Year Outcomes. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4146. [Google Scholar]
- Liu, X.-N.; Mi, S.-L.; Chen, Y.; Wang, Y. Corneal stromal mesenchymal stem cells: Reconstructing a bioactive cornea and repairing the corneal limbus and stromal microenvironment. Int. J. Ophthalmol. 2021, 14, 448–455. [Google Scholar] [CrossRef]
- Netto, M.V.; Mohan, R.R.; Sinha, S.; Sharma, A.; Dupps, W.; Wilson, S.E. Stromal haze, myofibroblasts, and surface irregularity after PRK. Exp. Eye Res. 2006, 82, 788–797. [Google Scholar] [CrossRef] [Green Version]
- Torricelli, A.A.M.; Santhanam, A.; Wu, J.; Singh, V.; Wilson, S.E. The corneal fibrosis response to epithelial-stromal injury. Exp. Eye Res. 2016, 142, 110–118. [Google Scholar] [CrossRef] [Green Version]
- Medeiros, C.S.; Marino, G.K.; Santhiago, M.R.; Wilson, S.E. The Corneal Basement Membranes and Stromal Fibrosis. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4044–4053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa, A.P.; Alves, M.; Furtado, J.M.F.; Adriano, L.; Nominato, L.F.; Dias, L.C.; Fantucci, M.Z.; de Andrade Batista Murashima, A.; Rocha, E.M. Corneal blindness in Plato’s cave: The acting forces to prevent and revert corneal opacity. Part I: Epidemiology and new physiopathological concepts. Arq. Bras. Oftalmol. 2020, 83, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Niederkorn, J.Y.; Larkin, D.F.P. Immune privilege of corneal allografts. Ocul. Immunol. Inflamm. 2010, 18, 162–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borderie, V.M.; Boëlle, P.-Y.; Touzeau, O.; Allouch, C.; Boutboul, S.; Laroche, L. Predicted long-term outcome of corneal transplantation. Ophthalmology 2009, 116, 2354–2360. [Google Scholar] [CrossRef] [PubMed]
- Klebe, S.; Coster, D.J.; Williams, K.A. Rejection and acceptance of corneal allografts. Curr. Opin. Organ Transplant. 2009, 14, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Pantanelli, S.M.; Sabesan, R.; Ching, S.S.T.; Yoon, G.; Hindman, H.B. Visual performance with wave aberration correction after penetrating, deep anterior lamellar, or endothelial keratoplasty. Investig. Ophthalmol. Vis. Sci. 2012, 53, 4797–4804. [Google Scholar] [CrossRef]
- Nielsen, E.; Hjortdal, J. Visual acuity and contrast sensitivity after posterior lamellar keratoplasty. Acta Ophthalmol. 2012, 90, 756–760. [Google Scholar] [CrossRef]
- Dupps, W.J.; Wilson, S.E. Biomechanics and Wound Healing in the Cornea. Exp. Eye Res. 2006, 83, 709–720. [Google Scholar] [CrossRef] [Green Version]
- Chawla, S.; Ghosh, S. Establishment of in vitro model of corneal scar pathophysiology. J. Cell. Physiol. 2018, 233, 3817–3830. [Google Scholar] [CrossRef]
- Cogswell, D.; Sun, M.; Greenberg, E.; Margo, C.E.; Espana, E.M. Creation and grading of experimental corneal scars in mice models. Ocul. Surf. 2021, 19, 53–62. [Google Scholar] [CrossRef]
- Sherwin, T.; McGhee, C.N.J. Corneal epithelial homeostasis. Ophthalmology 2010, 117, 190–191, author reply 191–192. [Google Scholar] [CrossRef] [PubMed]
- Altshuler, A.; Amitai-Lange, A.; Tarazi, N.; Dey, S.; Strinkovsky, L.; Hadad-Porat, S.; Bhattacharya, S.; Nasser, W.; Imeri, J.; Ben-David, G.; et al. Discrete limbal epithelial stem cell populations mediate corneal homeostasis and wound healing. Cell Stem Cell 2021, 28, 1248–1261.e8. [Google Scholar] [CrossRef] [PubMed]
- Keratocyte Biology—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/32442558/ (accessed on 18 June 2022).
