UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas Plecoglossicida in Large Yellow Croaker (Larimichthys crocea)
Abstract
:1. Introduction
2. Results
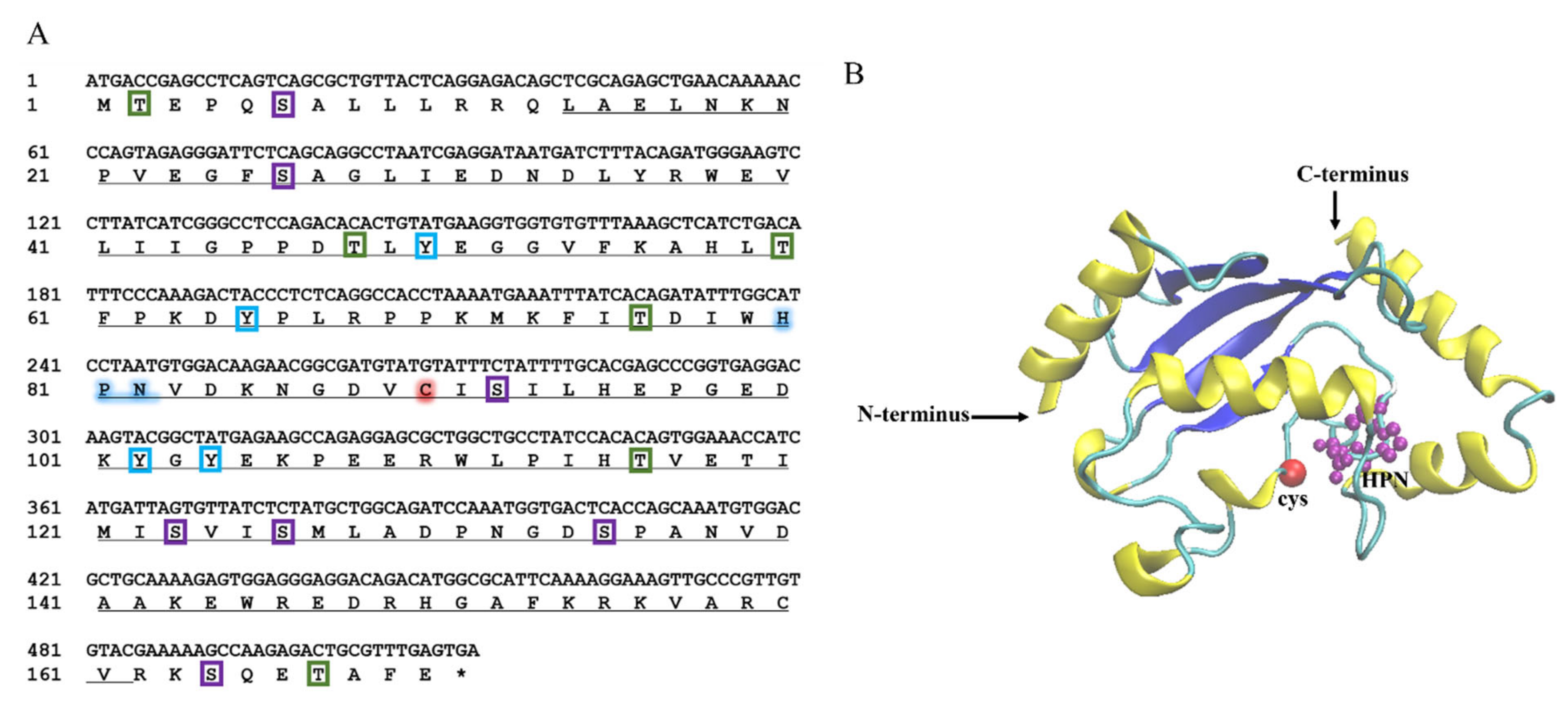
2.1. Sequence Characteristics of LcUbe2g1
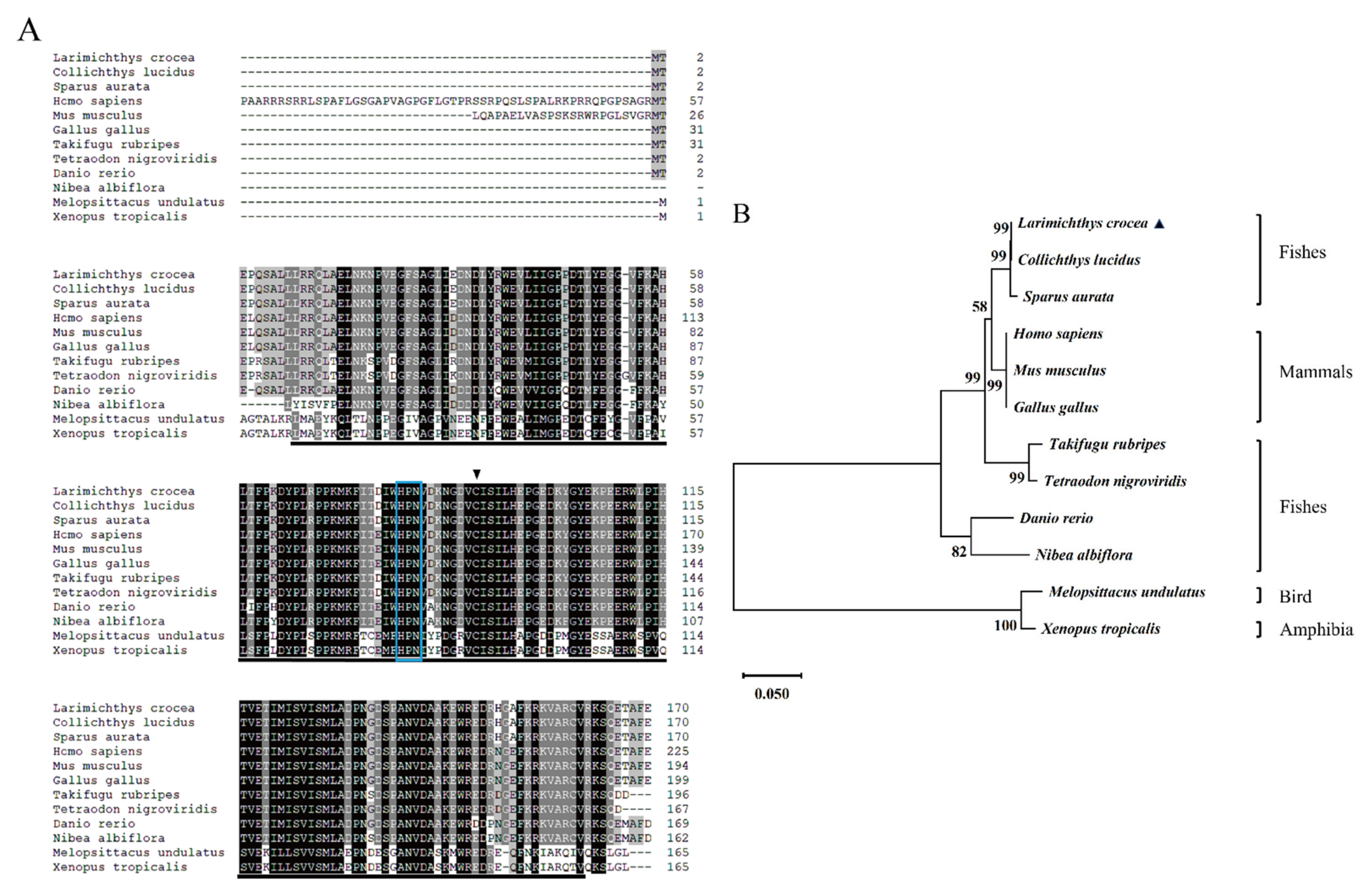
2.2. Tissue Expression and Subcellular Localization Analysis
2.3. Expression Analysis of LcUbe2g1 mRNA and Protein under P. plecoglossicida Infection
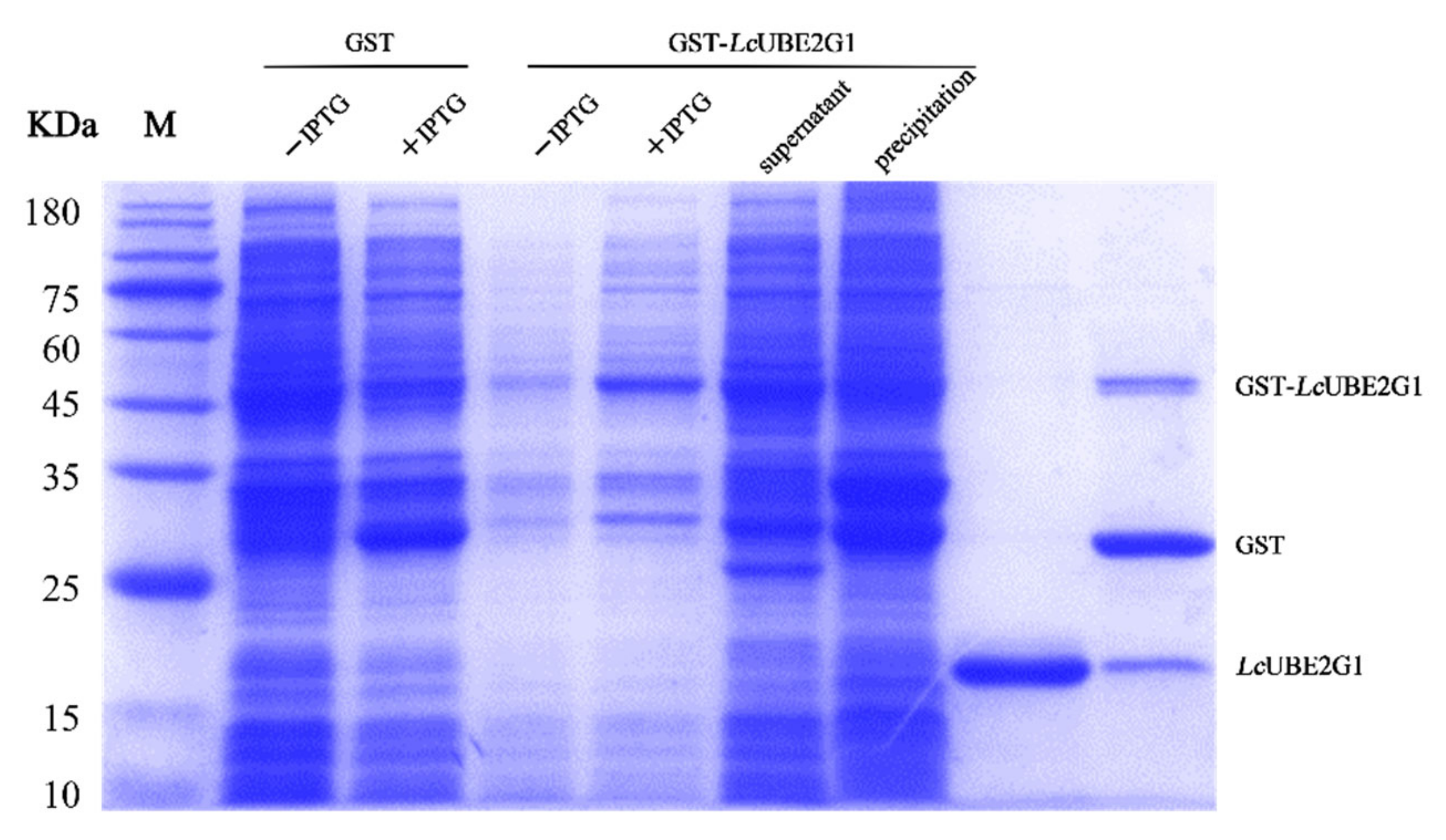
2.4. Recombinant LcUBE2G1 Protein
2.5. Interaction between LcUBE2G1 and Ub
2.6. Characterization of the LcUBE2G1 Interacting Proteins
3. Discussion
4. Materials and Methods
4.1. Pathogen Stimulation and Tissue Collection
4.2. RNA Extraction and cDNA Synthesis
4.3. Cloning and Sequence Analysis of LcUbe2g1
4.4. Construction of Expression Vector
4.5. Prokaryotic Expression and Purification of LcUBE2G1
4.6. Subcellular Localization of LcUBE2G1
4.7. Quantitative Real-Time PCR Analysis of LcUbe2g1
4.8. Immunocytochemistry Analysis
4.9. Immunohistochemistry Analysis
4.10. GST Pull-Down Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, F.; Song, Q.; Zhang, Y.; Wang, X.; Wang, Z. Molecular Characterization and Immune Responses of Rab5 in Large Yellow Croaker (Larimichthys crocea). Aquac. Fish. 2017, 2, 165–172. [Google Scholar] [CrossRef]
- Yao, C.; Kong, P.; Wang, Z.; Ji, P.; Cai, M.; Liu, X.; Han, X. Cloning and Expression Analysis of Two Alternative Splicing Toll-like Receptor 9 Isoforms A and B in Large Yellow Croaker, Pseudosciaena crocea. Fish Shellfish Immunol. 2008, 25, 648–656. [Google Scholar] [CrossRef]
