Electronic Structure and Solvation Effects from Core and Valence Photoelectron Spectroscopy of Serum Albumin
Abstract
:1. Introduction
2. Results and Discussion
2.1. Stability of the Protein in the Liquid Jet and under Irradiation
2.2. Core Ionization Study
2.3. Valence Band Edge Analysis
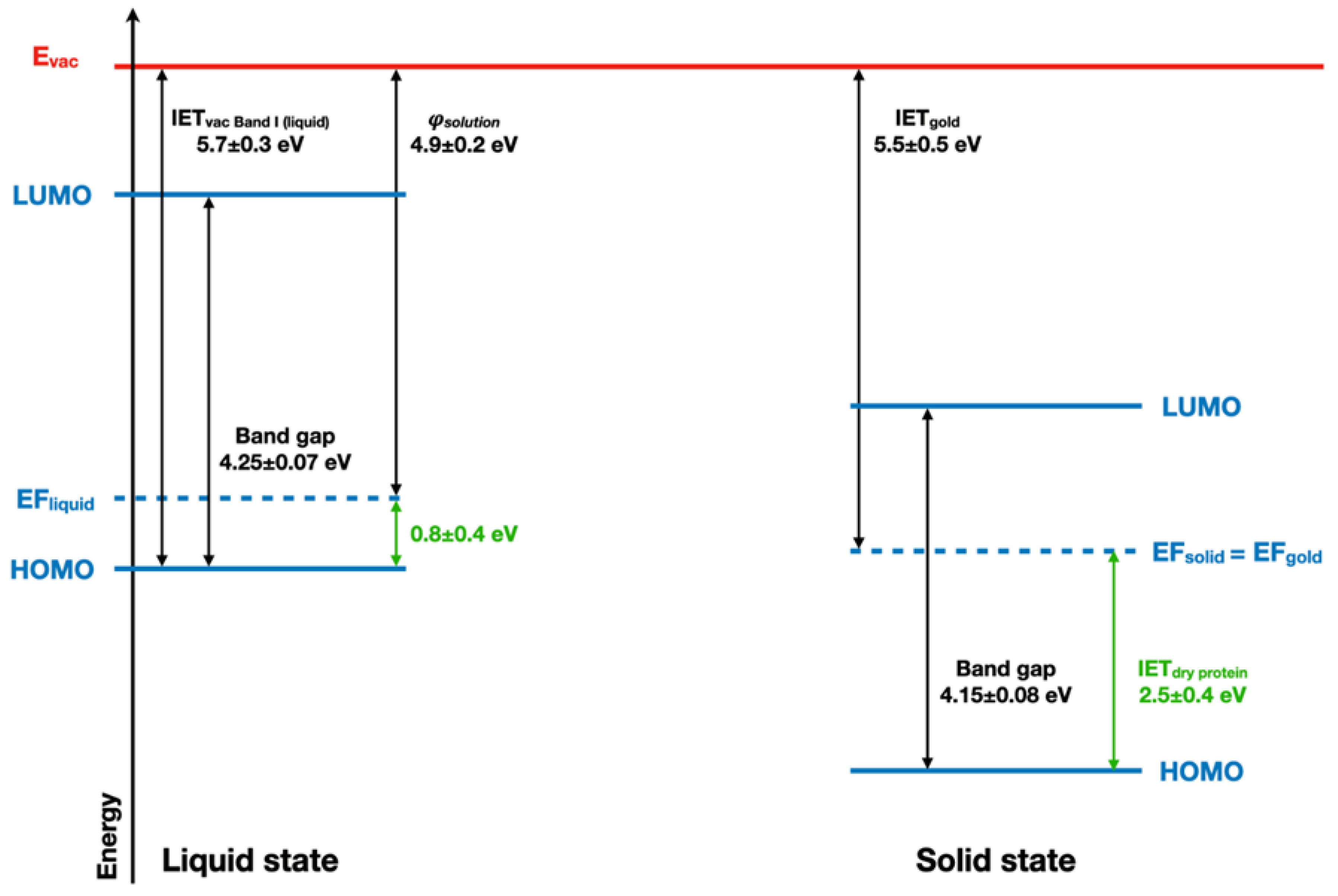
2.4. Implications of Ionization Energies and Work Functions of the Hydrated BSA
3. Materials and Methods
3.1. Chemicals
3.2. Photoelectron Spectroscopy
3.3. Structural Characterization
3.4. Computational Details
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sylvetsky, N. Toward Simple, Predictive Understanding of Protein-Ligand Interactions: Electronic Structure Calculations on Torpedo Californica Acetylcholinesterase Join Forces with the Chemist’s Intuition. Sci. Rep. 2020, 10, 9218. [Google Scholar] [CrossRef]
- Gogonea, V.; Shy, J.M.; Biswas, P.K. Electronic structure, ionization potential, and electron affinity of the enzyme cofactor (6R)-5,6,7,8-Tetrahydrobiopterin in the gas phase, solution, and protein environments. J. Phys. Chem. B 2006, 110, 22861–22871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, H.B.; Winkler, J.R. Electron tunneling through proteins. Q. Rev. Biophys. 2003, 36, 341–372. [Google Scholar] [CrossRef]
- De Vault, D.; Chance, B. Studies of Photosynthesis Using a Pulsed Laser: I. Temperature Dependence of Cytochrome Oxidation Rate in Chromatium. Evidence for Tunneling. Biophys. J. 1966, 6, 825–847. [Google Scholar] [CrossRef] [Green Version]
- Winkler, J.R.; Gray, H.B. Long-range electron tunneling. J. Am. Chem. Soc. 2014, 136, 2930–2939. [Google Scholar] [CrossRef]
- Beratan, D.N.; Onuchic, J.N. Electron tunneling pathways in proteins: Influences on the transfer rate. Photosynth. Res. 1989, 22, 173–186. [Google Scholar] [CrossRef]
- Faubel, M.; Schlemmer, S.; Tocnnies, J.P. A molecular beam study of the evaporation of water from a liquid jet. Z. Für Phys. D At. Mol. Clust. 1988, 10, 269–277. [Google Scholar] [CrossRef]
- Siegbahn, H.; Siegbahn, K. ESCA applied to liquids. J. Electron. Spectrosc. Relat. Phenom. 1973, 2, 319–325. [Google Scholar] [CrossRef]
- Yepes, D.; Seidel, R.; Winter, B.; Blumberger, J.; Jaque, P. Photoemission spectra and density functional theory calculations of 3d transition metal-aqua complexes (Ti-Cu) in aqueous solution. J. Phys. Chem. B 2014, 118, 6850–6863. [Google Scholar] [CrossRef] [PubMed]
- Jain, V.; Kjærvik, M.; Bahr, S.; Dietrich, P.; Meyer, M.; Thißen, A.; Linford, M.R. Bovine serum albumin, aqueous solution, by near-ambient pressure XPS. Surf. Sci. Spectra 2019, 26, 014027. [Google Scholar] [CrossRef]