- Weng, J.; Mohan, R.R.; Li, Q.; Wilson, S.E. IL-1 upregulates keratinocyte growth factor and hepatocyte growth factor mRNA and protein production by cultured stromal fibroblast cells: Interleukin-1 beta expression in the cornea. Cornea 1997, 16, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Ylä-Outinen, H.; Aaltonen, V.; Björkstrand, A.S.; Hirvonen, O.; Lakkakorpi, J.; Vähä-Kreula, M.; Laato, M.; Peltonen, J. Upregulation of tumor suppressor protein neurofibromin in normal human wound healing and in vitro evidence for platelet derived growth factor (PDGF) and transforming growth factor-beta1 (TGF-beta1) elicited increase in neurofibromin mRNA steady-state levels in dermal fibroblasts. J. Investig. Dermatol. 1998, 110, 232–237. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.J.; Mohan, R.R.; Mohan, R.R.; Wilson, S.E. Effect of PDGF, IL-1alpha, and BMP2/4 on corneal fibroblast chemotaxis: Expression of the platelet-derived growth factor system in the cornea. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1364–1372. [Google Scholar]
- Hong, J.W.; Liu, J.J.; Lee, J.S.; Mohan, R.R.; Mohan, R.R.; Woods, D.J.; He, Y.G.; Wilson, S.E. Proinflammatory chemokine induction in keratocytes and inflammatory cell infiltration into the cornea. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2795–2803. [Google Scholar]
- O’Brien, T.P.; Li, Q.; Ashraf, M.F.; Matteson, D.M.; Stark, W.J.; Chan, C.C. Inflammatory response in the early stages of wound healing after excimer laser keratectomy. Arch. Ophthalmol. 1998, 116, 1470–1474. [Google Scholar] [CrossRef] [Green Version]
- Wright, B.; Hopkinson, A.; Leyland, M.; Connon, C.J. The secretome of alginate-encapsulated limbal epithelial stem cells modulates corneal epithelial cell proliferation. PLoS ONE 2013, 8, e70860. [Google Scholar] [CrossRef]
- Hassell, J.R.; Kane, B.P.; Alexandrou, B.; Musselmann, K. IGF-II Is Present in the Cornea Stroma and Activates Keratocytes to Proliferate in vitro. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4814. [Google Scholar]
- Nagano, T.; Nakamura, M.; Nakata, K.; Yamaguchi, T.; Takase, K.; Okahara, A.; Ikuse, T.; Nishida, T. Effects of Substance P and IGF-1 in Corneal Epithelial Barrier Function and Wound Healing in a Rat Model of Neurotrophic Keratopathy. Investig. Ophthalmol. Vis. Sci. 2003, 44, 3810–3815. [Google Scholar] [CrossRef] [Green Version]
- Wilson, S.E.; He, Y.G.; Weng, J.; Zieske, J.D.; Jester, J.V.; Schultz, G.S. Effect of epidermal growth factor, hepatocyte growth factor, and keratinocyte growth factor, on proliferation, motility and differentiation of human corneal epithelial cells. Exp. Eye Res. 1994, 59, 665–678. [Google Scholar] [CrossRef]
- Li, D.Q.; Tseng, S.C. Differential regulation of keratinocyte growth factor and hepatocyte growth factor/scatter factor by different cytokines in human corneal and limbal fibroblasts. J. Cell. Physiol. 1997, 172, 361–372. [Google Scholar] [CrossRef]
- David, T.; Rieck, P.; Renard, G.; Hartmann, C.; Courtois, Y.; Pouliquen, Y. Corneal wound healing modulation using basic fibroblast growth factor after excimer laser photorefractive keratectomy. Cornea 1995, 14, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Hafezi, F.; Gatzioufas, Z.; Angunawela, R.; Ittner, L.M. Absence of IL-6 prevents corneal wound healing after deep excimer laser ablation in vivo. Eye 2018, 32, 156–157. [Google Scholar] [CrossRef] [PubMed]
