Changes in Metabolism and Mitochondrial Bioenergetics during Polyethylene-Induced Osteoclastogenesis
Abstract
:1. Introduction
2. Results
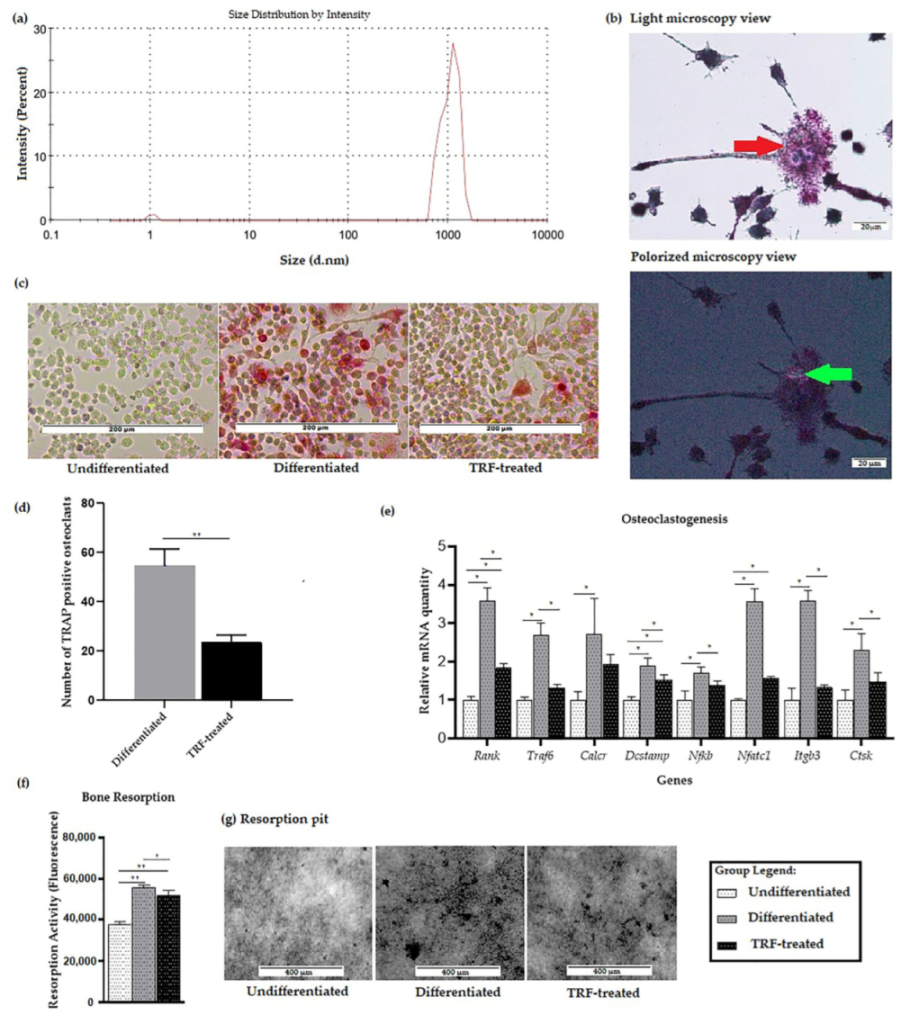
2.1. Confirmation of PE-Induced Osteoclast Differentiation and Activity
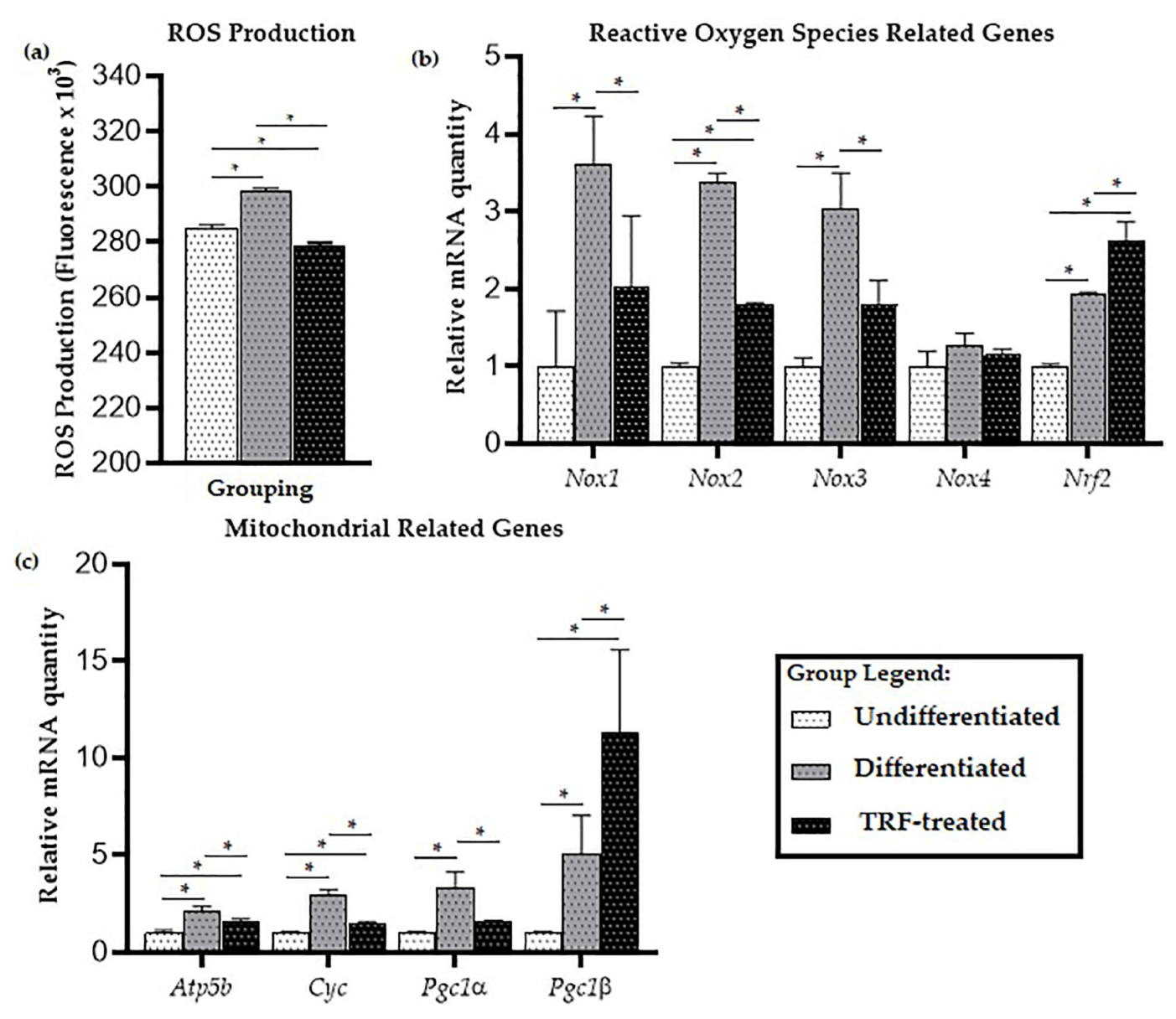
2.2. ROS Production
2.3. Mitochondrial-Related Genes Expression
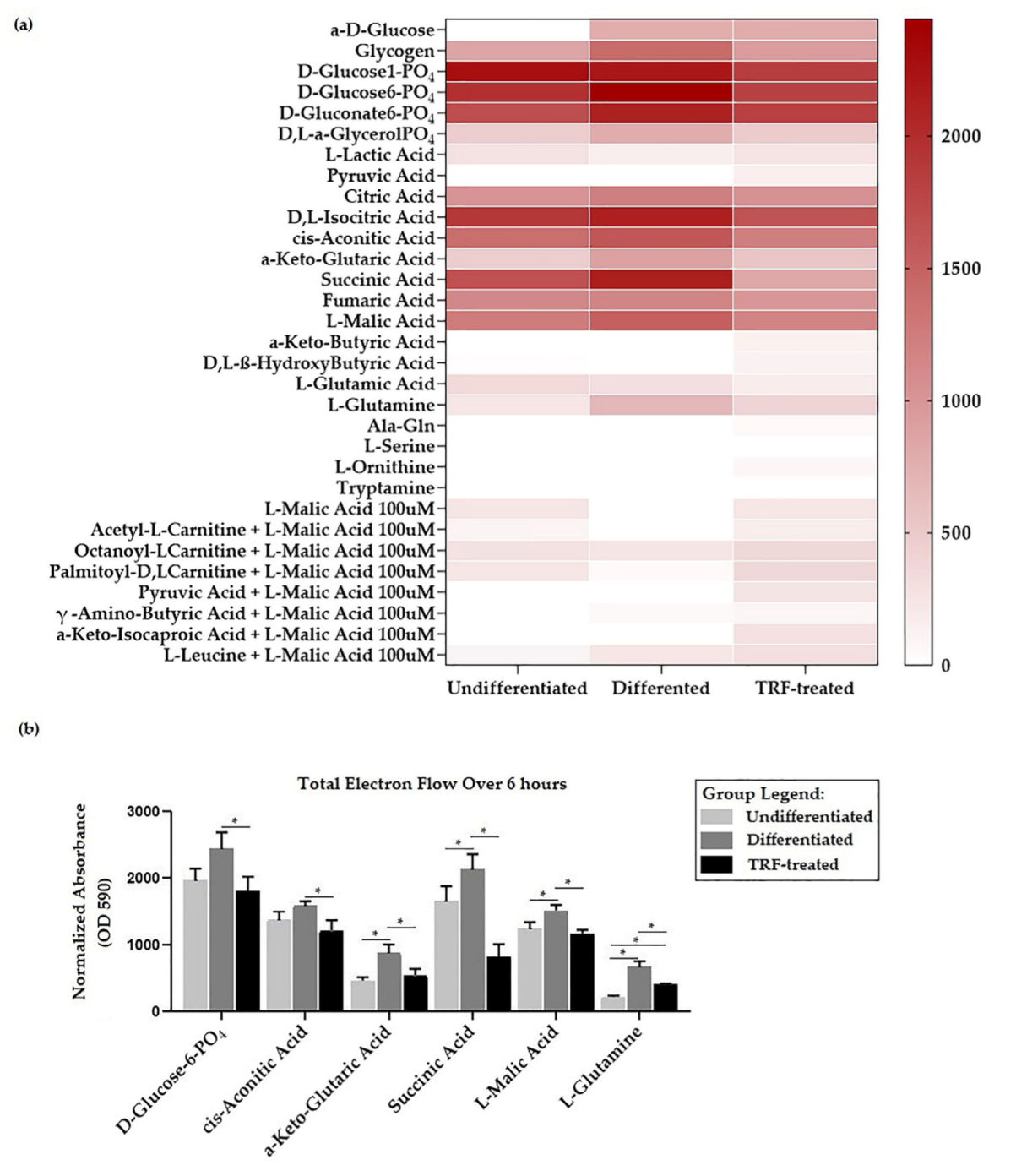
2.4. Cellular Metabolic Changes
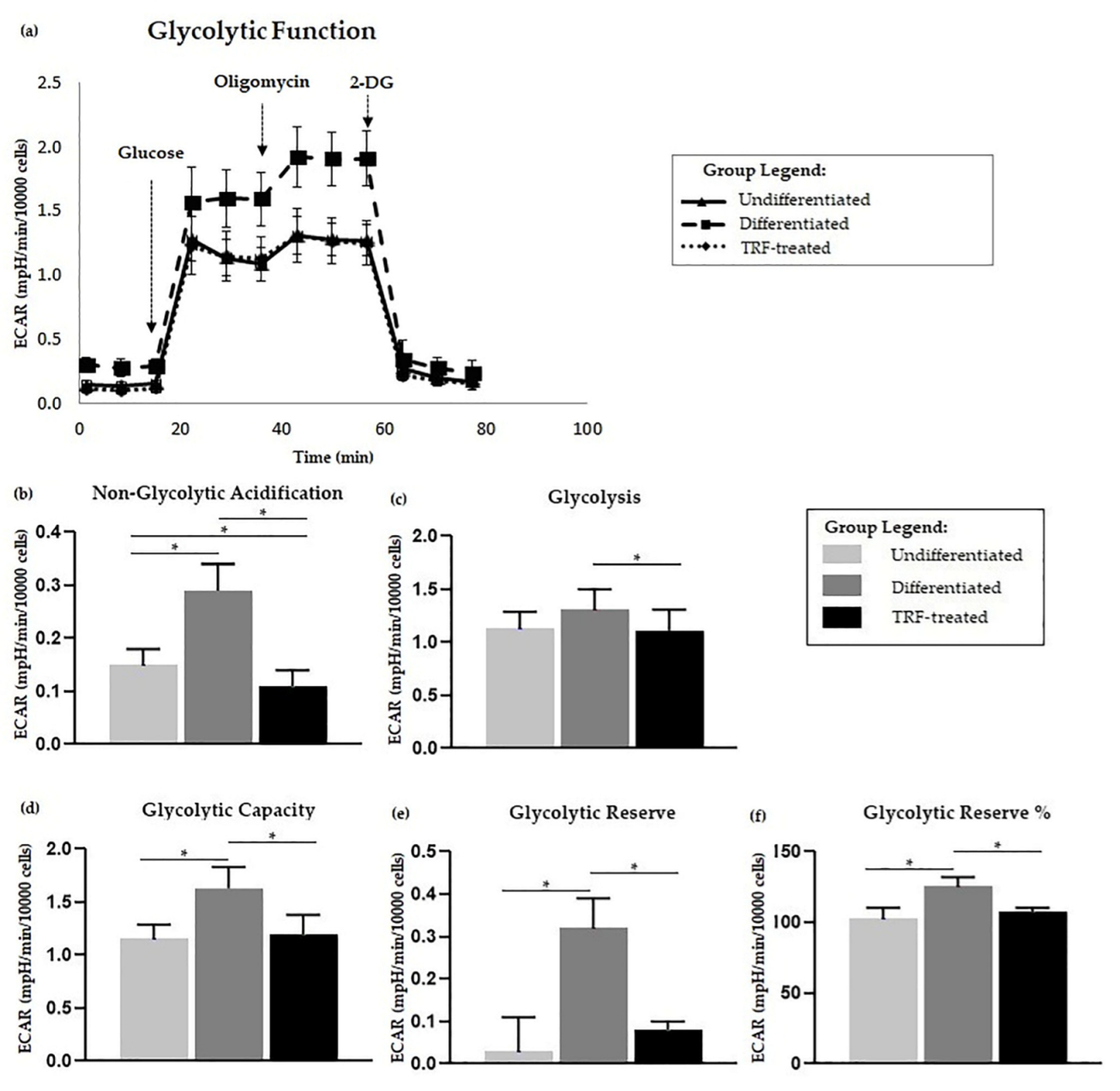
2.5. Glycolysis Stress Assay
2.6. Mitochondria Substrate Oxidation Profile
3. Discussion
4. Materials and Methods
4.1. Preparation of PE Particle
4.2. RAW264.7 Culture
4.3. Preparation of TRF
4.4. Gene Expression Study
4.5. Tartrate-Resistant Acid Phosphatase (TRAP) Staining
4.6. Bone Resorption Assay
4.7. MitoPlate Assay
4.8. Mitochondrial Stress Test
4.9. Glycolysis Stress Test
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; van Assen, A.H.G.; Reis, C.R.; Setroikromo, R.; van Merkerk, R.; Boersma, Y.L.; Cool, R.H.; Quax, W.J. Novel RANKL DE-loop mutants antagonize RANK-mediated osteoclastogenesis. FEBS J. 2017, 284, 2501–2512. [Google Scholar] [CrossRef] [Green Version]
- Simonet, W.S.; Lacey, D.L.; Dunstan, C.R.; Kelley, M.; Chang, M.S.; Lüthy, R.; Nguyen, H.Q.; Wooden, S.; Bennett, L.; Boone, T.; et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell 1997, 89, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Arai, F.; Miyamoto, T.; Ohneda, O.; Inada, T.; Sudo, T.; Brasel, K.; Miyata, T.; Anderson, D.M.; Suda, T. Commitment and differentiation of osteoclast precursor cells by the sequential expression of c-Fms and receptor activator of nuclear factor kappaB (RANK) receptors. J. Exp. Med. 1999, 190, 1741–1754. [Google Scholar] [CrossRef]
- Lee, Z.H.; Kim, H.H. Signal transduction by receptor activator of nuclear factor kappa B in osteoclasts. Biochem. Biophys. Res. Commun. 2003, 305, 211–214. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, N. Regulation of NFATc1 in Osteoclast Differentiation. J. Bone Metab. 2014, 21, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Takayanagi, H.; Kim, S.; Koga, T.; Nishina, H.; Isshiki, M.; Yoshida, H.; Saiura, A.; Isobe, M.; Yokochi, T.; Inoue, J.; et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 2002, 3, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, F.; Nishimura, R.; Matsubara, T.; Tanaka, S.; Inoue, J.; Reddy, S.V.; Hata, K.; Yamashita, K.; Hiraga, T.; Watanabe, T.; et al. Critical roles of c-Jun signaling in regulation of NFAT family and RANKL-regulated osteoclast differentiation. J. Clin. Investig. 2004, 114, 475–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkins, G.J.; Haynes, D.R.; Howie, D.W.; Findlay, D.M. Role of polyethylene particles in peri-prosthetic osteolysis: A review. World J. Orthop. 2011, 2, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Bonfield, W.; Kadoya, Y.; Yamac, T.; Freeman, M.A.; Scott, G.; Revell, P.A. The size and shape of particulate polyethylene wear debris in total joint replacements. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 1997, 211, 11–15. [Google Scholar] [CrossRef]
