Hematopoietic–Mesenchymal Signals Regulate the Properties of Mesenchymal Stem Cells
Abstract
:1. Introduction
2. Results
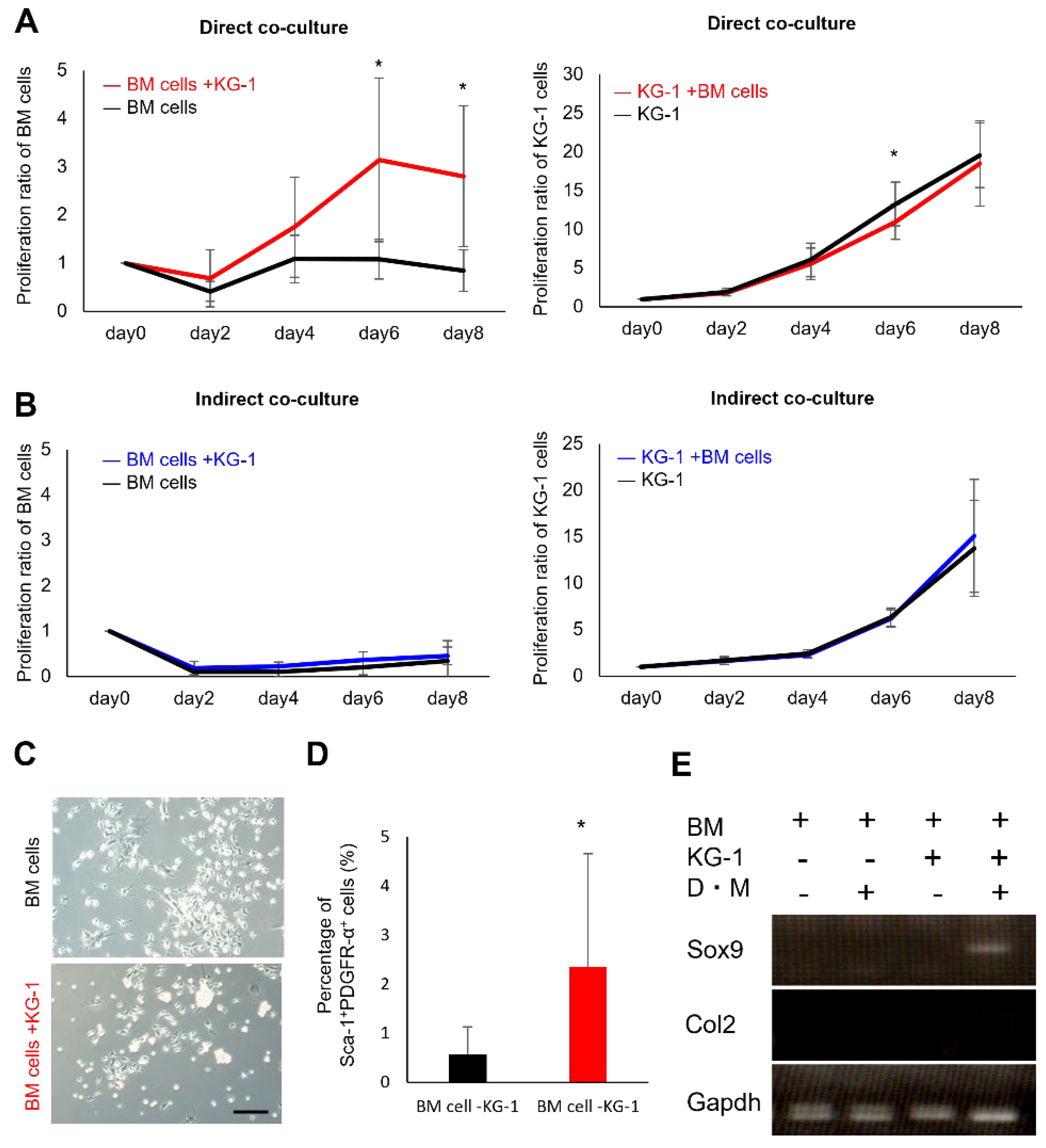
2.1. Proliferation and Chondrogenic Potential of Mesenchymal Cells Increase during Co-Culture with Hematopoietic Cells
2.2. Direct Mesenchymal-Hematopoietic Cell Interaction Promotes the Stemness of MSCs
2.3. Co-Culture with HSCs Promotes Collagen Synthesis and Adipogenic Differentiation
2.4. MSCs Lacking Mpl Tend to Have Enhanced Differentiation Activity
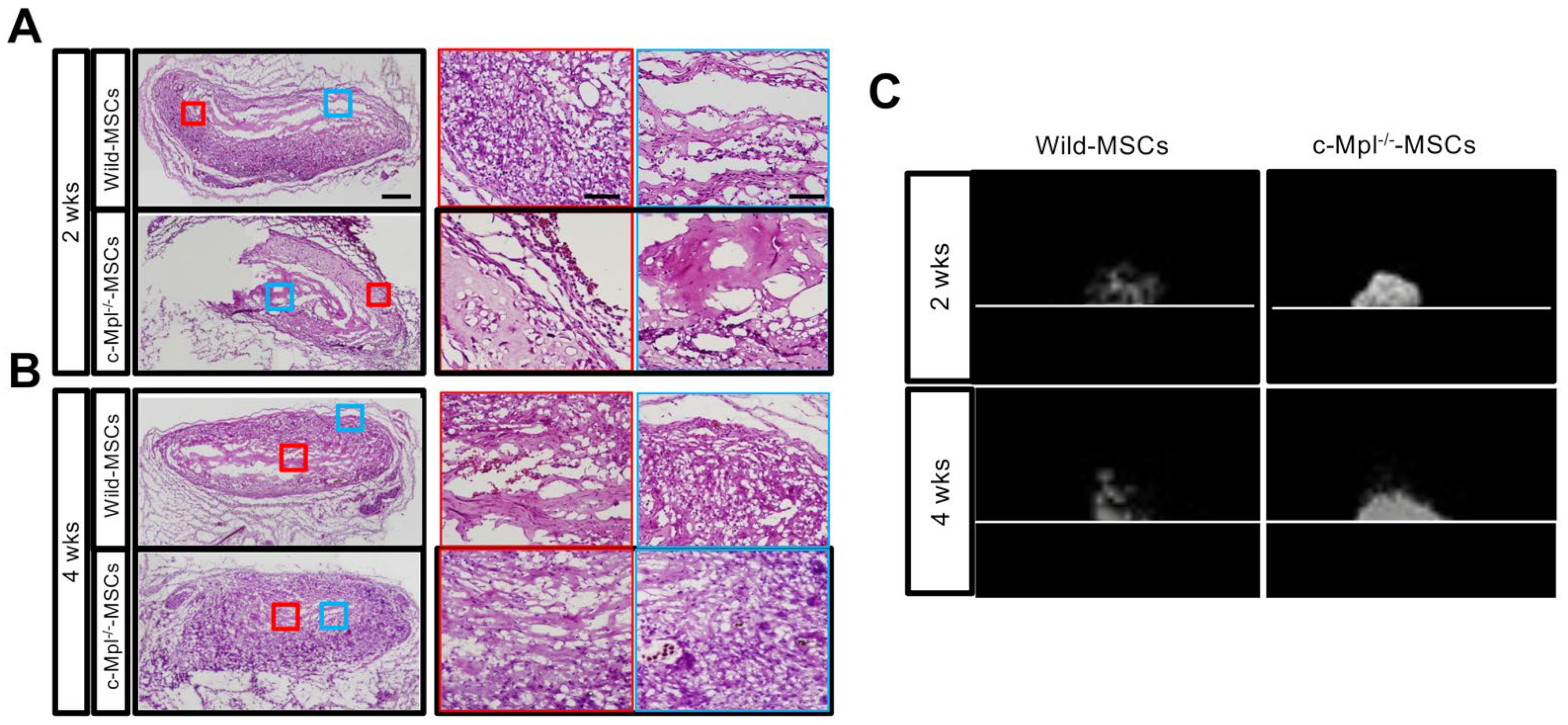
2.5. c-Mpl Gene Deficiency Affects the Maintenance of Ectopic Bone
2.6. The c-Mpl in HSCs Regulates MSC Stem Cell Properties
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Preparation of BM Cell Suspension
4.3. Antibody Staining and Flow Cytometry
4.4. Cell Culture
4.5. CFU-F Assay
4.6. Differentiation Cultures
4.7. Co-Culture of MSCs with Hematopoietic Cells
4.8. RNA Isolation and RT-PCR
4.9. Real-Time RT-PCR Analysis for Selected mRNAs
4.10. Microarray
4.11. GSEA of Microarray
4.12. In Vivo Bone and Stroma Assay
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Uejima, S.; Okada, K.; Kagami, H.; Taguchi, A.; Ueda, M. Bone marrow stromal cell therapy improves femoral bone mineral density and mechanical strength in ovariectomized rats. Cytotherapy 2008, 10, 479–489. [Google Scholar] [PubMed]
- Petrenko, Y.; Vackova, I.; Kekulova, K.; Chudickova, M.; Koci, Z.; Turnovcova, K.; Kupcova Skalnikova, H.; Vodicka, P.; Kubinova, S. A Comparative Analysis of Multipotent Mesenchymal Stromal Cells derived from Different Sources, with a Focus on Neuroregenerative Potential. Sci. Rep. 2020, 10, 4290. [Google Scholar] [PubMed]
- Mabuchi, Y.; Morikawa, S.; Harada, S.; Akazawa, C.; Okano, H.; Matsuzaki, Y. LNGFR(+)THY-1(+)VCAM-1(hi+) cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Rep. 2013, 1, 152–165. [Google Scholar]
- Suto, E.G.; Mabuchi, Y.; Suzuki, N.; Suzuki, K.; Ogata, Y.; Taguchi, M.; Muneta, T.; Sekiya, I.; Akazawa, C. Prospectively isolated mesenchymal stem/stromal cells are enriched in the CD73+ population and exhibit efficacy after transplantation. Sci. Rep. 2017, 7, 4838. [Google Scholar] [PubMed] [Green Version]