- Cole, N.; Bao, S.; Willcox, M.; Husband, A.J. Expression of Interleukin-6 in the Cornea in Response to Infection with Different Strains of Pseudomonas aeruginosa. Infect. Immun. 1999, 67, 2497–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, T.; Nakamura, M.; Mishima, H.; Otori, T.; Hikida, M. Interleukin 6 Facilitates Corneal Epithelial Wound Closure In Vivo. Arch. Ophthalmol. 1992, 110, 1292–1294. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Hirano, T.; Taga, T.; Kishimoto, T. Biology of multifunctional cytokines: IL 6 and related molecules (IL 1 and TNF). FASEB J. 1990, 4, 2860–2867. [Google Scholar] [CrossRef]
- Orozco Morales, M.L.; Marsit, N.M.; McIntosh, O.D.; Hopkinson, A.; Sidney, L.E. Anti-inflammatory potential of human corneal stroma-derived stem cells determined by a novel in vitro corneal epithelial injury model. World J. Stem Cells 2019, 11, 84–99. [Google Scholar] [CrossRef]
- Wilson, S.E. Corneal wound healing. Exp. Eye Res. 2020, 197, 108089. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, R.C.; Wilson, S.E. Fibrocytes, Wound Healing, and Corneal Fibrosis. Investig. Ophthalmol. Vis. Sci. 2020, 61, 28. [Google Scholar] [CrossRef] [Green Version]
- McKay, T.B.; Hutcheon, A.E.K.; Zieske, J.D. Biology of corneal fibrosis: Soluble mediators, integrins, and extracellular vesicles. Eye 2020, 34, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Kamil, S.; Mohan, R.R. Corneal stromal wound healing: Major regulators and therapeutic targets. Ocul. Surf. 2021, 19, 290–306. [Google Scholar] [CrossRef] [PubMed]
- Bazan, H.E.P. Cellular and molecular events in corneal wound healing: Significance of lipid signalling. Exp. Eye Res. 2005, 80, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Hertsenberg, A.J.; Shojaati, G.; Funderburgh, M.L.; Mann, M.M.; Du, Y.; Funderburgh, J.L. Corneal stromal stem cells reduce corneal scarring by mediating neutrophil infiltration after wounding. PLoS ONE 2017, 12, e0171712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, L.; Funderburgh, J.L.; Khandaker, I.; Geary, M.L.; Yang, T.; Basu, R.; Funderburgh, M.L.; Du, Y.; Yam, G.H.-F. The anti-scarring effect of corneal stromal stem cell therapy is mediated by transforming growth factor β3. Eye Vis. 2020, 7, 52. [Google Scholar] [CrossRef]
- Chameettachal, S.; Prasad, D.; Parekh, Y.; Basu, S.; Singh, V.; Bokara, K.K.; Pati, F. Prevention of Corneal Myofibroblastic Differentiation In Vitro Using a Biomimetic ECM Hydrogel for Corneal Tissue Regeneration. ACS Appl. Bio Mater. 2021, 4, 533–544. [Google Scholar] [CrossRef]
- Chandru, A.; Agrawal, P.; Ojha, S.K.; Selvakumar, K.; Shiva, V.K.; Gharat, T.; Selvam, S.; Thomas, M.B.; Damala, M.; Prasad, D.; et al. Human Cadaveric Donor Cornea Derived Extra Cellular Matrix Microparticles for Minimally Invasive Healing/Regeneration of Corneal Wounds. Biomolecules 2021, 11, 532. [Google Scholar] [CrossRef]
- Du, Y.; SundarRaj, N.; Funderburgh, M.L.; Harvey, S.A.; Birk, D.E.; Funderburgh, J.L. Secretion and Organization of a Cornea-like Tissue In Vitro by Stem Cells from Human Corneal Stroma. Investig. Ophthalmol. Vis. Sci. 2007, 48, 5038–5045. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Hayashida, Y.; Chen, M.-Y.; Xie, H.T.; Tseng, S.C.G. A New Isolation Method of Human Limbal Progenitor Cells by Maintaining Close Association with Their Niche Cells. Tissue Eng. Part C Methods 2011, 17, 537. [Google Scholar] [CrossRef] [Green Version]
- Yam, G.H.-F.; Yang, T.; Geary, M.L.; Santra, M.; Funderburgh, M.; Rubin, E.; Du, Y.; Sahel, J.A.; Jhanji, V.; Funderburgh, J.L. Human corneal stromal stem cells express anti-fibrotic microRNA-29a and 381-5p—A robust cell selection tool for stem cell therapy of corneal scarring. J. Adv. Res. 2022; in press. [Google Scholar] [CrossRef]
- Birk, D.E.; Fitch, J.M.; Linsenmayer, T.F. Organization of collagen types I and V in the embryonic chicken cornea. Investig. Ophthalmol. Vis. Sci. 1986, 27, 1470–1477. [Google Scholar]