- Zhang, D.L.; Han, F.; Yu, D.H.; Xiao, S.J.; Li, M.Y.; Chen, J.; Wang, Z.Y. Characterization of E3 Ubiquitin Ligase Neuregulin Receptor Degradation Protein-1 (Nrdp1) in the Large Yellow Croaker (Larimichthys crocea) and Its Immune Responses to Cryptocaryon Irritans. Gene 2015, 556, 98–105. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, L.; Li, C.; Shao, G.; Chen, X. Transcriptome Analysis Revealed Multiple Immune Processes and Energy Metabolism Pathways Involved in the Defense Response of the Large Yellow Croaker Larimichthys crocea against Pseudomonas Plecoglossicida. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100886. [Google Scholar] [CrossRef]
- Tang, Y.; Xin, G.; Zhao, L.-M.; Huang, L.-X.; Qin, Y.-X.; Su, Y.-Q.; Zheng, W.-Q.; Wu, B.; Lin, N.; Yan, Q.-P. Novel insights into host-pathogen interactions of large yellow croakers (Larimichthys crocea) and pathogenic bacterium Pseudomonas plecoglossicida using time-resolved dual RNA-seq of infected spleens. Zool. Res. 2020, 41, 314–327. [Google Scholar] [CrossRef]
- Nakamura, N. Ubiquitin System. Int. J. Mol. Sci. 2018, 19, E1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-S.; Qiu, X.-B. Ubiquitin at the Crossroad of Cell Death and Survival. Chin. J. Cancer 2013, 32, 640–647. [Google Scholar] [CrossRef] [Green Version]
- Lu, G.; Weng, S.; Matyskiela, M.; Zheng, X.; Fang, W.; Wood, S.; Surka, C.; Mizukoshi, R.; Lu, C.-C.; Mendy, D.; et al. UBE2G1 Governs the Destruction of Cereblon Neomorphic Substrates. eLife 2018, 7, e40958. [Google Scholar] [CrossRef]
- Chiba, T.; Tanaka, K. Cullin-Based Ubiquitin Ligase and Its Control by NEDD8-Conjugating System. Curr. Protein Pept. Sci. 2004, 5, 177–184. [Google Scholar] [CrossRef]
- Kirisako, T.; Kamei, K.; Murata, S.; Kato, M.; Fukumoto, H.; Kanie, M.; Sano, S.; Tokunaga, F.; Tanaka, K.; Iwai, K. A Ubiquitin Ligase Complex Assembles Linear Polyubiquitin Chains. EMBO J. 2006, 25, 4877–4887. [Google Scholar] [CrossRef]
