Large-Scale Identification of Multiple Classes of Host Defense Peptide-Inducing Compounds for Antimicrobial Therapy
Abstract
:1. Introduction
2. Results
2.1. HTS for Small-Molecule Compounds That Induce HDP Synthesis
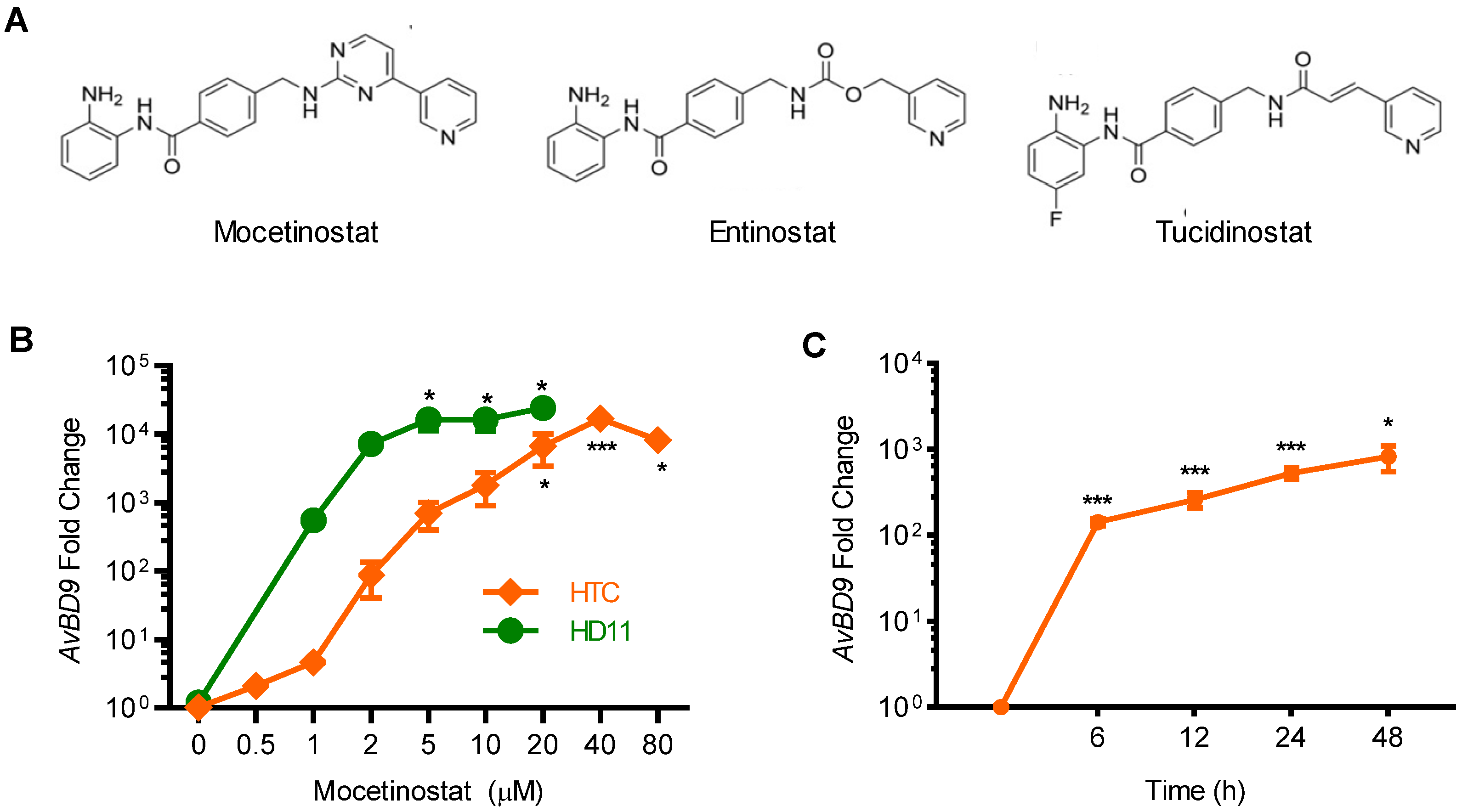
2.2. Confirmation of the AvBD9-Inducing Capacity of Mocetinostat in Chicken Macrophages
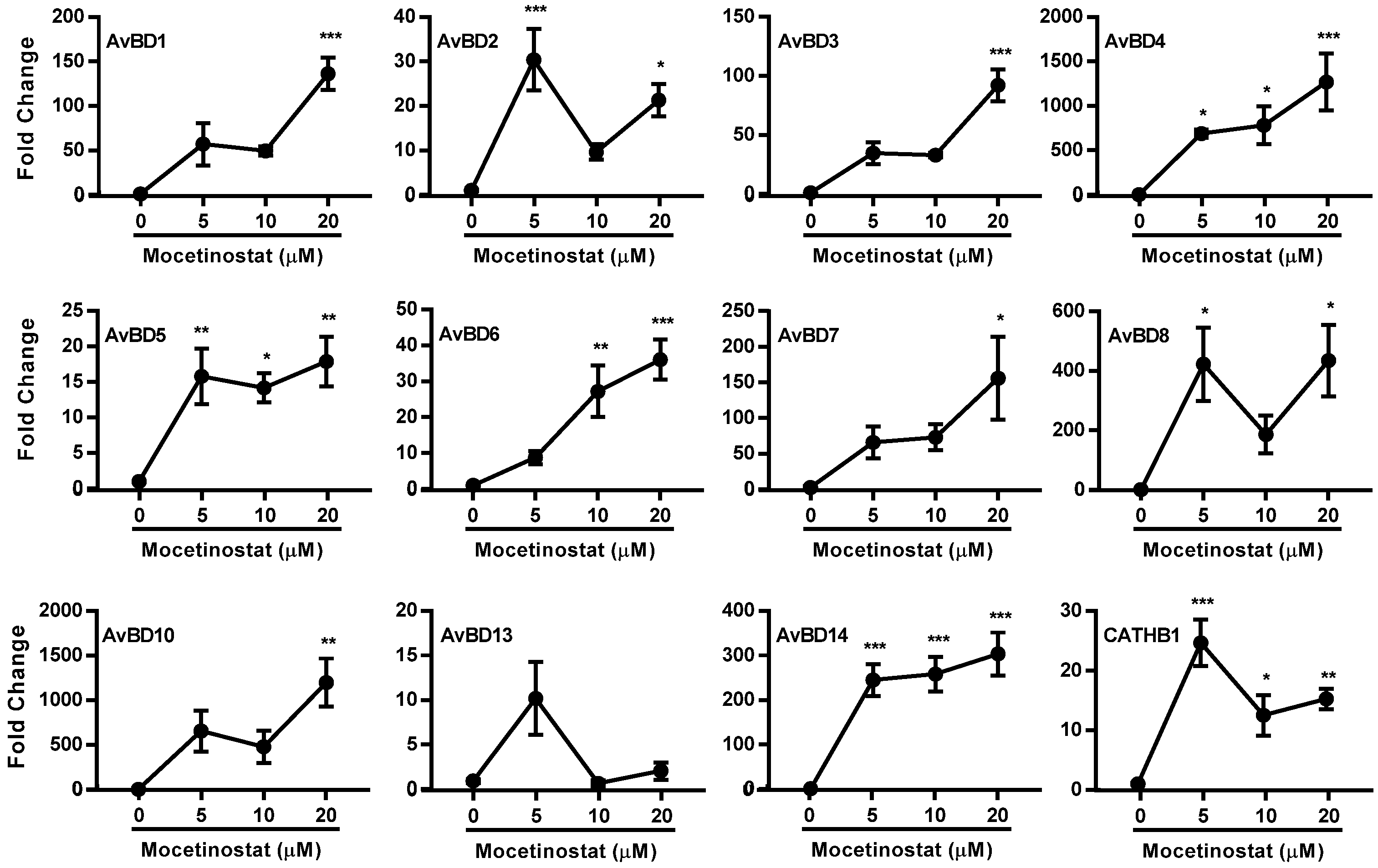
2.3. Induction of Multiple HDP and Barrier-Function Genes by Mocetinostat
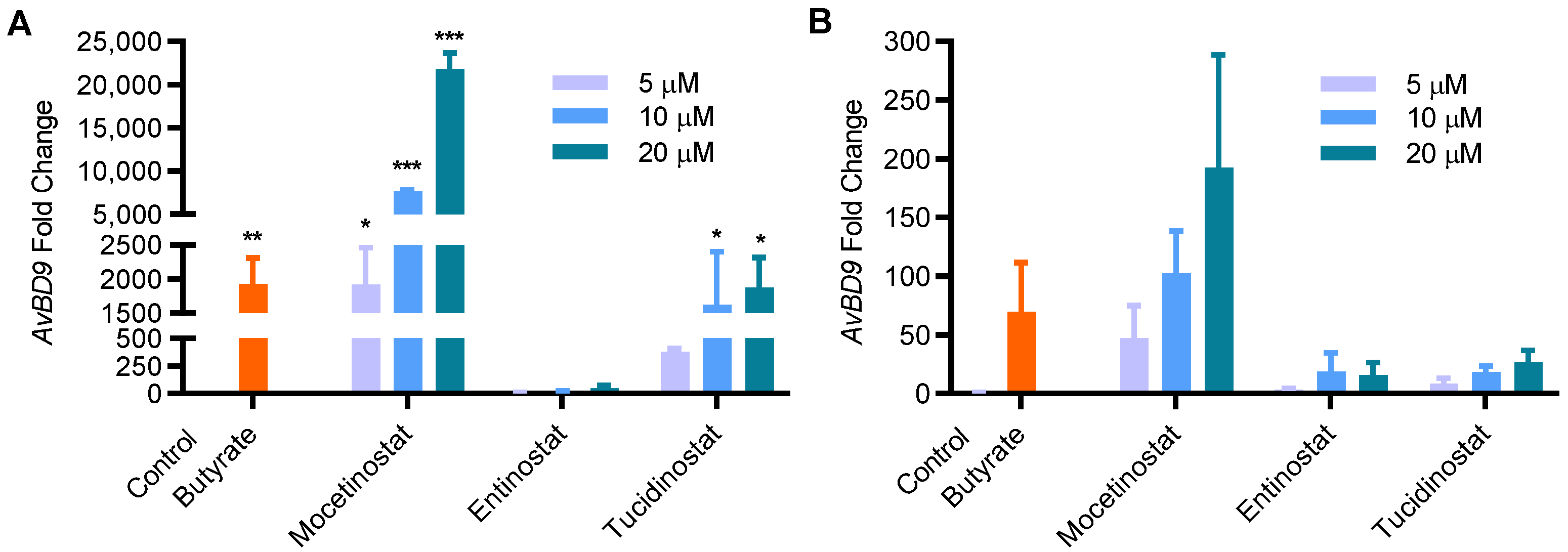
2.4. Comparison of HDP-Inducing Efficacy among Mocetinostat, Entinostat, and Tucidinostat
2.5. Mocetinostat Augments the Antibacterial Activity of Chicken Macrophages without Directly Killing Bacteria
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. HTS for HDP-Inducing Compounds
4.4. Validation of Hit Compounds
4.5. RNA Isolation and RT-qPCR
4.6. Chicken Intestinal Explant Culture
4.7. Antibacterial Assay of Chicken HTC Macrophages
4.8. Minimum Inhibitory Concentration (MIC) Assay
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| AKTi | protein kinase B inhibitor |
| AvBD9 | avian β-defensin 9 |
| CaChi | calcium-channel inhibitor |
| CLDN1 | claudin 1 |
| D2DRi | dopamine receptor D2 inhibitor |
| EGFRi | epidermal growth factor receptor antagonist |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| HDAC | histone deacetylase |
| HDACi | HDAC inhibitor |
| HDPs | host defense peptides |
| HER2 | human epidermal growth factor receptor 2 antagonist |
| HH1Ri | histamine H1 receptor inhibitor |
| HMT | histone methyltransferase inhibitor |
| HMT | HMT inhibitor |
| HTS | high-throughput screening |
| MIC | minimum inhibitory concentration |
| mTOR | mammalian target of rapamycin |
| MUC2 | mucin 2 |
| NF-κBi | inhibitor to nuclear factor kappa-light-chain-enhancer of activated B cells |
| NRi | noradrenaline reuptake inhibitor |
| p53a | p53 activator |
| PBMCs | peripheral blood mononuclear cells |
| PI3Ki | phosphatidylinositide 3-kinase inhibitor |
| RT-qPCR | reverse transcriptase-quantitative PCR |