- Ottosson, N.; Børve, K.J.; Spangberg, D.; Bergersen, H.; Sæthre, L.J.; Faubel, M.; Pokapanich, W.; Öhrwall, G.; Björneholm, O.; Winter, B. On the Origins of Core−Electron Chemical Shifts of Small Biomolecules in Aqueous Solution: Insights from Photoemission and ab Initio Calculations of Glycine aq. J. Am. Chem. Soc. 2011, 133, 3120–3130. [Google Scholar] [CrossRef] [PubMed]
- Ottosson, N.; Romanova, A.O.; Soderstrom, J.; Bjorneholm, O.; Ohrwall, G.; Fedorov, M.V. Molecular sinkers: X-ray photoemission and atomistic simulations of benzoic acid and benzoate at the aqueous solution/vapor interface. J. Phys. Chem. B 2012, 116, 13017–13023. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Vila, F.; Sterrer, M.; Thürmer, S.; Winter, B.; Ammann, M.; Rehr, J.J.; van Bokhoven, J.A. Electronic structures of formic acid (HCOOH) and formate (HCOO-) in aqueous solutions. J. Phys. Chem. Lett. 2012, 3, 1754–1759. [Google Scholar] [CrossRef] [PubMed]
- Makowski, M.J.; Galhenage, J.; Langford, J.; Hemminger, C. Liquid-Jet X-ray Photoelectron Spectra of TiO2 Nanoparticles in an Aqueous Electrolyte Solution. J. Phys. Chem. Lett. 2016, 7, 1732–1735. [Google Scholar] [CrossRef]
- Brown, M.A.; Jordan, I.; Redondo, A.B.; Kleibert, A.; Wörner, H.J.; van Bokhoven, J.A. In situ photoelectron spectroscopy at the liquid/nanoparticle interface. Surf. Sci. 2013, 610, 1–6. [Google Scholar] [CrossRef]
- Peters, T. All About Albumin; Elsevier: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Butler, J.; Jayson, G.G.; Swallow, A.J. The reaction between the superoxide anion radical and cytochrome c. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 1975, 408, 215–222. [Google Scholar] [CrossRef]
- D’Imprima, E.; Floris, D.; Joppe, M.; Sánchez, R.; Grininger, M.; Kühlbrandt, W. Protein denaturation at the air-water interface and how to prevent it. Elife 2019, 8, e42747. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shrestha, M.; Luo, M.; Sit, I.; Song, M.; Grassian, V.H.; Xiong, W. Salting up of Proteins at the Air/Water Interface. Langmuir 2019, 35, 13815–13820. [Google Scholar] [CrossRef]
- Feldhoff, R.C.; Peters, T. Fragments of Bovine Serum Albumin Produced by Limited Proteolysis. Isolation and Characterization of Peptic Fragments. Biochemistry 1975, 14, 4508–4514. [Google Scholar] [CrossRef]
- Gülseren, I.; Güzey, D.; Bruce, B.D.; Weiss, J. Structural and functional changes in ultrasonicated bovine serum albumin solutions. Sonochemistry 2007, 14, 173–183. [Google Scholar] [CrossRef]