- Illgen, R.L., 2nd; Bauer, L.M.; Hotujec, B.T.; Kolpin, S.E.; Bakhtiar, A.; Forsythe, T.M. Highly crosslinked vs conventional polyethylene particles: Relative in vivo inflammatory response. J. Arthroplast. 2009, 24, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Sieving, A.; Wu, B.; Mayton, L.; Nasser, S.; Wooley, P.H. Morphological characteristics of total joint arthroplasty-derived ultra-high molecular weight polyethylene (UHMWPE) wear debris that provoke inflammation in a murine model of inflammation. J. Biomed. Mater. Res. Part A 2003, 64, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Crotti, T.N.; Smith, M.D.; Findlay, D.M.; Zreiqat, H.; Ahern, M.J.; Weedon, H.; Hatzinikolous, G.; Capone, M.; Holding, C.; Haynes, D.R. Factors regulating osteoclast formation in human tissues adjacent to peri-implant bone loss: Expression of receptor activator NFkappaB, RANK ligand and osteoprotegerin. Biomaterials 2004, 25, 565–573. [Google Scholar] [CrossRef]
- Holding, C.A.; Findlay, D.M.; Stamenkov, R.; Neale, S.D.; Lucas, H.; Dharmapatni, A.S.; Callary, S.A.; Shrestha, K.R.; Atkins, G.J.; Howie, D.W.; et al. The correlation of RANK, RANKL and TNFalpha expression with bone loss volume and polyethylene wear debris around hip implants. Biomaterials 2006, 27, 5212–5219. [Google Scholar] [CrossRef] [PubMed]
- Nich, C.; Takakubo, Y.; Pajarinen, J.; Ainola, M.; Salem, A.; Sillat, T.; Rao, A.J.; Raska, M.; Tamaki, Y.; Takagi, M.; et al. Macrophages-Key cells in the response to wear debris from joint replacements. J. Biomed. Mater. Res. Part A 2013, 101, 3033–3045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alias, E.; Dharmapatni, A.S.; Holding, A.C.; Atkins, G.J.; Findlay, D.M.; Howie, D.W.; Crotti, T.N.; Haynes, D.R. Polyethylene particles stimulate expression of ITAM-related molecules in peri-implant tissues and when stimulating osteoclastogenesis in vitro. Acta Biomater. 2012, 8, 3104–3112. [Google Scholar] [CrossRef] [PubMed]
- Howie, D.W.; Neale, S.D.; Stamenkov, R.; McGee, M.A.; Taylor, D.J.; Findlay, D.M. Progression of acetabular periprosthetic osteolytic lesions measured with computed tomography. J. Bone Jt. Surg. 2007, 89, 1818–1825. [Google Scholar] [CrossRef]
- Lemma, S.; Sboarina, M.; Porporato, P.E.; Zini, N.; Sonveaux, P.; Di Pompo, G.; Baldini, N.; Avnet, S. Energy metabolism in osteoclast formation and activity. Int. J. Biochem. Cell Biol. 2016, 79, 168–180. [Google Scholar] [CrossRef]
- Li, D.Z.; Zhang, Q.X.; Dong, X.X.; Li, H.D.; Ma, X. Treatment with hydrogen molecules prevents RANKL-induced osteoclast differentiation associated with inhibition of ROS formation and inactivation of MAPK, AKT and NF-kappa B pathways in murine RAW264.7 cells. J. Bone Miner. Metab. 2014, 32, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.J.; Lees, R.L.; Trujillo, E.; Martín-Vasallo, P.; Heersche, J.N.; Mobasheri, A. ATPase pumps in osteoclasts and osteoblasts. Int. J. Biochem. Cell Biol. 2002, 34, 459–476. [Google Scholar] [CrossRef]
- Arnett, T.R.; Orriss, I.R. Metabolic properties of the osteoclast. Bone 2018, 115, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Baron, R.; Neff, L.; Tran Van, P.; Nefussi, J.R.; Vignery, A. Kinetic and cytochemical identification of osteoclast precursors and their differentiation into multinucleated osteoclasts. Am. J. Pathol. 1986, 122, 363–378. [Google Scholar] [PubMed]
- Miyazaki, T.; Iwasawa, M.; Nakashima, T.; Mori, S.; Shigemoto, K.; Nakamura, H.; Katagiri, H.; Takayanagi, H.; Tanaka, S. Intracellular and extracellular ATP coordinately regulate the inverse correlation between osteoclast survival and bone resorption. J. Biol. Chem. 2012, 287, 37808–37823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.M.; Jeong, D.; Kang, H.K.; Jung, S.Y.; Kang, S.S.; Min, B.M. Osteoclast precursors display dynamic metabolic shifts toward accelerated glucose metabolism at an early stage of RANKL-stimulated osteoclast differentiation. Cell. Physiol. Biochem. 2007, 20, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Kwak, H.B.; Lee, B.K.; Oh, J.; Yeon, J.T.; Choi, S.W.; Cho, H.J.; Lee, M.S.; Kim, J.J.; Bae, J.M.; Kim, S.H.; et al. Inhibition of osteoclast differentiation and bone resorption by rotenone, through down-regulation of RANKL-induced c-Fos and NFATc1 expression. Bone 2010, 46, 724–731. [Google Scholar] [CrossRef]