- Yokota, T.; McCourt, J.; Ma, F.; Ren, S.; Li, S.; Kim, T.-H.; Kurmangaliyev, Y.Z.; Nasiri, R.; Ahadian, S.; Nguyen, T.; et al. Type V Collagen in Scar Tissue Regulates the Size of Scar after Heart Injury. Cell 2020, 182, 545–562.e23. [Google Scholar] [CrossRef] [PubMed]
- Niyibizi, C.; Kavalkovich, K.; Yamaji, T.; Woo, S.L. Type V collagen is increased during rabbit medial collateral ligament healing. Knee Surg. Sports Traumatol. Arthrosc. 2000, 8, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, F.; Burillon, C.; Garrone, R. Human corneal fibrillogenesis. Collagen V structural analysis and fibrillar assembly by stromal fibroblasts in culture. Investig. Ophthalmol. Vis. Sci. 1996, 37, 1749–1760. [Google Scholar]
- McLaughlin, J.S.; Linsenmayer, T.F.; Birk, D.E. Type V collagen synthesis and deposition by chicken embryo corneal fibroblasts in vitro. J. Cell Sci. 1989, 94, 371–379. [Google Scholar] [CrossRef]
- DeNigris, J.; Yao, Q.; Birk, E.K.; Birk, D.E. Altered dermal fibroblast behavior in a collagen V haploinsufficient murine model of classic Ehlers-Danlos syndrome. Connect. Tissue Res. 2016, 57, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.H.; Jia, Y.Y.S.; Zeng, Y.M.; Li, Z.F.; Lin, J.S. Transcriptome analysis identifies the differentially expressed genes related to the stemness of limbal stem cells in mice. Gene 2021, 775, 145447. [Google Scholar] [CrossRef]
- Call, M.; Elzarka, M.; Kunesh, M.; Hura, N.; Birk, D.E.; Kao, W.W. Therapeutic efficacy of mesenchymal stem cells for the treatment of congenital and acquired corneal opacity. Mol. Vis. 2019, 25, 415–426. [Google Scholar]
- Pietruszewska, W.; Bojanowska-Poźniak, K.; Kobos, J. Matrix metalloproteinases MMP1, MMP2, MMP9 and their tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: An immunohistochemical study. Otolaryngol. Pol. Pol. Otolaryngol. 2016, 70, 32–43. [Google Scholar] [CrossRef]
- Visse, R.; Nagase, H. Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases. Circ. Res. 2003, 92, 827–839. [Google Scholar] [CrossRef] [Green Version]
- Huh, M.-I.; Lee, Y.-M.; Seo, S.-K.; Kang, B.-S.; Chang, Y.; Lee, Y.-S.; Fini, M.E.; Kang, S.-S.; Jung, J.-C. Roles of MMP/TIMP in regulating matrix swelling and cell migration during chick corneal development. J. Cell. Biochem. 2007, 101, 1222–1237. [Google Scholar] [CrossRef] [PubMed]
- Kwak, H.-J.; Park, M.-J.; Cho, H.; Park, C.-M.; Moon, S.-I.; Lee, H.-C.; Park, I.-C.; Kim, M.-S.; Rhee, C.H.; Hong, S.-I. Transforming growth factor-beta1 induces tissue inhibitor of metalloproteinase-1 expression via activation of extracellular signal-regulated kinase and Sp1 in human fibrosarcoma cells. Mol. Cancer Res. 2006, 4, 209–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivonen, S.-K.; Lazaridis, K.; Decock, J.; Chantry, A.; Edwards, D.R.; Kähäri, V.-M. TGF-β-Elicited Induction of Tissue Inhibitor of Metalloproteinases (TIMP)-3 Expression in Fibroblasts Involves Complex Interplay between Smad3, p38α, and ERK1/2. PLoS ONE 2013, 8, e57474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amjadi, S.; Mai, K.; McCluskey, P.; Wakefield, D. The Role of Lumican in Ocular Disease. ISRN Ophthalmol. 2013, 2013, 632302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, E.C.; Lin, M.; Liu, C.-Y.; Kao, W.W.-Y.; Perez, V.L.; Pearlman, E. Keratocan and Lumican Regulate Neutrophil Infiltration and Corneal Clarity in Lipopolysaccharide-induced Keratitis by Direct Interaction with CXCL1*. J. Biol. Chem. 2007, 282, 35502–35509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funderburgh, J.L. The Corneal Stroma. In Encyclopedia of the Eye; Dartt, D.A., Ed.; Academic Press: Oxford, UK, 2010; pp. 515–521. ISBN 978-0-12-374203-2. [Google Scholar]