- Leggio, C.; Galantini, L.; Pavel, N.V. About the albumin structure in solution: Cigar expanded form versus heart normal shape. Phys. Chem. Chem. Phys. 2008, 10, 6741–6750. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, G.; Goel, A.; Kleibert, A.; Cvetko, D.; Brown, M.A. Quantitative ionization energies and work functions of aqueous solutions. Phys. Chem. Chem. Phys. 2016, 18, 29506–29515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tissot, H.; Gallet, J.J.; Bournel, F.; Olivieri, G.; Silly, M.G.; Sirotti, F.; Boucly, A.; Rochet, F. The Electronic Structure of Saturated NaCl and NaI Solutions in Contact with a Gold Substrate. Top. Catal. 2016, 59, 605–620. [Google Scholar] [CrossRef]
- Thürmer, S.; Malerz, S.; Trinter, F.; Hergenhahn, U.; Lee, C.; Neumark, D.M.; Meijer, G.; Winter, B.; Wilkinson, L. Accurate Vertical Ionization Energy and Work Function Determinations of Liquid Water and Aqueous Solutions. Chem. Sci. 2021, 12, 10558–10582. [Google Scholar] [CrossRef] [PubMed]
- Björneholm, O.; Hansen, M.H.; Hodgson, A.; Liu, L.M.; Limmer, D.T.; Michaelides, A.; Pedevilla, P.; Rossmeisl, J.; Shen, H.; Tocci, G.; et al. Water at Interfaces. Chem. Rev. 2016, 116, 7698–7726. [Google Scholar] [CrossRef] [PubMed]
- Walz, M.M.; Caleman, C.; Werner, J.; Ekholm, V.; Lundberg, D.; Prisle, N.L.; Öhrwall, G.; Björneholm, O. Surface behavior of amphiphiles in aqueous solution: A comparison between different pentanol isomers. Phys. Chem. Chem. Phys. 2015, 17, 14036–14044. [Google Scholar] [CrossRef] [Green Version]
- Paynter, R.W.; Benalia, H. A time- and angle-resolved X-ray photoelectron spectroscopy study of polystyrene exposed to a nitrogen plasma. J. Electron. Spectrosc. Relat. Phenom. 2004, 136, 209–220. [Google Scholar] [CrossRef]
- Gelius, U.; Siegbahn, K. ESCA studies of molecular core and valence levels in the gas phase. Gen. Discuss. Faraday Soc. 1972, 54, 257–268. [Google Scholar] [CrossRef]
- Hernandez-Laguna, A.; Maruani, J.; McWeeny, R.; Wilson, S. Quantum Systems in Chemistry and Physics; Kluwer Academic Publishers: Dordrecht, The Netherland, 2002. [Google Scholar]
- Stasyuk, A.J.; Solà, M.; Voityuk, A.A. Reliable charge assessment on encapsulated fragment for endohedral systems. Sci. Rep. 2018, 8, 2882. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2017, 27, 112–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, B.; Weber, R.; Widdra, W.; Dittmar, M.; Faubel, M.; Hertel, V. Full Valence Band Photoemission from Liquid Water Using EUV Synchrotron Radiation. J. Phys. Chem. A 2004, 108, 2625–2632. [Google Scholar] [CrossRef]
- Wolak, M.A.; Balaeff, A.; Gutmann, S.; Helmrich, H.J.; Vosloo, R.; Beerbom, M.M.; Wierzbinski, E.; Waldeck, D.H.; Bezer, S.; Achim, C.; et al. Electronic structure of self-assembled peptide nucleic acid thin films. J. Phys. Chem. C 2011, 115, 17123–17135. [Google Scholar] [CrossRef]