- Ishii, K.A.; Fumoto, T.; Iwai, K.; Takeshita, S.; Ito, M.; Shimohata, N.; Aburatani, H.; Taketani, S.; Lelliott, C.J.; Vidal-Puig, A.; et al. Coordination of PGC-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nat. Med. 2009, 15, 259–266. [Google Scholar] [CrossRef]
- Zeng, R.; Faccio, R.; Novack, D.V. Alternative NF-κB Regulates RANKL-Induced Osteoclast Differentiation and Mitochondrial Biogenesis via Independent Mechanisms. J. Bone Miner. Res. 2015, 30, 2287–2299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indo, Y.; Takeshita, S.; Ishii, K.A.; Hoshii, T.; Aburatani, H.; Hirao, A.; Ikeda, K. Metabolic regulation of osteoclast differentiation and function. J. Bone Miner. Res. 2013, 28, 2392–2399. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lee, W.C.; Song, C.; Ye, L.; Abel, E.D.; Long, F. Both aerobic glycolysis and mitochondrial respiration are required for osteoclast differentiation. FASEB J. 2020, 34, 11058–11067. [Google Scholar] [CrossRef]
- Ahn, H.; Lee, K.; Kim, J.M.; Kwon, S.H.; Lee, S.H.; Lee, S.Y.; Jeong, D. Accelerated Lactate Dehydrogenase Activity Potentiates Osteoclastogenesis via NFATc1 Signaling. PLoS ONE 2016, 11, e0153886. [Google Scholar] [CrossRef] [Green Version]
- Agidigbi, T.S.; Kim, C. Reactive Oxygen Species in Osteoclast Differentiation and Possible Pharmaceutical Targets of ROS-Mediated Osteoclast Diseases. Int. J. Mol. Sci. 2019, 20, 3576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Thannickal, V.J.; Fanburg, B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000, 279, L1005–L1028. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, S.; Koenigstein, A.; Joseph, J.; Sun, L.; Kalyanaraman, B.; Zaidi, M.; Avadhani, N.G. Role of mitochondrial reactive oxygen species in osteoclast differentiation. Ann. N. Y. Acad. Sci. 2010, 1192, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Garrett, I.R.; Boyce, B.F.; Oreffo, R.O.; Bonewald, L.; Poser, J.; Mundy, G.R. Oxygen-derived free radicals stimulate osteoclastic bone resorption in rodent bone in vitro and in vivo. J. Clin. Investig. 1990, 85, 632–639. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.K.; Choi, Y.G.; Baik, J.Y.; Han, S.Y.; Jeong, D.W.; Bae, Y.S.; Kim, N.; Lee, S.Y. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood 2005, 106, 852–859. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Chang, E.J.; Kim, H.M.; Lee, S.B.; Kim, H.D.; Su Kim, G.; Kim, H.H. Antioxidant alpha-lipoic acid inhibits osteoclast differentiation by reducing nuclear factor-kappaB DNA binding and prevents in vivo bone resorption induced by receptor activator of nuclear factor-kappaB ligand and tumor necrosis factor-alpha. Free Radic. Biol. Med. 2006, 40, 1483–1493. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Choksi, S.; Qu, J.; Jang, J.; Choe, M.; Banfi, B.; Engelhardt, J.F.; Liu, Z.G. NADPH Oxidases Are Essential for Macrophage Differentiation. J. Biol. Chem. 2016, 291, 20030–20041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, H.; Yamamoto, H.; Tominaga, K.; Masuda, K.; Kawai, T.; Teshima-Kondo, S.; Rokutan, K. NADPH oxidase-derived reactive oxygen species are essential for differentiation of a mouse macrophage cell line (RAW264.7) into osteoclasts. J. Med. Investig. 2009, 56, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Peng, K.T.; Tsai, M.H.; Lee, C.W.; Chiang, Y.C.; Chen, P.C.; Chen, C.C.; Chang, C.H.; Shih, H.N.; Chang, P.J. Dysregulated expression of antioxidant enzymes in polyethylene particle-induced periprosthetic inflammation and osteolysis. PLoS ONE 2018, 13, e0202501. [Google Scholar] [CrossRef]
