Role of Glycolysis/Gluconeogenesis and HIF-1 Signaling Pathways in Rats with Dental Fluorosis Integrated Proteomics and Metabolomics Analysis
Abstract
:1. Introduction
2. Results
2.1. Identified Differentially Expressed Proteins and Metabolites
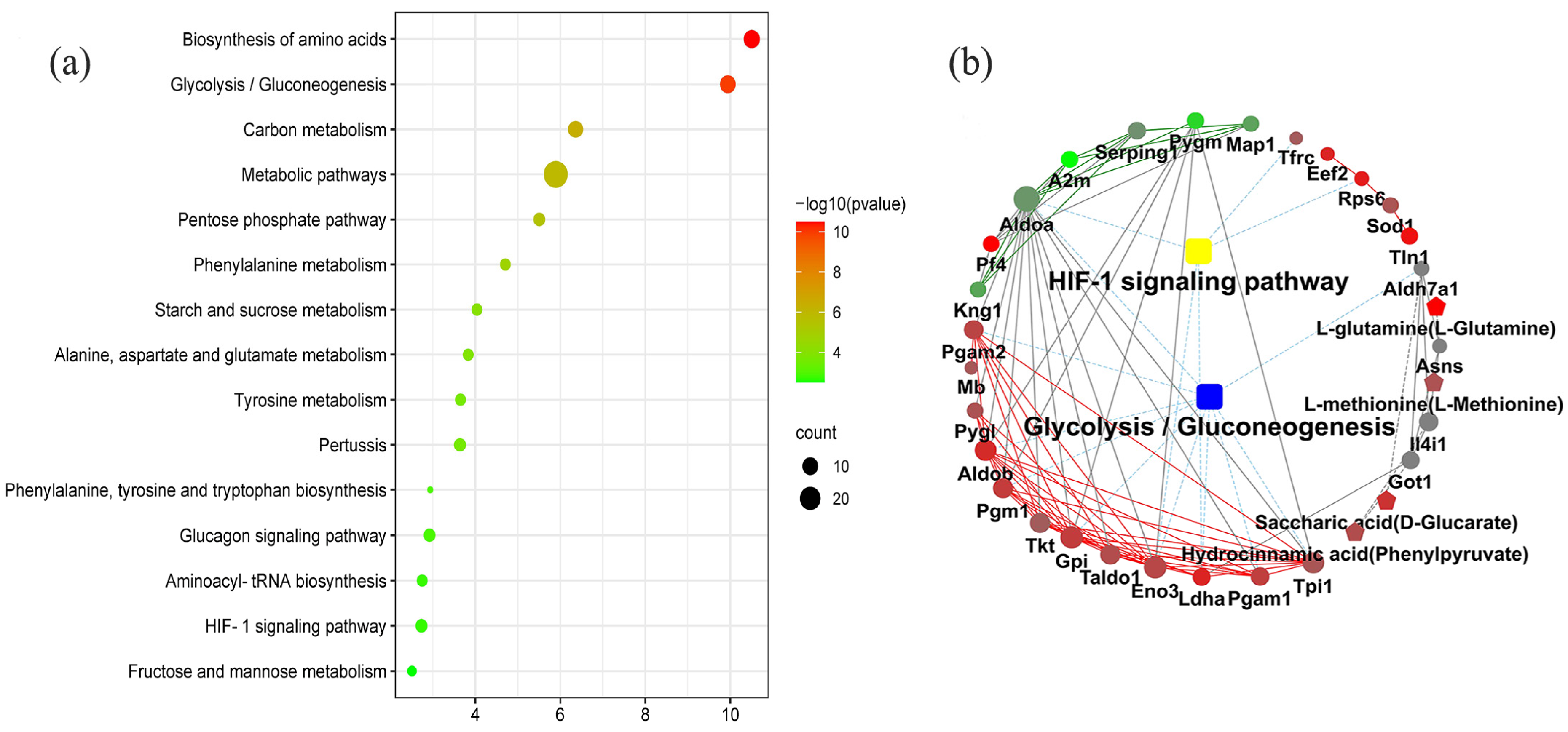
2.2. KEGG Pathway Enrichment Analysis
2.3. GO Function Enrichment Analysis
2.4. KEGG Markup Language (KGML) Network Diagram
2.5. Validation Analysis
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. Protein and Metabolite Extraction
4.2.1. Proteins Extraction
4.2.2. Metabolites Extraction
4.3. LC-MS/MS and GC-MS Analyses
4.3.1. Differentially Expressed Proteins
4.3.2. Differentially Expressed Metabolites
4.4. Identified Differentially Expressed Proteins and Metabolites
4.5. Integrated Proteomics and Metabolomics Analyses
4.6. Validation of Database
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, G.; Kumari, B.; Sinam, G.; Kriti; Kumar, N.; Mallick, S. Fluoride distribution and contamination in the water, soil and plants continuum and its remedial technologies, an Indian perspective—A review. Environ. Pollut. 2018, 239, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.; Amphlett, B.; Robbé, I.J. The long-term effects of water fluoridation on the human skeleton. J. Dent. Res. 2011, 90, 683–684. [Google Scholar] [CrossRef] [PubMed]
- Iheozor-Ejiofor, Z.; Worthington, H.V.; Walsh, T.; O’Malley, L.; Clarkson, J.E.; Macey, R.; Alam, R.; Tugwell, P.; Welch, V.; Glenny, A.M. Water fluoridation for the prevention of dental caries. Cochrane Database Syst. Rev. 2015, 2015, Cd010856. [Google Scholar] [CrossRef]
- Rezaee, T.; Bouxsein, M.L.; Karim, L. Increasing fluoride content deteriorates rat bone mechanical properties. Bone 2020, 136, 115369. [Google Scholar] [PubMed]
- Yadav, K.K.; Kumar, S.; Pham, Q.B.; Gupta, N.; Rezania, S.; Kamyab, H.; Yadav, S.; Vymazal, J.; Kumar, V.; Tri, D.Q.; et al. Fluoride contamination, health problems and remediation methods in Asian groundwater: A comprehensive review. Ecotoxicol. Environ. Saf. 2019, 182, 109362. [Google Scholar] [CrossRef]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Huang, L.E.; Arany, Z.; Livingston, D.M.; Bunn, H.F. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 1996, 271, 32253–32259. [Google Scholar] [CrossRef] [Green Version]
- Obach, M.; Navarro-Sabaté, A.; Caro, J.; Kong, X.; Duran, J.; Gómez, M.; Perales, J.C.; Ventura, F.; Rosa, J.L.; Bartrons, R. 6-Phosphofructo-2-kinase (pfkfb3) gene promoter contains hypoxia-inducible factor-1 binding sites necessary for transactivation in response to hypoxia. J. Biol. Chem. 2004, 279, 53562–53570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butterfield, D.A.; Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019, 20, 148–160. [Google Scholar] [CrossRef]