| SEM | standard error of the mean |
| TJP1 | tight junction protein 1 |
| TOPIIi | topoisomerase II inhibitor |
References
- Schrader, S.M.; Vaubourgeix, J.; Nathan, C. Biology of antimicrobial resistance and approaches to combat it. Sci. Transl. Med. 2020, 12, eaaz6992. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [PubMed]
- Zumla, A.; Rao, M.; Wallis, R.S.; Kaufmann, S.H.; Rustomjee, R.; Mwaba, P.; Vilaplana, C.; Yeboah-Manu, D.; Chakaya, J.; Ippolito, G.; et al. Host-directed therapies for infectious diseases: Current status, recent progress, and future prospects. Lancet Infect. Dis. 2016, 16, e47–e63. [Google Scholar] [CrossRef] [Green Version]
- Bergman, P.; Raqib, R.; Rekha, R.S.; Agerberth, B.; Gudmundsson, G.H. Host Directed Therapy against Infection by Boosting Innate Immunity. Front. Immunol. 2020, 11, 1209. [Google Scholar]
- Ting, D.S.J.; Beuerman, R.W.; Dua, H.S.; Lakshminarayanan, R.; Mohammed, I. Strategies in Translating the Therapeutic Potentials of Host Defense Peptides. Front. Immunol. 2020, 11, 983. [Google Scholar]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Sunkara, L.T. Avian antimicrobial host defense peptides: From biology to therapeutic applications. Pharmaceuticals 2014, 7, 220–247. [Google Scholar] [CrossRef] [Green Version]
- Cuperus, T.; Coorens, M.; van Dijk, A.; Haagsman, H.P. Avian host defense peptides. Dev. Comp. Immunol. 2013, 41, 352–369. [Google Scholar] [CrossRef]
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar]
- Rodriguez-Carlos, A.; Jacobo-Delgado, Y.M.; Santos-Mena, A.O.; Rivas-Santiago, B. Modulation of cathelicidin and defensins by histone deacetylase inhibitors: A potential treatment for multi-drug resistant infectious diseases. Peptides 2021, 140, 170527. [Google Scholar] [CrossRef]
- Wu, J.; Ma, N.; Johnston, L.J.; Ma, X. Dietary Nutrients Mediate Intestinal Host Defense Peptide Expression. Adv. Nutr. 2020, 11, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.; Ma, X.; Liu, Y.; Qiao, S.; Hou, Y.; Zhang, G. Dietary modulation of endogenous host defense peptide synthesis as an alternative approach to in-feed antibiotics. Anim. Nutr. 2018, 4, 160–169. [Google Scholar] [PubMed]