- Consentius, C.; Mirenska, A.; Jurisch, A.; Reinke, S.; Scharm, M.; Zenclussen, A.C.; Hennig, C.; Volk, H.D. In situ detection of CD73+ CD90+ CD105+ lineage: Mesenchymal stromal cells in human placenta and bone marrow specimens by chipcytometry. Cytom. A 2018, 93, 889–893. [Google Scholar]
- Moravcikova, E.; Meyer, E.M.; Corselli, M.; Donnenberg, V.S.; Donnenberg, A.D. Proteomic Profiling of Native Unpassaged and Culture-Expanded Mesenchymal Stromal Cells (MSC). Cytom. A 2018, 93, 894–904. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Jahagirdar, B.N.; Reinhardt, R.L.; Schwartz, R.E.; Largaespada, D.A.; Verfaillie, C.M. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 2002, 418, 41–49. [Google Scholar]
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010, 466, 829–834. [Google Scholar]
- Cordeiro Gomes, A.; Hara, T.; Lim, V.Y.; Herndler-Brandstetter, D.; Ikuta, K.; Pereira, J.P. Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity 2016, 45, 1219–1231. [Google Scholar]
- Méndez-Ferrer, S.; Bonnet, D.; Steensma, D.P.; Hasserjian, R.P.; Ghobrial, I.M.; Gribben, J.G.; Andreeff, M.; Krause, D.S. Bone marrow niches in haematological malignancies. Nat. Rev. Cancer 2020, 20, 285–298. [Google Scholar]
- Greenbaum, A.; Hsu, Y.M.; Day, R.B.; Schuettpelz, L.G.; Christopher, M.J.; Borgerding, J.N.; Nagasawa, T.; Link, D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 2013, 495, 227–230. [Google Scholar]
- Hanoun, M.; Arnal-Estapé, A.; Maryanovich, M.; Guha, C.; Frenette, P.S. Nestin+NG2+ Cells Form a Reserve Stem Cell Population in the Mouse Prostate. Stem Cell Rep. 2019, 12, 1201–1211. [Google Scholar]
- Zhao, M.; Tao, F.; Venkatraman, A.; Li, Z.; He, X.C.; Li, L. N-Cadherin-Expressing Bone and Marrow Stromal Progenitor Cells Maintain Reserve Hematopoietic Stem Cells. Cell Rep. 2019, 26, 652–669.e6. [Google Scholar]
- Isern, J.; Martín-Antonio, B.; Ghazanfari, R.; Scheding, S.; Urbano-Ispizúa, Á.; Méndez-Ferrer, S. Self-renewing human bone marrow mesenspheres promote hematopoietic stem cell expansion. Cell Rep. 2013, 3, 1714–1724. [Google Scholar]
- Boulais, P.E.; Mizoguchi, T.; Zimmerman, S.; Nakahara, F.; Vivié, J.; Mar, J.C.; van Oudenaarden, A.; Frenette, P.S. The Majority of CD45− Ter119− CD31− Bone Marrow Cell Fraction Is of Hematopoietic Origin and Contains Erythroid and Lymphoid Progenitors. Immunity 2018, 49, 627–639.e6. [Google Scholar]