- Sepunaru, L.; Refaely-abramson, S.; Lovrinc, R.; Gavrilov, Y.; Agrawal, P. Electronic transport via homopeptides: The role of side chains and secondary structure. J. Am. Chem. Soc. 2015, 137, 9617–9626. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.C.; Yu, Y.; Wu, Z.H.; Lin, P.C.; Chen, W.R.; Su, C.C.; Chen, M.S.; Li, Y.L.; Huang, T.P.; Lee, Y.Y.; et al. Molecular Basis of the Antioxidant Capability of Glutathione Unraveled via Aerosol VUV Photoelectron Spectroscopy. J. Phys. Chem. B 2016, 120, 10181–10191. [Google Scholar] [CrossRef] [PubMed]
- Baghbanzadeh, M.; Bowers, C.M.; Rappoport, D.; Żaba, T.; Gonidec, M.; Al-Sayah, M.H.; Cyganik, P.; Aspuru-Guzik, A.; Whitesides, G.M. Charge Tunneling along Short Oligoglycine Chains. Angew. Chem. 2015, 127, 14956–14960. [Google Scholar] [CrossRef] [Green Version]
- Ariyasu, S.; Hayashi, H.; Xing, B.; Chiba, S. Site-Specific Dual Functionalization of Cysteine Residue in Peptides and Proteins with 2-Azidoacrylates. Bioconj. Chem. 2017, 28, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Kraut, E.A.; Grant, R.W.; Waldrop, J.R.; Kowalczyk, S.P. Precise Determination of the Valence-Band Edge in X Ray Photoemission Spectra. Phys. Rev. Lett. 1980, 44, 1620. [Google Scholar] [CrossRef]
- Lever, G.; Cole, D.J.; Hine, N.D.M.; Haynes, P.D.; Payne, M.C. Electrostatic considerations affecting the calculated HOMO-LUMO gap in protein molecules. J. Phys. Condens. Matter 2013, 25, 152101. [Google Scholar] [CrossRef]
- Kühne, T.D.; Iannuzzi, M.; Del Ben, M.; Rybkin, V.V.; Seewald, P.; Stein, F.; Laino, T.; Khaliullin, R.Z.; Schütt, O.; Schiffmann, F.; et al. CP2K: An electronic structure and molecular dynamics software package-Quickstep: Efficient and accurate electronic structure calculations. J. Chem. Phys. 2020, 152, 19413. [Google Scholar] [CrossRef] [PubMed]
- Pradier, C.M.; Ka, F. Adsorption of Bovine Serum Albumin on Chromium and Molybdenum Surfaces Investigated by Fourier-Transform Infrared Reflection-Absorption Spectroscopy (FT-IRRAS) and X-ray Photoelectron Spectroscopy. J. Phys. Chem. B 2003, 107, 6766–6773. [Google Scholar] [CrossRef]
- Salaneck, W.R. Intermolecular relaxation energies in anthracene. Phys. Rev. Lett. 1978, 40, 60–63. [Google Scholar] [CrossRef]
- Kera, S.; Ueno, N. Photoelectron spectroscopy on the charge reorganization energy and small polaron binding energy of molecular film. J. Electron. Spectrosc. Relat. Phenom. 2015, 204, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Pawlik, S.; Bauer, M.; Aeschlimann, M. Lifetime difference of photoexcited electrons between intraband and interband transitions. Surf. Sci. 1997, 377, 206–209. [Google Scholar] [CrossRef]
- Green, J.C.; Decleva, P. Photoionization cross-sections: A guide to electronic structure. Coord. Chem. Rev. 2005, 249, 209–228. [Google Scholar] [CrossRef]