- Raqib, R.; Sarker, P.; Bergman, P.; Ara, G.; Lindh, M.; Sack, D.A.; Nasirul Islam, K.M.; Gudmundsson, G.H.; Andersson, J.; Agerberth, B. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc. Natl. Acad. Sci. USA 2006, 103, 9178–9183. [Google Scholar] [CrossRef] [Green Version]
- Al-Mamun, A.; Mily, A.; Sarker, P.; Tiash, S.; Navarro, A.; Akter, M.; Talukder, K.A.; Islam, M.F.; Agerberth, B.; Gudmundsson, G.H.; et al. Treatment with phenylbutyrate in a pre-clinical trial reduces diarrhea due to enteropathogenic Escherichia coli: Link to cathelicidin induction. Microbes Infect. 2013, 15, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Mily, A.; Rekha, R.S.; Kamal, S.M.; Arifuzzaman, A.S.; Rahim, Z.; Khan, L.; Haq, M.A.; Zaman, K.; Bergman, P.; Brighenti, S.; et al. Significant Effects of Oral Phenylbutyrate and Vitamin D3 Adjunctive Therapy in Pulmonary Tuberculosis: A Randomized Controlled Trial. PLoS ONE 2015, 10, e0138340. [Google Scholar] [CrossRef]
- Sarker, P.; Ahmed, S.; Tiash, S.; Rekha, R.S.; Stromberg, R.; Andersson, J.; Bergman, P.; Gudmundsson, G.H.; Agerberth, B.; Raqib, R. Phenylbutyrate counteracts Shigella mediated downregulation of cathelicidin in rabbit lung and intestinal epithelia: A potential therapeutic strategy. PLoS ONE 2011, 6, e20637. [Google Scholar] [CrossRef] [Green Version]
- Sarker, P.; Banik, A.; Stromberg, R.; Gudmundsson, G.H.; Raqib, R.; Agerberth, B. Treatment with Entinostat Heals Experimental Cholera by Affecting Physical and Chemical Barrier Functions of Intestinal Epithelia. Antimicrob. Agents Chemother. 2017, 61, e02516–e02570. [Google Scholar]
- Yang, Q.; Whitmore, M.A.; Robinson, K.; Lyu, W.; Zhang, G. Butyrate, Forskolin, and Lactose Synergistically Enhance Disease Resistance by Inducing the Expression of the Genes Involved in Innate Host Defense and Barrier Function. Antibiotics 2021, 10, 1175. [Google Scholar] [CrossRef]