- Nakamura-Ishizu, A.; Matsumura, T.; Stumpf, P.S.; MacArthur, B.D.; Suda, T. Thrombopoietin Metabolically Primes Hematopoietic Stem Cells to Megakaryocyte-Lineage Differentiation. Cell Rep. 2018, 25, 1772–1785.e6. [Google Scholar]
- Umemoto, T.; Yamato, M.; Ishihara, J.; Nakauchi, H.; Eto, K.; Okano, T. Integrin-αvβ3 regulates thrombopoietin-mediated maintenance of hematopoietic stem cells. Blood 2012, 119, 83–94. [Google Scholar]
- Kohlscheen, S.; Wintterle, S.; Schwarzer, A.; Baum, C.; Modlich, U. Inhibition of Thrombopoietin/Mpl Signaling in Adult Hematopoiesis Identifies New Candidates for Hematopoietic Stem Cell Maintenance. PLoS ONE 2015, 10, e0131866. [Google Scholar]
- Kovtonyuk, L.V.; Manz, M.G.; Takizawa, H. Enhanced thrombopoietin but not G-CSF receptor stimulation induces self-renewing hematopoietic stem cell divisions in vivo. Blood 2016, 127, 3175–3179. [Google Scholar]
- Nagahisa, H.; Nagata, Y.; Ohnuki, T.; Osada, M.; Nagasawa, T.; Abe, T.; Todokoro, K. Bone marrow stromal cells produce thrombopoietin and stimulate megakaryocyte growth and maturation but suppress proplatelet formation. Blood 1996, 87, 1309–1316. [Google Scholar]
- Kaushansky, K.; Lok, S.; Holly, R.D.; Broudy, V.C.; Lin, N.; Bailey, M.C.; Forstrom, J.W.; Buddle, M.M.; Oort, P.J.; Hagen, F.S. Promotion of megakaryocyte progenitor expansion and differentiation by the c-Mpl ligand thrombopoietin. Nature 1994, 369, 568–571. [Google Scholar]
- Decker, M.; Leslie, J.; Liu, Q.; Ding, L. Hepatic thrombopoietin is required for bone marrow hematopoietic stem cell maintenance. Science 2018, 360, 106–110. [Google Scholar]
- Schepers, K.; Pietras, E.M.; Reynaud, D.; Hsiao, E.C.; Passegué, E. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell 2013, 13, 285–299. [Google Scholar]
- Hoffmann, A.D.; Peterson, M.A.; Friedland-Little, J.M.; Anderson, S.A.; Moskowitz, I.P. Sonic hedgehog is required in pulmonary endoderm for atrial septation. Development 2009, 136, 1761–1770. [Google Scholar]
- Rothová, M.; Peterková, R.; Tucker, A.S. Fate map of the dental mesenchyme: Dynamic development of the dental papilla and follicle. Dev. Biol. 2012, 366, 244–254. [Google Scholar]