- Holmström, K.M.; Kostov, R.V.; Dinkova-Kostova, A.T. The multifaceted role of Nrf2 in mitochondrial function. Curr. Opin. Toxicol. 2016, 1, 80–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peh, H.Y.; Tan, W.S.; Liao, W.; Wong, W.S. Vitamin E therapy beyond cancer: Tocopherol versus tocotrienol. Pharmacol. Ther. 2016, 162, 152–169. [Google Scholar] [CrossRef] [PubMed]
- Jaafar, F.; Abdullah, A.; Makpol, S. Cellular Uptake and Bioavailability of Tocotrienol-Rich Fraction in SIRT1-Inhibited Human Diploid Fibroblasts. Sci. Rep. 2018, 8, 10471. [Google Scholar] [CrossRef] [PubMed]
- Nor Azman, N.H.E.; Goon, J.A.; Abdul Ghani, S.M.; Hamid, Z.; Wan Ngah, W.Z. Comparing Palm Oil, Tocotrienol-Rich Fraction and α-Tocopherol Supplementation on the Antioxidant Levels of Older Adults. Antioxidants 2018, 7, 74. [Google Scholar] [CrossRef] [Green Version]
- Khor, S.C.; Wan Ngah, W.Z.; Mohd Yusof, Y.A.; Abdul Karim, N.; Makpol, S. Tocotrienol-Rich Fraction Ameliorates Antioxidant Defense Mechanisms and Improves Replicative Senescence-Associated Oxidative Stress in Human Myoblasts. Oxidative Med. Cell. Longev. 2017, 2017, 3868305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuid, A.N.; Abdul Kadir, K.; Luke, D.A.; Soleiman, N.S. Tocotrienol offers better protection than tocopherol from free radical-induced damage of rat bone. Clin. Exp. Pharmacol. Physiol. 2005, 32, 761–770. [Google Scholar] [CrossRef]
- Goon, J.A.; Nor Azman, N.H.E.; Abdul Ghani, S.M.; Hamid, Z.; Wan Ngah, W.Z. Comparing palm oil tocotrienol rich fraction with α-tocopherol supplementation on oxidative stress in healthy older adults. Clin. Nutr. ESPEN 2017, 21, 1–12. [Google Scholar] [CrossRef]
- Jolly, J.J.; Mohd Fozi, N.F.; Chin, K.Y.; Wong, S.K.; Chua, K.H.; Alias, E.; Adnan, N.S.; Ima-Nirwana, S. Skeletal microenvironment system utilising bovine bone scaffold co-cultured with human osteoblasts and osteoclast-like cells. Exp. Ther. Med. 2021, 22, 680. [Google Scholar] [CrossRef]
- Fozi, N.F.M.; Jolly, J.J.; Hui, C.K.; Alias, E.; Yong, C.K.; Soelaiman, I.N. Comparing the effects of alpha-tocopherol and tocotrienol isomers on osteoblasts hFOB 1.19 cultured on bovine bone scaffold. Sains Malays. 2021, 50, 2319–2328. [Google Scholar] [CrossRef]
- Maniam, S.; Mohamed, N.; Shuid, A.N.; Soelaiman, I.N. Palm tocotrienol exerted better antioxidant activities in bone than α-tocopherol. Basic Clin. Pharmacol. Toxicol. 2008, 103, 55–60. [Google Scholar] [CrossRef]
- Hapidin, H.; Othman, F.; Soleiman, I.N.; Shuid, A.N.; Mohamed, N. Beneficial effects of tocotrienol and tocopherol on bone histomorphometric parameters in Sprague–Dawley male rats after nicotine cessation. Calcif. Tissue Int. 2009, 84, 65–74. [Google Scholar]
- Yahaya, M.F.; Zainodin, A.; Pupathy, R.; Min, E.O.H.; Bakar, N.; Zamri, N.A.; Ismail, H.; Mohd Ramli, E. The effect of palm tocotrienol on surface osteoblast and osteoclast in excess glucocorticoid osteoporotic rat model. Sains Malays. 2018, 47, 2731–2739. [Google Scholar] [CrossRef]