- Robinson, K.; Yang, Q.; Li, H.; Zhang, L.; Aylward, B.; Arsenault, R.J.; Zhang, G. Butyrate and Forskolin Augment Host Defense, Barrier Function, and Disease Resistance without Eliciting Inflammation. Front. Nutr. 2021, 8, 778424. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, B.; Robinson, K.; Belem, T.; Lyu, W.; Deng, Z.; Ramanathan, R.; Zhang, G. Butyrate in combination with forskolin alleviates necrotic enteritis, increases feed efficiency, and improves carcass composition of broilers. J. Anim. Sci. Biotechnol. 2022, 13, 3. [Google Scholar] [CrossRef]
- Nylen, F.; Miraglia, E.; Cederlund, A.; Ottosson, H.; Stromberg, R.; Gudmundsson, G.H.; Agerberth, B. Boosting innate immunity: Development and validation of a cell-based screening assay to identify LL-37 inducers. Innate Immun. 2014, 20, 364–376. [Google Scholar] [CrossRef] [Green Version]
- Ottosson, H.; Nylen, F.; Sarker, P.; Miraglia, E.; Bergman, P.; Gudmundsson, G.H.; Raqib, R.; Agerberth, B.; Stromberg, R. Potent Inducers of Endogenous Antimicrobial Peptides for Host Directed Therapy of Infections. Sci. Rep. 2016, 6, 36692. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Wang, J.; Lyu, W.; Wieneke, X.; Matts, R.; Ma, X.; Zhang, G. Development of a Cell-Based High-Throughput Screening Assay to Identify Porcine Host Defense Peptide-Inducing Compounds. J. Immunol. Res. 2018, 2018, 5492941. [Google Scholar] [CrossRef]
- Lyu, W.; Deng, Z.; Sunkara, L.T.; Becker, S.; Robinson, K.; Matts, R.; Zhang, G. High Throughput Screening for Natural Host Defense Peptide-Inducing Compounds as Novel Alternatives to Antibiotics. Front. Cell. Infect. Microbiol. 2018, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lyu, W.; Zhang, W.; Chen, Y.; Luo, F.; Wang, Y.; Ji, H.; Zhang, G. Discovery of natural products capable of inducing porcine host defense peptide gene expression using cell-based high throughput screening. J. Anim. Sci. Biotechnol. 2021, 12, 14. [Google Scholar] [CrossRef]