- Garg, P.; Ceccarelli, D.F.; Keszei, A.F.A.; Kurinov, I.; Sicheri, F.; Sidhu, S.S. Structural and Functional Analysis of Ubiquitin-Based Inhibitors That Target the Backsides of E2 Enzymes. J. Mol. Biol. 2020, 432, 952–966. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Heride, C.; Urbé, S. The Demographics of the Ubiquitin System. Trends Cell Biol. 2015, 25, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Somasagara, R.R.; Tripathi, K.; Spencer, S.M.; Clark, D.W.; Barnett, R.; Bachaboina, L.; Scalici, J.; Rocconi, R.P.; Piazza, G.A.; Palle, K. Rad6 Upregulation Promotes Stem Cell-like Characteristics and Platinum Resistance in Ovarian Cancer. Biochem. Biophys. Res. Commun. 2016, 469, 449–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, T.; Deng, L.; Lu, X.; Pan, W.; Wu, Q.; Dai, J. Ubiquitin-Conjugating Enzyme UBE2J1 Negatively Modulates Interferon Pathway and Promotes RNA Virus Infection. Virol. J. 2018, 15, 132. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Ikeda, H.; Taira, N.; Hatoh, S.; Naito, M.; Doihara, H. Overexpression of UbcH10 Alternates the Cell Cycle Profile and Accelerate the Tumor Proliferation in Colon Cancer. BMC Cancer 2009, 9, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Cheng, Q.; Song, X.; Wang, H.; Wang, X.; Wang, L.; Zhu, B.; Song, L. A Vital Ubiquitin-Conjugating Enzyme CgUbe2g1 Participated in Regulation of Immune Response of Pacific Oyster Crassostrea Gigas. Dev. Comp. Immunol. 2019, 91, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Chen, Z.J. The Role of Ubiquitylation in Immune Defence and Pathogen Evasion. Nat. Rev. Immunol. 2012, 12, 35–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinngrebe, J.; Montinaro, A.; Peltzer, N.; Walczak, H. Ubiquitin in the Immune System. EMBO Rep. 2014, 15, 28–45. [Google Scholar] [CrossRef]