- Radzi, N.F.M.; Ismail, N.A.S.; Alias, E. Tocotrienols Regulate Bone Loss through Suppression on Osteoclast Differentiation and Activity: A Systematic Review. Curr. Drug Targets 2018, 19, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.K.; Mohamad Hazir, N.S.; Alias, E. Impacts of Hypoxia on Osteoclast Formation and Activity: Systematic Review. Int. J. Mol. Sci. 2021, 22, 146. [Google Scholar] [CrossRef]
- Haynes, D.R.; Crotti, T.N.; Potter, A.E.; Loric, M.; Atkins, G.J.; Howie, D.W.; Findlay, D.M. The osteoclastogenic molecules RANKL and RANK are associated with periprosthetic osteolysis. J. Bone Jt. Surgery. Br. Vol. 2001, 83, 902–911. [Google Scholar] [CrossRef]
- Song, C.; Yang, X.; Lei, Y.; Zhang, Z.; Smith, W.; Yan, J.; Kong, L. Evaluation of efficacy on RANKL induced osteoclast from RAW264.7 cells. J. Cell. Physiol. 2019, 234, 11969–11975. [Google Scholar] [CrossRef]
- Green, T.R.; Fisher, J.; Stone, M.; Wroblewski, B.M.; Ingham, E. Polyethylene particles of a ‘critical size’ are necessary for the induction of cytokines by macrophages in vitro. Biomaterials 1998, 19, 2297–2302. [Google Scholar] [CrossRef]
- Ren, W.; Yang, S.Y.; Fang, H.W.; Hsu, S.; Wooley, P.H. Distinct gene expression of receptor activator of nuclear factor-kappaB and rank ligand in the inflammatory response to variant morphologies of UHMWPE particles. Biomaterials 2003, 24, 4819–4826. [Google Scholar] [CrossRef]
- Yao, S.; Liu, D.; Pan, F.; Wise, G.E. Effect of vascular endothelial growth factor on RANK gene expression in osteoclast precursors and on osteoclastogenesis. Arch. Oral Biol. 2006, 51, 596–602. [Google Scholar] [CrossRef]
- Shen, Z.; Crotti, T.N.; McHugh, K.P.; Matsuzaki, K.; Gravallese, E.M.; Bierbaum, B.E.; Goldring, S.R. The role played by cell-substrate interactions in the pathogenesis of osteoclast-mediated peri-implant osteolysis. Arthritis Res. Ther. 2006, 8, R70. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Crotti, T.N.; Flannery, M.R.; Matsuzaki, K.; Goldring, S.R.; McHugh, K.P. A novel promoter regulates calcitonin receptor gene expression in human osteoclasts. Biochim. Biophys. Acta 2007, 1769, 659–667. [Google Scholar] [CrossRef]
- Matsumoto, M.; Kogawa, M.; Wada, S.; Takayanagi, H.; Tsujimoto, M.; Katayama, S.; Hisatake, K.; Nogi, Y. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J. Biol. Chem. 2004, 279, 45969–45979. [Google Scholar] [CrossRef] [Green Version]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef]
- Kim, K.; Lee, S.H.; Kim, J.H.; Choi, Y.; Kim, N. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP). Mol. Endocrinol. 2008, 22, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Koulouvaris, P.; Ly, K.; Ivashkiv, L.B.; Bostrom, M.P.; Nestor, B.J.; Sculco, T.P.; Purdue, P.E. Expression profiling reveals alternative macrophage activation and impaired osteogenesis in periprosthetic osteolysis. J. Orthop. Res. 2008, 26, 106–116. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, Y.; Dusting, G.J. NADPH oxidase-mediated redox signaling: Roles in cellular stress response, stress tolerance, and tissue repair. Pharmacol. Rev. 2011, 63, 218–242. [Google Scholar] [CrossRef] [Green Version]
- Kovac, S.; Angelova, P.R.; Holmström, K.M.; Zhang, Y.; Dinkova-Kostova, A.T.; Abramov, A.Y. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim. Biophys. Acta 2015, 1850, 794–801. [Google Scholar] [CrossRef] [Green Version]