- Deng, Z.; Lyu, W.; Zhang, G. High-Throughput Identification of Epigenetic Compounds to Enhance Chicken Host Defense Peptide Gene Expression. Antibiotics 2022, 11, 933. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Harper, B.; Hughes, I.E. A comparison in rabbit isolated hearts of the dysrhythmogenic potential of amitriptyline, maprotiline and mianserin in relation to their ability to block noradrenaline uptake. Br. J. Pharmacol. 1977, 59, 651–660. [Google Scholar] [CrossRef] [Green Version]
- Canonica, G.W.; Blaiss, M. Antihistaminic, anti-inflammatory, and antiallergic properties of the nonsedating second-generation antihistamine desloratadine: A review of the evidence. World Allergy Organ. J. 2011, 4, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, V.F.; Garcia, M.L.; Himmel, D.; Reuben, J.P.; Lam, Y.K.; Pan, J.X.; Han, G.Q.; Kaczorowski, G.J. Interaction of tetrandrine with slowly inactivating calcium channels. Characterization of calcium channel modulation by an alkaloid of Chinese medicinal herb origin. J. Biol. Chem. 1988, 263, 2238–2244. [Google Scholar] [CrossRef]
- Gurova, K.V.; Hill, J.E.; Guo, C.; Prokvolit, A.; Burdelya, L.G.; Samoylova, E.; Khodyakova, A.V.; Ganapathi, R.; Ganapathi, M.; Tararova, N.D.; et al. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 17448–17453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, J.R.; Sawada, K.; Nishibori, M.; Zhang, X.; Cheng, X. Two polymorphic forms of human histamine methyltransferase: Structural, thermal, and kinetic comparisons. Structure 2001, 9, 837–849. [Google Scholar] [CrossRef] [Green Version]
- Creese, I.; Burt, D.R.; Snyder, S.H. Dopamine receptor binding predicts clinical and pharmacological potencies of antischizophrenic drugs. Science 1976, 192, 481–483. [Google Scholar] [CrossRef]
- Blum, K.A.; Advani, A.; Fernandez, L.; Van Der Jagt, R.; Brandwein, J.; Kambhampati, S.; Kassis, J.; Davis, M.; Bonfils, C.; Dubay, M.; et al. Phase II study of the histone deacetylase inhibitor MGCD0103 in patients with previously treated chronic lymphocytic leukaemia. Br. J. Haematol. 2009, 147, 507–514. [Google Scholar] [CrossRef]