- Allan, C.J.; Gelius, D.U.; Allison, D.A.; Johansson, G.; Siegbahn, H.; Siegbahn, K. ESCA studies of CO2, CS2 and COS. J. Electron. Spectrosc. Relat. Phenom. 1972, 1, 131–151. [Google Scholar] [CrossRef]
- Yeh, J.-J. Atomic Calculation of Photoionization Cross-Sections and Asymmetry Parameters; Gordon and Breach Science Publishes: Philadelphia, PA, USA, 1993. [Google Scholar]
- Yeh, J.J.; Lindau, I. Copyright 0 1985 by Academic Press, Inc. At. Data Nucl. Data Tables 1985, 32, 1–155. [Google Scholar] [CrossRef]
- Franchini, C.; Reticcioli, M.; Setvin, M.; Diebold, U. Polarons in materials. Nat. Rev. Mater. 2021, 6, 560–586. [Google Scholar] [CrossRef]
- Nicolas, C.; Miron, C. Lifetime broadening of core-excited and -ionized states. J. Electron. Spectrosc. Relat. Phenom. 2012, 185, 267–272. [Google Scholar] [CrossRef]
- Chuev, G.N.; Lakhno, V.D. A polaron model for electron transfer in globular proteins. J. Biol. 1993, 163, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Płodzień, M.; Sowiński, T.; Kokkelmans, S. Simulating polaron biophysics with Rydberg atoms. Sci. Rep. 2018, 8, 9247. [Google Scholar] [CrossRef]
- Mallamace, F.; Baglioni, P.; Corsaro, C.; Chen, S.H.; Mallamace, D.; Vasi, C.; Stanley, H.E. The influence of water on protein properties. J. Chem. Phys. 2014, 141, 165104. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, B. Electrical conductivity of proteins. Nature 1962, 193, 364–365. [Google Scholar] [CrossRef] [PubMed]
- Gathercole, L.J.; Miles, M.J.; McMaster, T.J.; Holmes, D.F. Scanning probe microscopy of collagen I and pN-collagen I assemblies and the relevance to scanning tunnelling microscopy contrast generation in proteins. J. Chem. Soc. Faraday Trans. 1993, 89, 2589–2594. [Google Scholar] [CrossRef]
- Reali, M.; Saini, P.; Santato, C. Electronic and protonic transport in bio-sourced materials: A new perspective on semiconductivity. Mater. Adv. 2021, 2, 15–31. [Google Scholar] [CrossRef]
- For, E.; Positive, P.; Transport, H. Evidence for Possible Positive Hole Transport. J. Radioanal. Nucl. Chem. 1993, 171, 443–450. [Google Scholar]
- Fasman, G.D. Practical Handbook of Biochemistry and Molecular Biology; CRC Press: Boston, MA, USA, 1989. [Google Scholar]
- Malerz, S.; Trinter, F.; Hergenhahn, U.; Ghrist, A.; Ali, H.; Nicolas, C.; Saak, C.M.; Richter, C.; Hartweg, S.; Nahon, L.; et al. Low-energy constraints on photoelectron spectra measured from liquid water and aqueous solutions. Phys. Chem. Chem. Phys. 2021, 23, 8246–8260. [Google Scholar] [CrossRef] [PubMed]
- Truong, S.Y.; Yencha, A.J.; Juarez, A.M.; Cavanagh, S.J.; Bolognesi, P.; King, G.C. Threshold photoelectron spectroscopy of H2O and D2O over the photon energy range 12-40 eV. Chem. Phys. 2009, 355, 183–193. [Google Scholar] [CrossRef]