- Bedard, K.; Krause, K.H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Sithole, C.; Pieterse, C.; Howard, K.; Kasonga, A. GPR120 Inhibits RANKL-Induced Osteoclast Formation and Resorption by Attenuating Reactive Oxygen Species Production in RAW264.7 Murine Macrophages. Int. J. Mol. Sci. 2021, 22, 544. [Google Scholar] [CrossRef]
- Ke, K.; Sul, O.J.; Chung, S.W.; Suh, J.H.; Choi, H.S. Lack of NOD2 attenuates ovariectomy-induced bone loss via inhibition of osteoclasts. J. Endocrinol. 2017, 235, 85–96. [Google Scholar] [CrossRef]
- Rahman, M.M.; El Jamali, A.; Halade, G.V.; Ouhtit, A.; Abou-Saleh, H.; Pintus, G. Nox2 Activity Is Required in Obesity-Mediated Alteration of Bone Remodeling. Oxidative Med. Cell. Longev. 2018, 2018, 6054361. [Google Scholar] [CrossRef]
- Kang, I.S.; Kim, C. NADPH oxidase gp91(phox) contributes to RANKL-induced osteoclast differentiation by upregulating NFATc1. Sci. Rep. 2016, 6, 38014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Görlach, A.; Dimova, E.Y.; Petry, A.; Martínez-Ruiz, A.; Hernansanz-Agustín, P.; Rolo, A.P.; Palmeira, C.M.; Kietzmann, T. Reactive oxygen species, nutrition, hypoxia and diseases: Problems solved? Redox Biol. 2015, 6, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, S.; Lee, H.; Yang, Y.; Jeong, W. Nrf2 deficiency induces oxidative stress and promotes RANKL-induced osteoclast differentiation. Free Radic. Biol. Med. 2013, 65, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Atia, A.; Alrawaiq, N.S.; Abdullah, A. The Effect of Tocotrienol-Rich Fraction on the Expression of Glutathione S-Transferase Isoenzymes in Mice Liver. Sains Malays. 2018, 47, 2799–2809. [Google Scholar] [CrossRef]
- Casati, L.; Pagani, F.; Limonta, P.; Vanetti, C.; Stancari, G.; Sibilia, V. Beneficial effects of δ-tocotrienol against oxidative stress in osteoblastic cells: Studies on the mechanisms of action. Eur. J. Nutr. 2020, 59, 1975–1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arany, Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr. Opin. Genet. Dev. 2008, 18, 426–434. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Kim, K. Exercise-induced PGC-1α transcriptional factors in skeletal muscle. Integr. Med. Res. 2014, 3, 155–160. [Google Scholar] [CrossRef] [Green Version]
- Buccoliero, C.; Dicarlo, M.; Pignataro, P.; Gaccione, F.; Colucci, S.; Colaianni, G.; Grano, M. The Novel Role of PGC1α in Bone Metabolism. Int. J. Mol. Sci. 2021, 22, 4670. [Google Scholar] [CrossRef]
- Sánchez-de-Diego, C.; Artigas, N.; Pimenta-Lopes, C.; Valer, J.A.; Torrejon, B.; Gama-Pérez, P.; Villena, J.A.; Garcia-Roves, P.M.; Rosa, J.L.; Ventura, F. Glucose Restriction Promotes Osteocyte Specification by Activating a PGC-1α-Dependent Transcriptional Program. iScience 2019, 15, 79–94. [Google Scholar] [CrossRef]
- Yu, B.; Huo, L.; Liu, Y.; Deng, P.; Szymanski, J.; Li, J.; Luo, X.; Hong, C.; Lin, J.; Wang, C.Y. PGC-1α Controls Skeletal Stem Cell Fate and Bone-Fat Balance in Osteoporosis and Skeletal Aging by Inducing TAZ. Cell Stem Cell 2018, 23, 193–209.e195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Austin, S.; St-Pierre, J. PGC1α and mitochondrial metabolism—Emerging concepts and relevance in ageing and neurodegenerative disorders. J. Cell Sci. 2012, 125, 4963–4971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Z.; Wei, W.; Yang, M.; Du, Y.; Wan, Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 2014, 20, 483–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, A.; Shen, G.; Wang, X.; Liu, G.; Yang, F.; Chen, B.; Wang, M.; Xu, Y. Hepcidin-induced reduction in iron content and PGC-1β expression negatively regulates osteoclast differentiation to play a protective role in postmenopausal osteoporosis. Aging 2021, 13, 11296–11314. [Google Scholar] [CrossRef]