- Espana, E.M.; He, H.; Kawakita, T.; Di Pascuale, M.A.; Raju, V.K.; Liu, C.-Y.; Tseng, S.C.G. Human Keratocytes Cultured on Amniotic Membrane Stroma Preserve Morphology and Express Keratocan. Investig. Ophthalmol. Vis. Sci. 2003, 44, 5136–5141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, S.; Yokoo, S.; Yanagi, Y.; Usui, T.; Yokota, C.; Mimura, T.; Araie, M.; Yamagami, S.; Amano, S. Sphere Formation and Expression of Neural Proteins by Human Corneal Stromal Cells In Vitro. Investig. Ophthalmol. Vis. Sci. 2005, 46, 1620–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakita, T.; Espana, E.M.; He, H.; Hornia, A.; Yeh, L.-K.; Ouang, J.; Liu, C.-Y.; Tseng, S.C.G. Keratocan expression of murine keratocytes is maintained on amniotic membrane by downregulating tgf-β signaling. J. Biol. Chem. 2005, 280, 27085–27092. [Google Scholar] [CrossRef] [Green Version]
- Pouw, A.E.; Greiner, M.A.; Coussa, R.G.; Jiao, C.; Han, I.C.; Skeie, J.M.; Fingert, J.H.; Mullins, R.F.; Sohn, E.H. Cell-Matrix Interactions in the Eye: From Cornea to Choroid. Cells 2021, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Brem, H.; Kodra, A.; Golinko, M.S.; Entero, H.; Stojadinovic, O.; Wang, V.M.; Sheahan, C.M.; Weinberg, A.D.; Woo, S.L.C.; Ehrlich, H.P.; et al. Mechanism of sustained release of vascular endothelial growth factor in accelerating experimental diabetic healing. J. Investig. Dermatol. 2009, 129, 2275–2287. [Google Scholar] [CrossRef] [Green Version]
- Losi, P.; Briganti, E.; Errico, C.; Lisella, A.; Sanguinetti, E.; Chiellini, F.; Soldani, G. Fibrin-based scaffold incorporating VEGF- and bFGF-loaded nanoparticles stimulates wound healing in diabetic mice. Acta Biomater. 2013, 9, 7814–7821. [Google Scholar] [CrossRef]
- Yao, L.; Li, Z.; Su, W.; Li, Y.; Lin, M.; Zhang, W.; Liu, Y.; Wan, Q.; Liang, D. Role of mesenchymal stem cells on cornea wound healing induced by acute alkali burn. PLoS ONE 2012, 7, e30842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.-Y.; Chang, J.-H.; Lee, H.; Azar, D.T. Proangiogenic Interactions of Vascular Endothelial MMP14 With VEGF Receptor 1 in VEGFA-Mediated Corneal Angiogenesis. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3313–3322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepp, M.A.; Menko, A.S. Immune responses to injury and their links to eye disease. Transl. Res. J. Lab. Clin. Med. 2021, 236, 52–71. [Google Scholar] [CrossRef]
- Frank, S.; Hübner, G.; Breier, G.; Longaker, M.T.; Greenhalgh, D.G.; Werner, S. Regulation of vascular endothelial growth factor expression in cultured keratinocytes. Implications for normal and impaired wound healing. J. Biol. Chem. 1995, 270, 12607–12613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, R.; Omura, T.; Yoshiyama, M.; Hayashi, T.; Inamoto, S.; Koh, K.-R.; Ohta, K.; Izumi, Y.; Nakamura, Y.; Akioka, K.; et al. Vascular endothelial growth factor-expressing mesenchymal stem cell transplantation for the treatment of acute myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Li, C.; Yu, L. Gambogic acid inhibits spinal cord injury and inflammation through suppressing the p38 and Akt signaling pathways. Mol. Med. Rep. 2018, 17, 2026–2032. [Google Scholar] [CrossRef]
- Michaels, J.; Dobryansky, M.; Galiano, R.D.; Bhatt, K.A.; Ashinoff, R.; Ceradini, D.J.; Gurtner, G.C. Topical vascular endothelial growth factor reverses delayed wound healing secondary to angiogenesis inhibitor administration. Wound Repair Regen. 2005, 13, 506–512. [Google Scholar] [CrossRef]
- Hollborn, M.; Stathopoulos, C.; Steffen, A.; Wiedemann, P.; Kohen, L.; Bringmann, A. Positive feedback regulation between MMP-9 and VEGF in human RPE cells. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4360–4367. [Google Scholar] [CrossRef] [Green Version]