- Randles, L.; Walters, K.J. Ubiquitin and Its Binding Domains. Front. Biosci. 2012, 17, 2140–2157. [Google Scholar] [CrossRef] [Green Version]
- El Hokayem, J.; Amadei, C.; Obeid, J.-P.; Nawaz, Z. Ubiquitination of Nuclear Receptors. Clin. Sci. 2017, 131, 917–934. [Google Scholar] [CrossRef]
- Li, J.; Chai, Q.-Y.; Liu, C.H. The Ubiquitin System: A Critical Regulator of Innate Immunity and Pathogen-Host Interactions. Cell Mol. Immunol. 2016, 13, 560–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.-S.; Shenderov, K.; Huang, N.-N.; Kabat, J.; Abu-Asab, M.; Fitzgerald, K.A.; Sher, A.; Kehrl, J.H. Activation of Autophagy by Inflammatory Signals Limits IL-1β Production by Targeting Ubiquitinated Inflammasomes for Destruction. Nat. Immunol. 2012, 13, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Cook, B.W.; Shaw, G.S. Architecture of the Catalytic HPN Motif Is Conserved in All E2 Conjugating Enzymes. Biochem. J. 2012, 445, 167–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, S.; Wu, L.; Luo, M.; Xu, T.; Wu, X. Characterization and Function of an E2-17 KDa (UBE2D) in an Invertebrate Haliotis Diversicolor Supertexta. Fish Shellfish Immunol. 2013, 34, 1496–1504. [Google Scholar] [CrossRef]
- Hu, Q.; Chen, S. Cloning, Genomic Structure and Expression Analysis of Ubc9 in the Course of Development in the Half-Smooth Tongue Sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2013, 165, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Huang, T.; Zhang, L.; Zhou, Y.; Luo, H.; Xu, H.; Wang, X. Dysregulation of Ubiquitin-Proteasome System in Neurodegenerative Diseases. Front. Aging Neurosci. 2016, 8, 303. [Google Scholar] [CrossRef]