- Jo, Y.J.; Lee, H.I.; Kim, N.; Hwang, D.; Lee, J.; Lee, G.R.; Hong, S.E.; Lee, H.; Kwon, M.; Kim, N.Y.; et al. Cinchonine inhibits osteoclast differentiation by regulating TAK1 and AKT, and promotes osteogenesis. J. Cell. Physiol. 2021, 236, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rohatgi, N.; Veis, D.J.; Schilling, J.; Teitelbaum, S.L.; Zou, W. PGC1β Organizes the Osteoclast Cytoskeleton by Mitochondrial Biogenesis and Activation. J. Bone Miner. Res. 2018, 33, 1114–1125. [Google Scholar] [CrossRef]
- Seo, H.; Lee, I.; Chung, H.S.; Bae, G.-U.; Chang, M.; Song, E.; Kim, M.J. ATP5B regulates mitochondrial fission and fusion in mammalian cells. Anim. Cells Syst. 2016, 20, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, C.A.; Fisher-Wellman, K.H.; Neufer, P.D. From OCR and ECAR to energy: Perspectives on the design and interpretation of bioenergetics studies. J. Biol. Chem. 2021, 297, 101140. [Google Scholar] [CrossRef]
- Nanayakkara, G.K.; Wang, H.; Yang, X. Proton leak regulates mitochondrial reactive oxygen species generation in endothelial cell activation and inflammation—A novel concept. Arch. Biochem. Biophys. 2019, 662, 68–74. [Google Scholar] [CrossRef]
- Yamamoto, H.; Morino, K.; Mengistu, L.; Ishibashi, T.; Kiriyama, K.; Ikami, T.; Maegawa, H. Amla Enhances Mitochondrial Spare Respiratory Capacity by Increasing Mitochondrial Biogenesis and Antioxidant Systems in a Murine Skeletal Muscle Cell Line. Oxidative Med. Cell. Longev. 2016, 2016, 1735841. [Google Scholar] [CrossRef] [Green Version]
- Carbognin, E.; Betto, R.M.; Soriano, M.E.; Smith, A.G.; Martello, G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 2016, 35, 618–634. [Google Scholar] [CrossRef]
- Marchetti, P.; Fovez, Q.; Germain, N.; Khamari, R.; Kluza, J. Mitochondrial spare respiratory capacity: Mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 13106–13124. [Google Scholar] [CrossRef]
- Williams, J.P.; Blair, H.C.; McDonald, J.M.; McKenna, M.A.; Jordan, S.E.; Williford, J.; Hardy, R.W. Regulation of osteoclastic bone resorption by glucose. Biochem. Biophys. Res. Commun. 1997, 235, 646–651. [Google Scholar] [CrossRef]
- Ryu, W.I.; Cohen, B.M.; Sonntag, K.C. Hypothesis and Theory: Characterizing Abnormalities of Energy Metabolism Using a Cellular Platform as a Personalized Medicine Approach for Alzheimer’s Disease. Front. Cell Dev. Biol. 2021, 9, 697578. [Google Scholar] [CrossRef]
- Ikeda, K.; Takeshita, S. The role of osteoclast differentiation and function in skeletal homeostasis. J. Biochem. 2016, 159, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Radzki, R.P.; Bienko, M.; Pierzynowski, S.G. Anti-osteopenic effect of alpha-ketoglutarate sodium salt in ovariectomized rats. J. Bone Miner. Metab. 2012, 30, 651–659. [Google Scholar] [CrossRef]
- Oh, S.J.; Gu, D.R.; Jin, S.H.; Park, K.H.; Lee, S.H. Cytosolic malate dehydrogenase regulates RANKL-mediated osteoclastogenesis via AMPK/c-Fos/NFATc1 signaling. Biochem. Biophys. Res. Commun. 2016, 475, 125–132. [Google Scholar] [CrossRef]
- Slane, B.G.; Aykin-Burns, N.; Smith, B.J.; Kalen, A.L.; Goswami, P.C.; Domann, F.E.; Spitz, D.R. Mutation of succinate dehydrogenase subunit C results in increased O2 −, oxidative stress, and genomic instability. Cancer Res. 2006, 66, 7615–7620. [Google Scholar] [CrossRef] [Green Version]