- Jiang, P.; Du, W.; Wu, M. Regulation of the pentose phosphate pathway in cancer. Protein Cell 2014, 5, 592–602. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.C.; Glenny, A.-M.; Tsang, B.W.; Lo, E.C.; Worthington, H.V.; Marinho, V.C. Topical fluoride as a cause of dental fluorosis in children. Cochrane Database Syst. Rev. 2010, 1, CD007693. [Google Scholar] [CrossRef] [PubMed]
- Güner, Ş.; Uyar-Bozkurt, S.; Haznedaroğlu, E.; Menteş, A. Dental Fluorosis and Catalase Immunoreactivity of the Brain Tissues in Rats Exposed to High Fluoride Pre- and Postnatally. Biol. Trace Elem. Res. 2016, 174, 150–157. [Google Scholar] [CrossRef]
- Fejerskov, O.; Larsen, M.J.; Richards, A.; Baelum, V. Dental tissue effects of fluoride. Adv. Dent. Res. 1994, 8, 15–31. [Google Scholar] [CrossRef]
- DenBesten, P.K.; Yan, Y.; Featherstone, J.D.B.; Hilton, J.F.; Smith, C.E.; Li, W. Effects of fluoride on rat dental enamel matrix proteinases. Arch. Oral Biol. 2002, 47, 763–770. [Google Scholar] [CrossRef]
- Bartlett, J.D.; Simmer, J.P.; Xue, J.; Margolis, H.C.; Moreno, E.C. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene 1996, 183, 123–128. [Google Scholar] [CrossRef]
- Adamek, E.; Pawłowska-Góral, K.; Bober, K. In vitro and in vivo effects of fluoride ions on enzyme activity. Ann. Acad. Med. Stetin. 2005, 51, 69–85. [Google Scholar]
- Wei, W.; Gao, Y.; Wang, C.; Zhao, L.; Sun, D. Excessive fluoride induces endoplasmic reticulum stress and interferes enamel proteinases secretion. Environ. Toxicol. 2013, 28, 332–341. [Google Scholar] [CrossRef]
- Sasaki, T.; Tominaga, H.; Higashi, S. Microvascular architecture of the enamel organ in the rat-incisor maturation zone. Scanning and transmission electron microscopic studies. Acta Anat. 1984, 118, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Zhang, X.; Li, W.; Wang, J.; Sun, Z.; Niu, R. Fluoride exposure altered metabolomic profile in rat serum. Chemosphere 2020, 258, 127387. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, S.E.; O’Neill, L.A.J. HIF1α and metabolic reprogramming in inflammation. J. Clin. Investig. 2016, 126, 3699–3707. [Google Scholar] [CrossRef] [Green Version]
- Formenti, F.; Constantin-Teodosiu, D.; Emmanuel, Y.; Cheeseman, J.; Dorrington, K.L.; Edwards, L.M.; Humphreys, S.M.; Lappin, T.R.J.; McMullin, M.F.; McNamara, C.J.; et al. Regulation of human metabolism by hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA 2010, 107, 12722–12727. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Bosch-Marce, M.; Shimoda, L.A.; Tan, Y.S.; Baek, J.H.; Wesley, J.B.; Gonzalez, F.J.; Semenza, G.L. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008, 283, 10892–10903. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-w.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006, 3, 187–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massari, F.; Ciccarese, C.; Santoni, M.; Iacovelli, R.; Mazzucchelli, R.; Piva, F.; Scarpelli, M.; Berardi, R.; Tortora, G.; Lopez-Beltran, A.; et al. Metabolic phenotype of bladder cancer. Cancer Treat. Rev. 2016, 45, 46–57. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordet, J.P.; Maire, P.; Giallongo, A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [Green Version]
- Fantin, V.R.; St-Pierre, J.; Leder, P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 2006, 9, 425–434. [Google Scholar] [CrossRef] [Green Version]
- Feo, S.; Antona, V.; Barbieri, G.; Passantino, R.; Calì, L.; Giallongo, A. Transcription of the human beta enolase gene (ENO-3) is regulated by an intronic muscle-specific enhancer that binds myocyte-specific enhancer factor 2 proteins and ubiquitous G-rich-box binding factors. Mol. Cell Biol. 1995, 15, 5991–6002. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-W.; Dang, C.V. Multifaceted roles of glycolytic enzymes. Trends Biochem. Sci. 2005, 30, 142–150. [Google Scholar] [CrossRef]
- Liu, P.; Sun, S.-J.; Ai, Y.-J.; Feng, X.; Zheng, Y.-M.; Gao, Y.; Zhang, J.-Y.; Zhang, L.; Sun, Y.-P.; Xiong, Y.; et al. Elevated nuclear localization of glycolytic enzyme TPI1 promotes lung adenocarcinoma and enhances chemoresistance. Cell Death Dis. 2022, 13, 205. [Google Scholar] [CrossRef]
- Orosz, F.; Oláh, J.; Ovádi, J. Triosephosphate isomerase deficiency: New insights into an enigmatic disease. Biochim. Biophys. Acta 2009, 1792, 1168–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, K.; Ma, L.; Gu, H.; Li, J.; Lei, S. Fluoride induced endoplasmic reticulum stress and calcium overload in ameloblasts. Arch. Oral Biol. 2016, 69, 95–101. [Google Scholar] [CrossRef]
- Zhang, Z.; Shi, Y.; Zheng, H.; Zhou, Z.; Wu, Z.; Shen, D.; Wang, Y.; Zhang, Y.; Wang, Z.; Fu, B. A Hydroxypropyl Methylcellulose Film Loaded with AFCP Nanoparticles for Inhibiting Formation of Enamel White Spot Lesions. Int. J. Nanomed. 2021, 16, 7623–7637. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Chen, Y.; Ren, Y.; Feng, Y.; Long, S. GLUT1 biological function and inhibition: Research advances. Future Med. Chem. 2021, 13, 1227–1243. [Google Scholar] [CrossRef]
- Mohamed, M.S.; Bishr, M.K.; Almutairi, F.M.; Ali, A.G. Inhibitors of apoptosis: Clinical implications in cancer. Apoptosis 2017, 22, 1487–1509. [Google Scholar] [CrossRef]
- Ganapathy-Kanniappan, S.; Geschwind, J.-F.H. Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol. Cancer 2013, 12, 152. [Google Scholar] [CrossRef] [Green Version]