- Florentinus-Mefailoski, A.; Bowden, P.; Scheltens, P.; Killestein, J.; Teunissen, C.; Marshall, J.G. The Plasma Peptides of Alzheimer’s Disease. Clin. Proteomics 2021, 18, 17. [Google Scholar] [CrossRef]
- Vijayasimha, K.; Tran, M.V.; Leestemaker-Palmer, A.L.; Dolan, B.P. Direct Conjugation of NEDD8 to the N-Terminus of a Model Protein Can Induce Degradation. Cells 2021, 10, 854. [Google Scholar] [CrossRef]
- Xirodimas, D.P. Novel Substrates and Functions for the Ubiquitin-like Molecule NEDD8. Biochem. Soc. Trans. 2008, 36, 802–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Cheng, J.; Shi, T.; Yeh, E.T.H. Neddylation of a Breast Cancer-Associated Protein Recruits a Class III Histone Deacetylase That Represses NFkappaB-Dependent Transcription. Nat. Cell Biol. 2006, 8, 1171–1177. [Google Scholar] [CrossRef]




| Species | Common Name | GenBank Accession Numbers | Identity (%) |
|---|---|---|---|
| Larimichthys crocea | Large yellow croaker | ON081957.1 | 100% |
| Sparus aurata | Sparus aurata | XP_030294309.1 | 99.41% |
| Homo sapiens | Human | AAH26288.2 | 97.06% |
| Mus musculus | Mouse | EDL12691.1 | 97.06% |
| Gallus gallus | Red Junglefowl | NP_001074352.1 | 97.06% |
| Tetraodon nigroviridis | Spotted green pufferfish | CAF98540.1 | 94.01% |
| Takifugu rubripes | Tiger Puffer | XP_003970748.1 | 93.98% |
| Nibea albiflora | Yellow drum | KAG8011671.1 | 86.62% |
| Brachydanio rerio var | Danio rerio | AAH71506.1 | 86.47% |
| Xenopus tropicalis | Xenopus tropicalis | NP_001017198.1 | 53.80% |
| Melopsittacus undulatus | Budgerigar | XP_005151071.1 | 53.17% |
| Primer Name | Sequences (5′–3′) | Annealing Temperature (°C) | Purpose |
|---|---|---|---|
| P1(LcUbe2g1-cF) | ATGACCGAGCCTCAGTCAGCGCTGT | 62 | cDNA cloning |
| P2(LcUbe2g1-cR) | TCACTCAAACGCAGTCTCTTGGCTT | 58 | |