- Faubel, M.; Steiner, B.; Toennies, J.P. Photoelectron spectroscopy of liquid water, some alcohols, and pure nonane in free micro jets. J. Chem. Phys. 1997, 106, 9013–9031. [Google Scholar] [CrossRef]
- Winter, B.; Aziz, E.F.; Hergenhahn, U.; Faubel, M.; Hertel, I.V. Hydrogen bonds in liquid water studied by photoelectron spectroscopy. J. Chem. Phys. 2007, 126, 124504. [Google Scholar] [CrossRef]
- Nishizawa, K.; Kurahashi, N.; Sekiguchi, K.; Mizuno, T.; Ogi, Y.; Horio, T.; Oura, M.; Kosugi, N.; Suzuki, T. High-resolution soft X-ray photoelectron spectroscopy of liquid water. Phys. Chem. Chem. Phys. 2011, 13, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Fairley, N.; Fernandez, V.; Richard-Plouet, M.; Guillot-Deudon, C.; Walton, J.; Smith, E.; Flahaut, D.; Greiner, M.; Biesinger, M.; Tougaard, S.; et al. Systematic and collaborative approach to problem solving using X-ray photoelectron spectroscopy. Appl. Surf. Sci. Adv. 2021, 5, 100112. [Google Scholar] [CrossRef]
- Tougaard, S. Quantitative analysis of the inelastic background in surface electron spectroscopy. Surf. Interface Anal. 1988, 11, 453–472. [Google Scholar] [CrossRef]
- Tougaard, S. Universality classes of inelastic electron scattering cross-sections. Surf. Interface Anal. 1997, 25, 137–154. [Google Scholar] [CrossRef]
- Réfrégiers, M.; Wien, F.; Ta, H.P.; Premvardhan, L.; Bac, S.; Jamme, F.; Rouam, V.; Lagarde, B.; Polack, F.; Giorgetta, J.L.; et al. DISCO synchrotron-radiation circular-dichroism endstation at SOLEIL. J. Synchrotron Radiat. 2012, 19, 831–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miles, A.J.; Wallace, B.A. CDtoolX, a downloadable software package for processing and analyses of circular dichroism spectroscopic data. Protein Sci. 2018, 27, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl Acad Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1278–1289. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Persson, F.; Söderhjelm, P.; Halle, B. The geometry of protein hydration. J. Chem. Phys. 2018, 148, 215101. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, A.C.; Laage, D. Water Dynamics in Protein Hydration Shells: The Molecular Origins of the Dynamical Perturbation. J. Phys. Chem. B 2014, 118, 7715. [Google Scholar] [CrossRef]
- Goedecker, S.; Teter, M. Separable dual-space Gaussian pseudopotentials. Phys. Rev. B-Condens. Matter Mater. Phys. 1996, 54, 1703–1710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VandeVondele, J.; Hutter, J. Gaussian basis sets for accurate calculations on molecular systems in gas and condensed phases. J. Chem. Phys. 2007, 127, 114105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez Ramírez, L.; Boucly, A.; Saudrais, F.; Bournel, F.; Gallet, J.J.; Maisonhaute, E.; Milosavljević, A.R.; Nicolas, C.; Rochet, F. The Fermi Level as an Energy Reference in Liquid Jet X-Ray Photoelectron Spectroscopy Studies of Aqueous Solutions. Phys. Chem. Chem. Phys. 2021, 23, 16224–16233. [Google Scholar] [CrossRef] [PubMed]