- Dröse, S. Differential effects of complex II on mitochondrial ROS production and their relation to cardioprotective pre- and postconditioning. Biochim. Biophys. Acta 2013, 1827, 578–587. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Xie, C.; Li, X.; Yang, J.; Yu, T.; Zhang, R.; Zhang, T.; Saxena, D.; Snyder, M.; Wu, Y.; et al. Succinate and its G-protein-coupled receptor stimulates osteoclastogenesis. Nat. Commun. 2017, 8, 15621. [Google Scholar] [CrossRef] [PubMed]
- Sartori, M.; Vincenzi, F.; Ravani, A.; Cepollaro, S.; Martini, L.; Varani, K.; Fini, M.; Tschon, M. RAW 264.7 co-cultured with ultra-high molecular weight polyethylene particles spontaneously differentiate into osteoclasts: An in vitro model of periprosthetic osteolysis. J. Biomed. Mater. Res. Part A 2017, 105, 510–520. [Google Scholar] [CrossRef]
- Taciak, B.; Białasek, M.; Braniewska, A.; Sas, Z.; Sawicka, P.; Kiraga, Ł.; Rygiel, T.; Król, M. Evaluation of phenotypic and functional stability of RAW 264.7 cell line through serial passages. PLoS ONE 2018, 13, e0198943. [Google Scholar] [CrossRef] [PubMed]

| Gene | Sense (5′→3′) | Antisense (5′→3′) |
|---|---|---|
| Traf6 | AAA GCG AGA GAT TCT TTC CCT G | ACTGGGGACAATTCACTAGAGC |
| Rank | GCC CAG TCT CAT CGT TCT GC | GCA AGC ATC ATT GAC CCA ATT C |
| Ctsk | TGG AGT TGA CTT CCG CAA TCC | CCC ACA TCC TGC TGT TGA GAA T |
| Atp5b | AGT TGC TGA GGT CTT CAC GG | GGA GAT GGT CAT ATT CAC CTG C |
| Nox 1 | AAG CCA TCC TCA CAA TTG TTC C | AGG ATC CAC TTC CAA GAC TCA G |
| Nox 2 | TTC CAG TGC GTG TTG CTC G | GTG CAA TTG TGT GGA TGG CG |
| Nox 3 | AACAAGTGTGTGCTGTAGAGG | TCCAGGTTGAACAAGTGTGCC |
| Nox 4 | TAC TTC CAA GAT GAA CCA TGC C | GGA ATC GTT CTG TCC AGT CTC C |
| Nfatc1 | ACA TGC GAG CCA TCA TCG AC | TGT GAA CTC GGA AGA CCA GC |
| Gapdh | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGT |
| Pgc1a | CTTGTGTCAAGGTGGGC | TGAGGTGCTTATCGAGTTCCG |
| Pgc1b | CTTGTGTCAAGGTGGATGGC | TGAGGTGCTTATGCAGTTCCG |
| Cyc | GAA CAA GTG TGG TTG CAC CG | AGCTTCGGACTCGAAGACAG |
| Nrf2 | AGT GGATCCGCCAGCTACTC | GCAAGCGACTCATGGTCATC |
| Calcr | CACTGCTAAGGAGAGCCAGC | TGAGGCGCAGAAGTAAGCAC |
| Itgb3 | GTGGAAGAGCCTGAGTGTCC | AGATGAGCAGAGTAGCAAGGC |
| Dcstamp | ACGTGGAGAGCAAGGAACC | TCTCAGACACACTGAGACGTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamad Hazir, N.S.; Yahaya, N.H.M.; Zawawi, M.S.F.; Damanhuri, H.A.; Mohamed, N.; Alias, E. Changes in Metabolism and Mitochondrial Bioenergetics during Polyethylene-Induced Osteoclastogenesis. Int. J. Mol. Sci. 2022, 23, 8331. https://doi.org/10.3390/ijms23158331
Mohamad Hazir NS, Yahaya NHM, Zawawi MSF, Damanhuri HA, Mohamed N, Alias E. Changes in Metabolism and Mitochondrial Bioenergetics during Polyethylene-Induced Osteoclastogenesis. International Journal of Molecular Sciences. 2022; 23(15):8331. https://doi.org/10.3390/ijms23158331
Chicago/Turabian StyleMohamad Hazir, Nur Shukriyah, Nor Hamdan Mohamad Yahaya, Muhamad Syahrul Fitri Zawawi, Hanafi Ahmad Damanhuri, Norazlina Mohamed, and Ekram Alias. 2022. "Changes in Metabolism and Mitochondrial Bioenergetics during Polyethylene-Induced Osteoclastogenesis" International Journal of Molecular Sciences 23, no. 15: 8331. https://doi.org/10.3390/ijms23158331
APA StyleMohamad Hazir, N. S., Yahaya, N. H. M., Zawawi, M. S. F., Damanhuri, H. A., Mohamed, N., & Alias, E. (2022). Changes in Metabolism and Mitochondrial Bioenergetics during Polyethylene-Induced Osteoclastogenesis. International Journal of Molecular Sciences, 23(15), 8331. https://doi.org/10.3390/ijms23158331