- Batlevi, C.L.; Crump, M.; Andreadis, C.; Rizzieri, D.; Assouline, S.E.; Fox, S.; van der Jagt, R.H.C.; Copeland, A.; Potvin, D.; Chao, R.; et al. A phase 2 study of mocetinostat, a histone deacetylase inhibitor, in relapsed or refractory lymphoma. Br. J. Haematol. 2017, 178, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Grivas, P.; Mortazavi, A.; Picus, J.; Hahn, N.M.; Milowsky, M.I.; Hart, L.L.; Alva, A.; Bellmunt, J.; Pal, S.K.; Bambury, R.M.; et al. Mocetinostat for patients with previously treated, locally advanced/metastatic urothelial carcinoma and inactivating alterations of acetyltransferase genes. Cancer 2019, 125, 533–540. [Google Scholar] [CrossRef]
- Choy, E.; Ballman, K.; Chen, J.; Dickson, M.A.; Chugh, R.; George, S.; Okuno, S.; Pollock, R.; Patel, R.M.; Hoering, A.; et al. SARC018_SPORE02: Phase II Study of Mocetinostat Administered with Gemcitabine for Patients with Metastatic Leiomyosarcoma with Progression or Relapse following Prior Treatment with Gemcitabine-Containing Therapy. Sarcoma 2018, 2018, 2068517. [Google Scholar] [CrossRef] [Green Version]
- Beug, H.; von Kirchbach, A.; Doderlein, G.; Conscience, J.F.; Graf, T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 1979, 18, 375–390. [Google Scholar] [CrossRef]
- Miraglia, E.; Nylen, F.; Johansson, K.; Arner, E.; Cebula, M.; Farmand, S.; Ottosson, H.; Stromberg, R.; Gudmundsson, G.H.; Agerberth, B.; et al. Entinostat up-regulates the CAMP gene encoding LL-37 via activation of STAT3 and HIF-1alpha transcription factors. Sci. Rep. 2016, 6, 33274. [Google Scholar] [CrossRef] [PubMed]
- Sunkara, L.T.; Achanta, M.; Schreiber, N.B.; Bommineni, Y.R.; Dai, G.; Jiang, W.; Lamont, S.; Lillehoj, H.S.; Beker, A.; Teeter, R.G.; et al. Butyrate enhances disease resistance of chickens by inducing antimicrobial host defense peptide gene expression. PLoS ONE 2011, 6, e27225. [Google Scholar] [CrossRef]
- National Committee for Clinical Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 6th ed.; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2003. [Google Scholar]