- Heintz-Buschart, A.; Wilmes, P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, Y.; Chu, Q.; Zhang, P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019, 447, 41–47. [Google Scholar] [CrossRef]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.-K.; Amini, B.; Andersen, E.; Andersson, A.-C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene Names | Description | Accession | p-Value | Fold Change |
|---|---|---|---|---|
| Ldha | l-lactate dehydrogenase A chain | P04642 | 2.14 × 10−2 | 1.74 |
| Aldob | Fructose-bisphosphate aldolase | Q66HT1 | 2.63 × 10−11 | 1.67 |
| Pgm1 | Phosphoglucomutase-1 | P38652 | 1.12 × 10−10 | 1.50 |
| Gpi | Glucose-6-phosphate isomerase | Q6P6V0 | 5.78 × 10−8 | 1.50 |
| Pgam1 | Phosphoglycerate mutase 1 | P25113 | 6.13 × 10−7 | 1.48 |
| Pgam2 | Phosphoglycerate mutase 2 | P16290 | 2.20 × 10−9 | 1.43 |
| Eno3 | Beta-enolase | P15429 | 1.35 × 10−7 | 1.40 |
| Tpi1 | Triosephosphate isomerase | P48500 | 2.95 × 10−4 | 1.29 |
| Aldoa | Fructose-bisphosphate aldolase A | P05065 | 3.32 × 10−11 | 0.69 |
| Tfrc | transferrin receptor | Q99376 | 1.03 × 10−9 | 1.25 |
| Rps6 | ribosomal protein s6 | P62755 | 1.18 × 10−2 | 1.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ba, Y.; Yang, S.; Yu, S.; Hou, X.; Du, Y.; Gao, M.; Zuo, J.; Sun, L.; Fu, X.; Li, Z.; et al. Role of Glycolysis/Gluconeogenesis and HIF-1 Signaling Pathways in Rats with Dental Fluorosis Integrated Proteomics and Metabolomics Analysis. Int. J. Mol. Sci. 2022, 23, 8266. https://doi.org/10.3390/ijms23158266
Ba Y, Yang S, Yu S, Hou X, Du Y, Gao M, Zuo J, Sun L, Fu X, Li Z, et al. Role of Glycolysis/Gluconeogenesis and HIF-1 Signaling Pathways in Rats with Dental Fluorosis Integrated Proteomics and Metabolomics Analysis. International Journal of Molecular Sciences. 2022; 23(15):8266. https://doi.org/10.3390/ijms23158266
Chicago/Turabian StyleBa, Yue, Shuo Yang, Shuiyuan Yu, Xiangbo Hou, Yuhui Du, Minghui Gao, Juan Zuo, Lei Sun, Xiaoli Fu, Zhiyuan Li, and et al. 2022. "Role of Glycolysis/Gluconeogenesis and HIF-1 Signaling Pathways in Rats with Dental Fluorosis Integrated Proteomics and Metabolomics Analysis" International Journal of Molecular Sciences 23, no. 15: 8266. https://doi.org/10.3390/ijms23158266
APA StyleBa, Y., Yang, S., Yu, S., Hou, X., Du, Y., Gao, M., Zuo, J., Sun, L., Fu, X., Li, Z., Huang, H., Zhou, G., & Yu, F. (2022). Role of Glycolysis/Gluconeogenesis and HIF-1 Signaling Pathways in Rats with Dental Fluorosis Integrated Proteomics and Metabolomics Analysis. International Journal of Molecular Sciences, 23(15), 8266. https://doi.org/10.3390/ijms23158266