- Olson, E.N.; Schneider, M.D. Sizing up the heart: Development redux in disease. Genes Dev. 2003, 17, 1937–1956. [Google Scholar]
- Tthesleff, I. Homeobox genes and growth tactors in regulation of craniofacial and tooth morphogenesis. Acta Odontol. Scand. 1995, 53, 129–134. [Google Scholar]
- Mitsiadis, T.A.; Lardelli, M.; Lendahl, U.; Thesleff, I. Expression of Notch 1,2 and 3 is regulated by epithelial-mesenchymal interactions and retinoic and in the developing mouse tooth and associated with determination of ameloblast cell fate. J. Cell Biol. 1995, 130, 407–418. [Google Scholar] [PubMed]
- Morikawa, S.; Mabuchi, Y.; Kubota, Y.; Suda, T.; Okano, H.; Matsuzaki, Y. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 2009, 206, 2483–2496. [Google Scholar] [PubMed]
- Houlihan, D.D.; Mabuchi, Y.; Morikawa, S.; Niibe, K.; Araki, D.; Suzuki, S.; Okano, H.; Matsuzaki, Y. Isolation of mouse mesenchymal stem cells on the basis of expression of Sca-1 and PDGFR-α. Nat. Protoc. 2012, 7, 2103–2111. [Google Scholar]
- Matsunaga, T.; Shirasawa, H.; Tanabe, M.; Ohnuma, N.; Takahashi, H.; Simizu, B. Expression of alternatively spliced src messenger RNAs related to neuronal differentiation in human neuroblastomas. Cancer Res. 1993, 53, 3179–3185. [Google Scholar]
- Jaworski, J.; Kapitein, L.C.; Gouveia, S.M.; Dortland, B.R.; Wulf, P.S.; Grigoriev, I.; Camera, P.; Spangler, S.A.; Di Stefano, P.; Demmers, J.; et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 2009, 61, 85–100. [Google Scholar]
- Gambardella, L.; McManus, S.A.; Moignard, V.; Sebukhan, D.; Delaune, A.; Andrews, S.; Bernard, W.G.; Morrison, M.A.; Riley, P.R.; Göttgens, B.; et al. BNC1 regulates cell heterogeneity in human pluripotent stem cell-derived epicardium. Development 2019, 146, 174441. [Google Scholar]
- Mahoney, M.G.; Tang, W.; Xiang, M.M.; Moss, S.B.; Gerton, G.L.; Stanley, J.R.; Tseng, H. Translocation of the zinc finger protein basonuclin from the mouse germ cell nucleus to the midpiece of the spermatozoon during spermiogenesis. Biol. Reprod. 1998, 59, 388–394. [Google Scholar]
- Sgodda, M.; Aurich, H.; Kleist, S.; Aurich, I.; König, S.; Dollinger, M.M.; Fleig, W.E.; Christ, B. Hepatocyte differentiation of mesenchymal stem cells from rat peritoneal adipose tissue in vitro and in vivo. Exp. Cell Res. 2007, 313, 2875–2886. [Google Scholar]