- Shvedunova, M.; Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell. Biol. 2022, 23, 329–349. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.E.; English, D.M.; Cowley, S.M. Acetylation & Co: An expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. 2019, 63, 97–107. [Google Scholar] [PubMed] [Green Version]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef]
- Zughaibi, T.A.; Suhail, M.; Tarique, M.; Tabrez, S. Targeting PI3K/Akt/mTOR Pathway by Different Flavonoids: A Cancer Chemopreventive Approach. Int. J. Mol. Sci. 2021, 22, 12455. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Wu, W.; Sun, M.; Bilotta, A.J.; Yao, S.; Xiao, Y.; Huang, X.; Eaves-Pyles, T.D.; Golovko, G.; et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal. Immunol. 2018, 11, 752–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, N.; Moradei, O.; Raeppel, S.; Leit, S.; Frechette, S.; Gaudette, F.; Paquin, I.; Bernstein, N.; Bouchain, G.; Vaisburg, A.; et al. Discovery of N-(2-aminophenyl)-4-[(4-pyridin-3-ylpyrimidin-2-ylamino)methyl]benzamide (MGCD0103), an orally active histone deacetylase inhibitor. J. Med. Chem. 2008, 51, 4072–4075. [Google Scholar] [CrossRef]
- Khan, N.; Jeffers, M.; Kumar, S.; Hackett, C.; Boldog, F.; Khramtsov, N.; Qian, X.; Mills, E.; Berghs, S.C.; Carey, N.; et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem. J. 2008, 409, 581–589. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.C.S.; Chan, A.H.Y.; Ganesan, A. Thirty Years of HDAC Inhibitors: 2020 Insight and Hindsight. J. Med. Chem. 2020, 63, 12460–12484. [Google Scholar] [CrossRef]
- Lee, J.L.; Wang, M.J.; Chen, J.Y. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J. Cell Biol. 2009, 185, 949–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albanese, A.; Daly, L.A.; Mennerich, D.; Kietzmann, T.; See, V. The Role of Hypoxia-Inducible Factor Post-Translational Modifications in Regulating Its Localisation, Stability, and Activity. Int. J. Mol. Sci. 2020, 22, 268. [Google Scholar] [CrossRef]