| P3(LcUbe2g1-GST-F) | CCCCTGGGATCCCCGGAATTCATGACCGAGCCTCAGTCAGCG | 75 | Prokaryotic expression |
| P4(LcUbe2g1-GST-R) | GTCACGATGCGGCCGCTCGAGTCACTCAAACGCAGTCTCTTGG | 74 | |
| P5(LcUbe2g1-GFP-F) | CTACCGGACTCAGATCTCGAGATGACCGAGCCTCAGTCAGCG | 73 | Subcellular localization |
| P6(LcUbe2g1-GFP-R) | GTACCGTCGACTGCAGAATTCCCTCAAACGCAGTCTCTTGG | 71 | |
| β-actin-QF | TTATGAAGGCTATGCCCTGCC | 54 | qPCR-PCR |
| β-actin-QR | TGAAGGAGTAGCCACGCTCTGT | 56 | |
| LcUbe2g1-QF | CAAATGTGGACGCTGCAAAAG | 53 | qPCR-PCR |
| LcUbe2g1-QR | CCTGACGACAGGGTGGAAGA | 56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, J.; Li, W.; Wang, B.; Zhang, S.; Xiao, Y.; Han, F.; Wang, Z. UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas Plecoglossicida in Large Yellow Croaker (Larimichthys crocea). Int. J. Mol. Sci. 2022, 23, 8298. https://doi.org/10.3390/ijms23158298
Peng J, Li W, Wang B, Zhang S, Xiao Y, Han F, Wang Z. UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas Plecoglossicida in Large Yellow Croaker (Larimichthys crocea). International Journal of Molecular Sciences. 2022; 23(15):8298. https://doi.org/10.3390/ijms23158298
Chicago/Turabian StylePeng, Jia, Wanbo Li, Bi Wang, Sen Zhang, Yao Xiao, Fang Han, and Zhiyong Wang. 2022. "UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas Plecoglossicida in Large Yellow Croaker (Larimichthys crocea)" International Journal of Molecular Sciences 23, no. 15: 8298. https://doi.org/10.3390/ijms23158298
APA StylePeng, J., Li, W., Wang, B., Zhang, S., Xiao, Y., Han, F., & Wang, Z. (2022). UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas Plecoglossicida in Large Yellow Croaker (Larimichthys crocea). International Journal of Molecular Sciences, 23(15), 8298. https://doi.org/10.3390/ijms23158298