- Van Setten, G.B. Vascular endothelial growth factor (VEGF) in normal human corneal epithelium: Detection and physiological importance. Acta Ophthalmol. Scand. 1997, 75, 649–652. [Google Scholar] [CrossRef]
- Ambati, B.K.; Nozaki, M.; Singh, N.; Takeda, A.; Jani, P.D.; Suthar, T.; Albuquerque, R.J.C.; Richter, E.; Sakurai, E.; Newcomb, M.T.; et al. Corneal avascularity is due to soluble VEGF receptor-1. Nature 2006, 443, 993–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.H.-K.; Tsai, R.J.-F.; Chu, W.-K.; Kao, C.-H.; Chen, J.-K. Inhibition of Vascular Endothelial Cell Morphogenesis in Cultures by Limbal Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1822–1828. [Google Scholar]
- Eslani, M.; Putra, I.; Shen, X.; Hamouie, J.; Afsharkhamseh, N.; Besharat, S.; Rosenblatt, M.I.; Dana, R.; Hematti, P.; Djalilian, A.R. Corneal Mesenchymal Stromal Cells Are Directly Antiangiogenic via PEDF and sFLT-1. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5507–5517. [Google Scholar] [CrossRef] [PubMed]
- Damala, M.; Swioklo, S.; Koduri, M.A.; Mitragotri, N.S.; Basu, S.; Connon, C.J.; Singh, V. Encapsulation of human limbus-derived stromal/mesenchymal stem cells for biological preservation and transportation in extreme Indian conditions for clinical use. Sci. Rep. 2019, 9, 16950. [Google Scholar] [CrossRef]




| Gene Ontology ID | Biological Process | Genes Involved | False Discovery Ratio |
|---|---|---|---|
| GO:0032964 | Collagen biosynthetic process | COL5A1, COL1A1 | 0.0028 |
| GO:1905048 | Regulation of metallopeptidase activity | TIMP1, TIMP2, STAT3 | 0.00013 |
| GO:0070102 | Interleukin-6-mediated signaling pathway | IL-6, STAT3, | 0.0061 |
| GO:0030199 | Collagen fibril organization | COL5A1, COL1A1, COL3A1, LUM | 3.5 × 10−5 |
| GO:0035633 | Maintenance of blood-brain barrier | VEGF, IL-6 | 0.0171 |
| GO:0048661 | Positive regulation of smooth muscle cell proliferation | MMP9, IL-6, IL-13, IL-10 | 0.00021 |
| GO:0042060 | Wound healing | COL5A1, COL1A1, COL3A1, TIMP1, HIF1A, VEGFA, IL-6, TGFB-1 | 3.4 × 10−6 |
| GO:0060485 | Mesenchyme development | ACTA2/SMA, TGFB1, HIF1A | 0.0299 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavakkoli, F.; Damala, M.; Koduri, M.A.; Gangadharan, A.; Rai, A.K.; Dash, D.; Basu, S.; Singh, V. Transcriptomic Profiling of Human Limbus-Derived Stromal/Mesenchymal Stem Cells—Novel Mechanistic Insights into the Pathways Involved in Corneal Wound Healing. Int. J. Mol. Sci. 2022, 23, 8226. https://doi.org/10.3390/ijms23158226
Tavakkoli F, Damala M, Koduri MA, Gangadharan A, Rai AK, Dash D, Basu S, Singh V. Transcriptomic Profiling of Human Limbus-Derived Stromal/Mesenchymal Stem Cells—Novel Mechanistic Insights into the Pathways Involved in Corneal Wound Healing. International Journal of Molecular Sciences. 2022; 23(15):8226. https://doi.org/10.3390/ijms23158226
Chicago/Turabian StyleTavakkoli, Fatemeh, Mukesh Damala, Madhuri Amulya Koduri, Abhilash Gangadharan, Amit K. Rai, Debasis Dash, Sayan Basu, and Vivek Singh. 2022. "Transcriptomic Profiling of Human Limbus-Derived Stromal/Mesenchymal Stem Cells—Novel Mechanistic Insights into the Pathways Involved in Corneal Wound Healing" International Journal of Molecular Sciences 23, no. 15: 8226. https://doi.org/10.3390/ijms23158226
APA StyleTavakkoli, F., Damala, M., Koduri, M. A., Gangadharan, A., Rai, A. K., Dash, D., Basu, S., & Singh, V. (2022). Transcriptomic Profiling of Human Limbus-Derived Stromal/Mesenchymal Stem Cells—Novel Mechanistic Insights into the Pathways Involved in Corneal Wound Healing. International Journal of Molecular Sciences, 23(15), 8226. https://doi.org/10.3390/ijms23158226