- Xu, D.; Wang, Z.; Xia, Y.; Shao, F.; Xia, W.; Wei, Y.; Li, X.; Qian, X.; Lee, J.H.; Du, L.; et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 2020, 580, 530–535. [Google Scholar]
- Sun, J.; Nishiyama, N.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013, 14, 219. [Google Scholar]
- Hardcastle, T.J.; Kelly, K.A. baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinform. 2010, 11, 422. [Google Scholar]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar]
- Olson, T.S.; Caselli, A.; Otsuru, S.; Hofmann, J.T.; Williams, R.; Paolucci, P.; Dominici, M.; Horwitz, E.M. Megakaryocytes promote murine osteoblastic HSC niche expansion and stem cell engraftment after radioablative conditioning. Blood 2013, 121, 5238–5249. [Google Scholar] [CrossRef] [Green Version]
- Kimura, S.; Roberts, A.W.; Metcalf, D.; Alexander, W.S. Hematopoietic stem cell deficiencies in mice lacking c-Mpl, the receptor for thrombopoietin. Proc. Natl. Acad. Sci. USA 1998, 95, 1195–1200. [Google Scholar] [CrossRef] [Green Version]
- Ihara, K.; Ishii, E.; Eguchi, M.; Takada, H.; Suminoe, A.; Good, R.A.; Hara, T. Identification of mutations in the c-mpl gene in congenital amegakaryocytic thrombocytopenia. Proc. Natl. Acad. Sci. USA 1999, 96, 3132–3136. [Google Scholar] [CrossRef] [Green Version]
- Kaushansky, K. The molecular mechanisms that control thrombopoiesis. J. Clin. Investig. 2005, 115, 3339–3347. [Google Scholar] [CrossRef]
- Pardanani, A.D.; Levine, R.L.; Lasho, T.; Pikman, Y.; Mesa, A.R.; Wadleigh, M.; Steensma, P.D.; Elliott, A.M.; Wolanskyj, P.A.; Hogan, J.W.; et al. MPL515 mutations in myeloproliferative and other myeloid disorders: A study of 1182 patients. Blood 2006, 108, 3472–3476. [Google Scholar] [CrossRef] [Green Version]
- Alexander, W.S.; Roberts, A.W.; Nicola, N.A.; Li, R.; Metcalf, D. Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood 1996, 87, 2162–2170. [Google Scholar] [CrossRef] [Green Version]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Kanazawa, S.; Okada, H.; Hojo, H.; Ohba, S.; Iwata, J.; Komura, M.; Hikita, A.; Hoshi, K. Mesenchymal stromal cells in the bone marrow niche consist of multi-populations with distinct transcriptional and epigenetic properties. Sci. Rep. 2021, 11, 15811. [Google Scholar] [CrossRef]