- Tauc, J.; Grigorovici, R.; Vancu, A. Optical Properties and Electronic Structure of Amorphous Germanium. Physica Status Solidi (b) 1966, 15, 627–637. [Google Scholar] [CrossRef]
- Muñoz, A.I.; Mischler, S. Electrochimica Acta Electrochemical Quartz Crystal Microbalance and X-Ray Photoelectron Spectroscopy Study of Cathodic Reactions in Bovine Serum Albumin Containing Solutions on a Physical Vapour Deposition-CoCrMo Biomedical Alloy. Electrochim. Acta 2015, 180, 96–103. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, X.; Dou, C.; Liu, H.; Wang, S.; Shen, P. Structural Studies on Metal-Serum Albumin. IV. The Interaction of Zn(II), Cd(II) and Hg(II) with HSA and BSA. Biophys. Chem. 1992, 42, 201–211. [Google Scholar] [PubMed]
- Debiemme-chouvy, C.; Haskouri, S.; Cachet, H. Study by XPS of the Chlorination of Proteins Aggregated onto Tin Dioxide during Electrochemical Production of Hypochlorous Acid. Appl. Surf. Sci. 2007, 253, 5506–5510. [Google Scholar] [CrossRef]
- Chen, W.; Wu, X.; Car, R. X-Ray Absorption Signatures of the Molecular Environment in Water and Ice. Phys. Rev. Lett. 2010, 017802, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boonaert, C.J.P.; Dufre, Y.F.; Derclaye, S.R.; Rouxhet, P.G. Adhesion of Lactococcus Lactis to Model Substrata: Direct Study of the Interface. Colloids Surf. B Biointerfaces 2001, 22, 171–182. [Google Scholar] [CrossRef]
- Torres Bautista, B.E.; Carvalho, M.L.; Seyeux, A.; Zanna, S.; Cristiani, P.; Tribollet, B.; Marcus, P.; Frateur, I. Effect of Protein Adsorption on the Corrosion Behavior of 70Cu-30Ni Alloy in Artificial Seawater. Bioelectrochemistry 2014, 97, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Fears, K.P. Measuring the PK/PI of Biomolecules Using X-Ray Photoelectron Spectroscopy. Anal. Chem. 2014, 86, 8526–8529. [Google Scholar] [CrossRef] [PubMed]
- Valero-Vidal, C.; Igual-Muñoz, A.; Olsson, C.-O.A.; Mischler, S. Adsorption of BSA on Passivated CoCrMo PVD Alloy: An EQCM and XPS Investigation. J. Electrochem. Soc. 2014, 161, C294–C301. [Google Scholar] [CrossRef]
- Frateur, I.; Lartundo-Rojas, L.; Méthivier, C.; Galtayries, A.; Marcus, P. Influence of Bovine Serum Albumin in Sulphuric Acid Aqueous Solution on the Corrosion and the Passivation of an Iron-Chromium Alloy. Electrochim. Acta 2006, 51, 1550–1557. [Google Scholar] [CrossRef]