- Albright, E.R.; Kalejta, R.F. Myeloblastic cell lines mimic some but not all aspects of human cytomegalovirus experimental latency defined in primary CD34+ cell populations. J. Virol. 2013, 8717, 9802–9812. [Google Scholar] [CrossRef] [Green Version]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2015, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdallah, B.M.; Ditzel, N.; Kassem, M. Assessment of bone formation capacity using in vivo transplantation assays: Procedure and tissue analysis. Methods Mol. Biol. 2008, 455, 89–100. [Google Scholar] [PubMed]




| Gapdh | FW | 5′- CCACTAACATCAAATGGGGTGAGG -3′ |
| RV | 5′- TACTTGGCAGGTTTCTCCAGGC -3′ | |
| Hprt | FW | 5′- TCAGTCAACGGGGGACATAAA -3′ |
| RV | 5′- GGGGCTGTACTGCTTAACCAG -3′ | |
| Col2a1 | FW | 5′- TTGAGACAGCACGACGTGGAG -3′ |
| RV | 5′- AGCCAFFTTGCCATCGCCATA -3′ | |
| Sox9 | FW | 5′- TCTCCTAATGCTATCTTCAAGGCG -3′ |
| RV | 5′- TGCTCAGTTCACCGATGTCCAC -3′ | |
| Alp | FW | 5′- CACAATATCAAGGATATCGACGTGA -3′ |
| RV | 5′- ACATCAGTTCTGTTCTTCGGGTACA -3′ | |
| Bglap | FW | 5′- GGGCAATAAGGTAGTGAACAG -3′ |
| RV | 5′- GCAFCACAFFTCCTAAATAGT -3′ | |
| Adipoq | FW | 5′- TGTTCCTCTTAATCCTGCCCA -3′ |
| RV | 5′- CCAACCTGCACAAGTTCCCTT -3′ | |
| Pparg | FW | 5′- ACCACTCGCATTCCTTTGAC -3′ |
| RV | 5′- TGGGTCAGCTCTTGTGAATG -3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanazawa, S.; Okada, H.; Riu, D.; Mabuchi, Y.; Akazawa, C.; Iwata, J.; Hoshi, K.; Hikita, A. Hematopoietic–Mesenchymal Signals Regulate the Properties of Mesenchymal Stem Cells. Int. J. Mol. Sci. 2022, 23, 8238. https://doi.org/10.3390/ijms23158238
Kanazawa S, Okada H, Riu D, Mabuchi Y, Akazawa C, Iwata J, Hoshi K, Hikita A. Hematopoietic–Mesenchymal Signals Regulate the Properties of Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2022; 23(15):8238. https://doi.org/10.3390/ijms23158238
Chicago/Turabian StyleKanazawa, Sanshiro, Hiroyuki Okada, Dan Riu, Yo Mabuchi, Chihiro Akazawa, Junichi Iwata, Kazuto Hoshi, and Atsuhiko Hikita. 2022. "Hematopoietic–Mesenchymal Signals Regulate the Properties of Mesenchymal Stem Cells" International Journal of Molecular Sciences 23, no. 15: 8238. https://doi.org/10.3390/ijms23158238