- Swaidan, A.; Borthakur, P.; Boruah, P.K.; Das, M.R.; Barras, A.; Hamieh, S.; Toufaily, J.; Hamieh, T.; Szunerits, S.; Boukherroub, R. A Facile Preparation of CuS-BSA Nanocomposite as Enzyme Mimics: Application for Selective and Sensitive Sensing of Cr(VI) Ions. Sens. Actuators B Chem. 2019, 294, 253–262. [Google Scholar] [CrossRef]
- Sanner, M.F.; Olson, A.J.; Spehner, J.C. Reduced Surface: An Efficient Way to Compute Molecular Surfaces. Biopolymers 1996, 38, 305–320. [Google Scholar] [CrossRef]

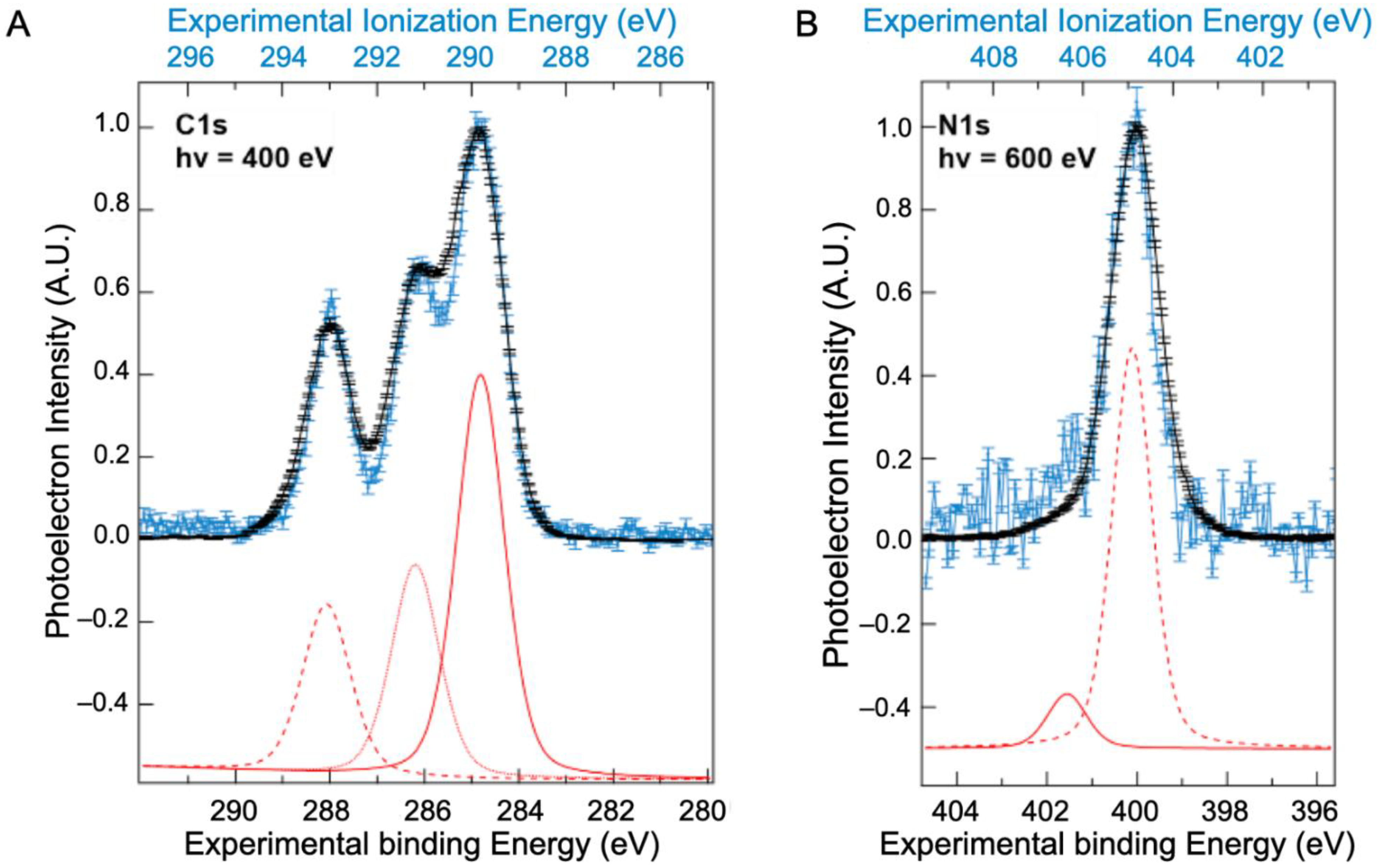
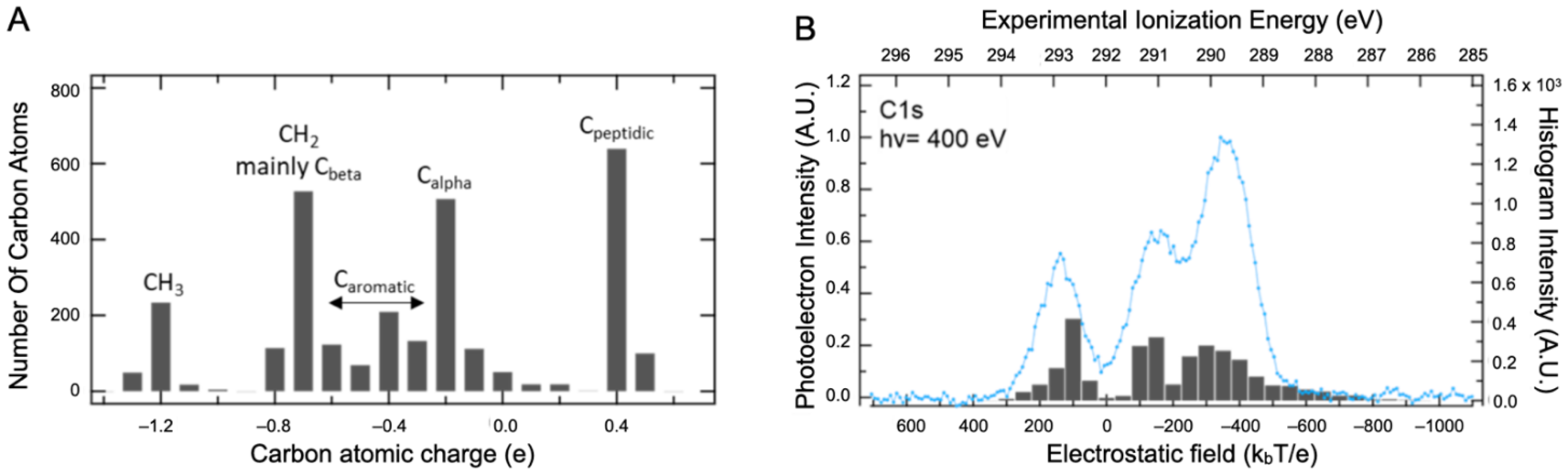
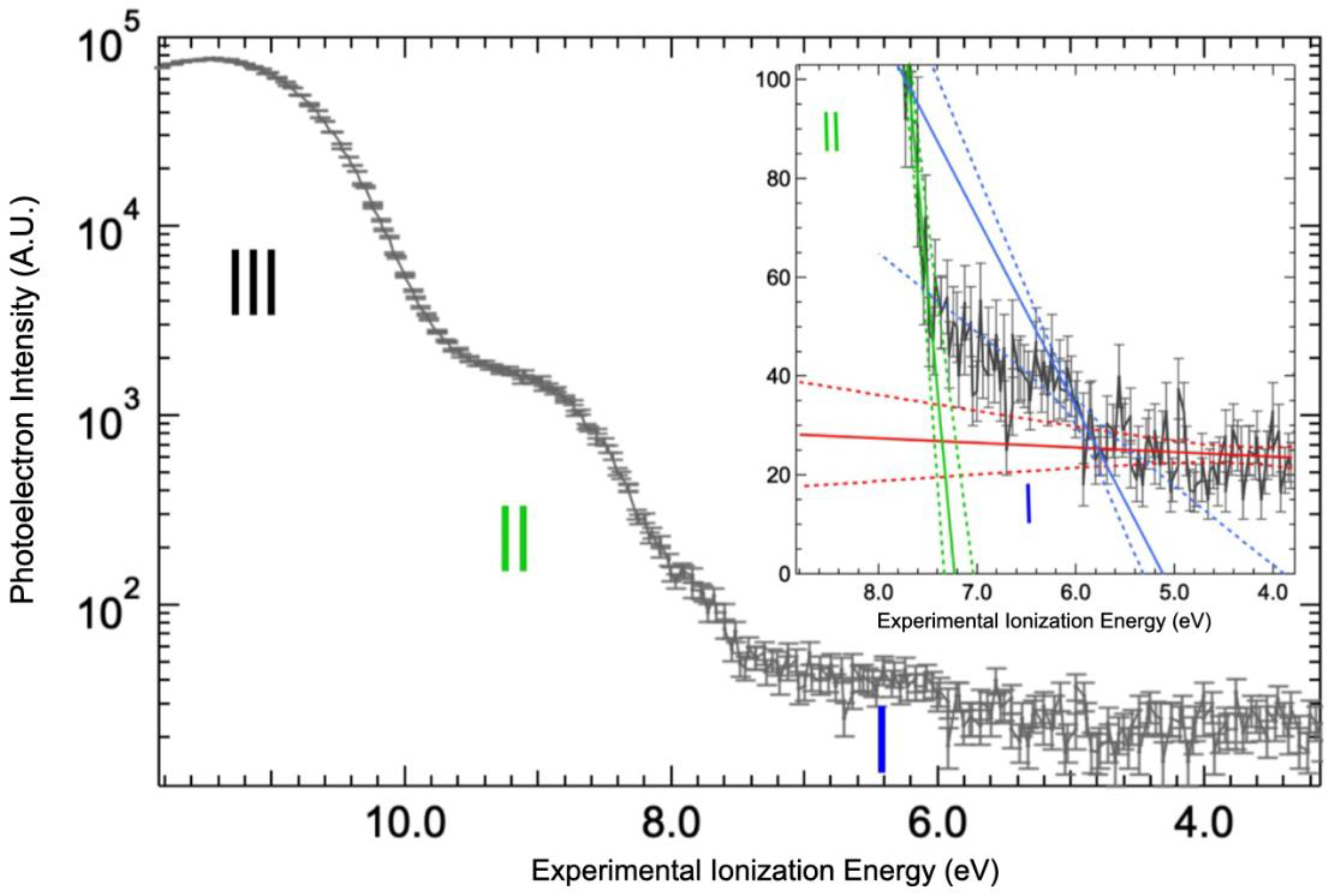
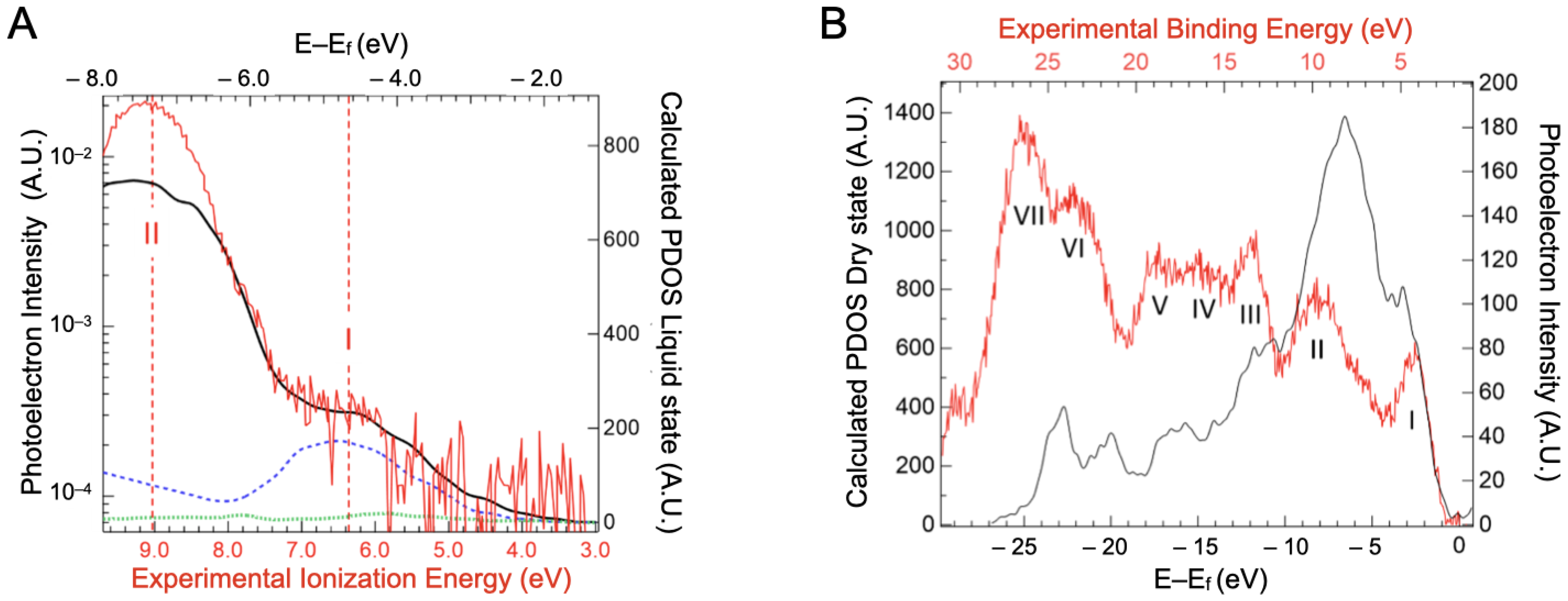
| Solid State | Liquid State | Liquid State | C 1s Attribution and %At [10] | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (This Study) | (This Study) | (NAP-XPS) [10] | ||||||||
| Position (±0.35 eV) | %At | FWHM (eV) | Position (±0.13 eV) | %At | FWHM (eV) | Position (±1.7 eV) | FWHM (eV) | |||
| C1s | 284.9 | 47 | 1.18 | 289.8 | 52.5 | 0.93 | 285 | 1.47 | C-C/C-H | 46 |
| 286.2 | 29 | 1.18 | 291.1 | 26 | 0.93 | 286.2 | 1.47 | C-O/C-N | 28 | |
| 288.0 | 24 | 1.18 | 293.0 | 21.5 | 0.93 | 287.4 | 1.47 | N-C=O | 24 | |
| 288.6 | 1.47 | O-C=O | 2 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renault, J.-P.; Huart, L.; Milosavljević, A.R.; Bozek, J.D.; Palaudoux, J.; Guigner, J.-M.; Marichal, L.; Leroy, J.; Wien, F.; Hervé Du Penhoat, M.-A.; et al. Electronic Structure and Solvation Effects from Core and Valence Photoelectron Spectroscopy of Serum Albumin. Int. J. Mol. Sci. 2022, 23, 8227. https://doi.org/10.3390/ijms23158227
Renault J-P, Huart L, Milosavljević AR, Bozek JD, Palaudoux J, Guigner J-M, Marichal L, Leroy J, Wien F, Hervé Du Penhoat M-A, et al. Electronic Structure and Solvation Effects from Core and Valence Photoelectron Spectroscopy of Serum Albumin. International Journal of Molecular Sciences. 2022; 23(15):8227. https://doi.org/10.3390/ijms23158227
Chicago/Turabian StyleRenault, Jean-Philippe, Lucie Huart, Aleksandar R. Milosavljević, John D. Bozek, Jerôme Palaudoux, Jean-Michel Guigner, Laurent Marichal, Jocelyne Leroy, Frank Wien, Marie-Anne Hervé Du Penhoat, and et al. 2022. "Electronic Structure and Solvation Effects from Core and Valence Photoelectron Spectroscopy of Serum Albumin" International Journal of Molecular Sciences 23, no. 15: 8227. https://doi.org/10.3390/ijms23158227
APA StyleRenault, J.-P., Huart, L., Milosavljević, A. R., Bozek, J. D., Palaudoux, J., Guigner, J.-M., Marichal, L., Leroy, J., Wien, F., Hervé Du Penhoat, M.-A., & Nicolas, C. (2022). Electronic Structure and Solvation Effects from Core and Valence Photoelectron Spectroscopy of Serum Albumin. International Journal of Molecular Sciences, 23(15), 8227. https://doi.org/10.3390/ijms23158227