- Cesena, T.I.; Cardinaux, J.R.; Kwok, R.; Schwartz, J. CCAAT/enhancer-binding protein (C/EBP) beta is acetylated at multiple lysines: Acetylation of C/EBPbeta at lysine 39 modulates its ability to activate transcription. J. Biol. Chem. 2007, 282, 956–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rath, N.C.; Parcells, M.S.; Xie, H.; Santin, E. Characterization of a spontaneously transformed chicken mononuclear cell line. Vet. Immunol. Immunopathol. 2003, 96, 93–104. [Google Scholar] [CrossRef]
- Yang, Q.; Fong, L.A.; Lyu, W.; Sunkara, L.T.; Xiao, K.; Zhang, G. Synergistic Induction of Chicken Antimicrobial Host Defense Peptide Gene Expression by Butyrate and Sugars. Front. Microbiol. 2021, 12, 781649. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Sunkara, L.T.; Zeng, X.; Curtis, A.R.; Zhang, G. Cyclic AMP synergizes with butyrate in promoting beta-defensin 9 expression in chickens. Mol. Immunol. 2014, 57, 171–180. [Google Scholar] [CrossRef]
- Schauber, J.; Dorschner, R.A.; Yamasaki, K.; Brouha, B.; Gallo, R.L. Control of the innate epithelial antimicrobial response is cell-type specific and dependent on relevant microenvironmental stimuli. Immunology 2006, 118, 509–519. [Google Scholar] [CrossRef]
- Xiao, Y.; Dai, H.; Bommineni, Y.R.; Soulages, J.L.; Gong, Y.X.; Prakash, O.; Zhang, G. Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken. FEBS J. 2006, 273, 2581–2593. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Herrera, A.I.; Bommineni, Y.R.; Soulages, J.L.; Prakash, O.; Zhang, G. The central kink region of fowlicidin-2, an alpha-helical host defense peptide, is critically involved in bacterial killing and endotoxin neutralization. J. Innate Immun. 2009, 1, 268–280. [Google Scholar] [CrossRef]
- Bommineni, Y.R.; Dai, H.; Gong, Y.X.; Soulages, J.L.; Fernando, S.C.; Desilva, U.; Prakash, O.; Zhang, G. Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities. FEBS J. 2007, 274, 418–428. [Google Scholar] [CrossRef] [PubMed]
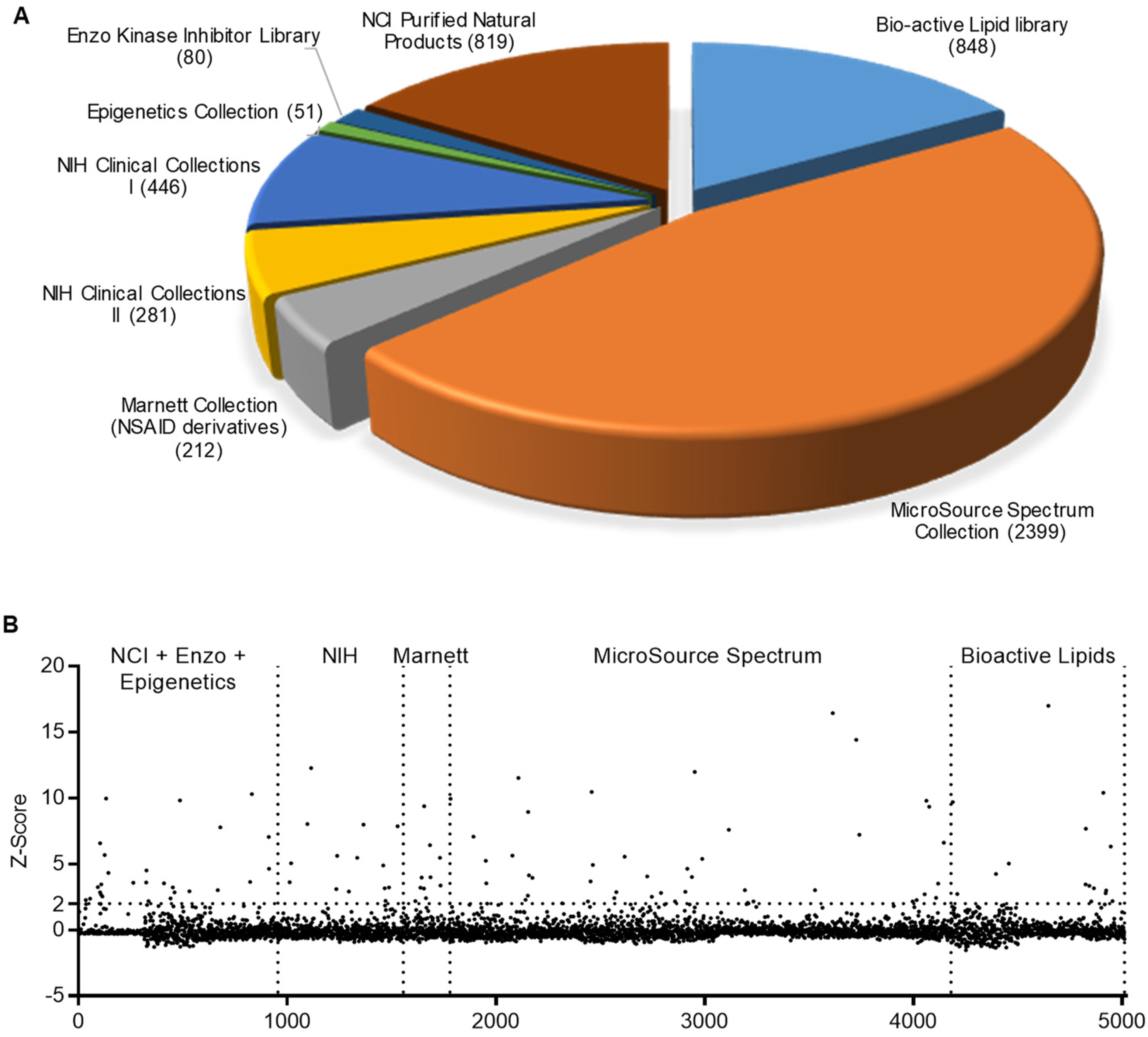
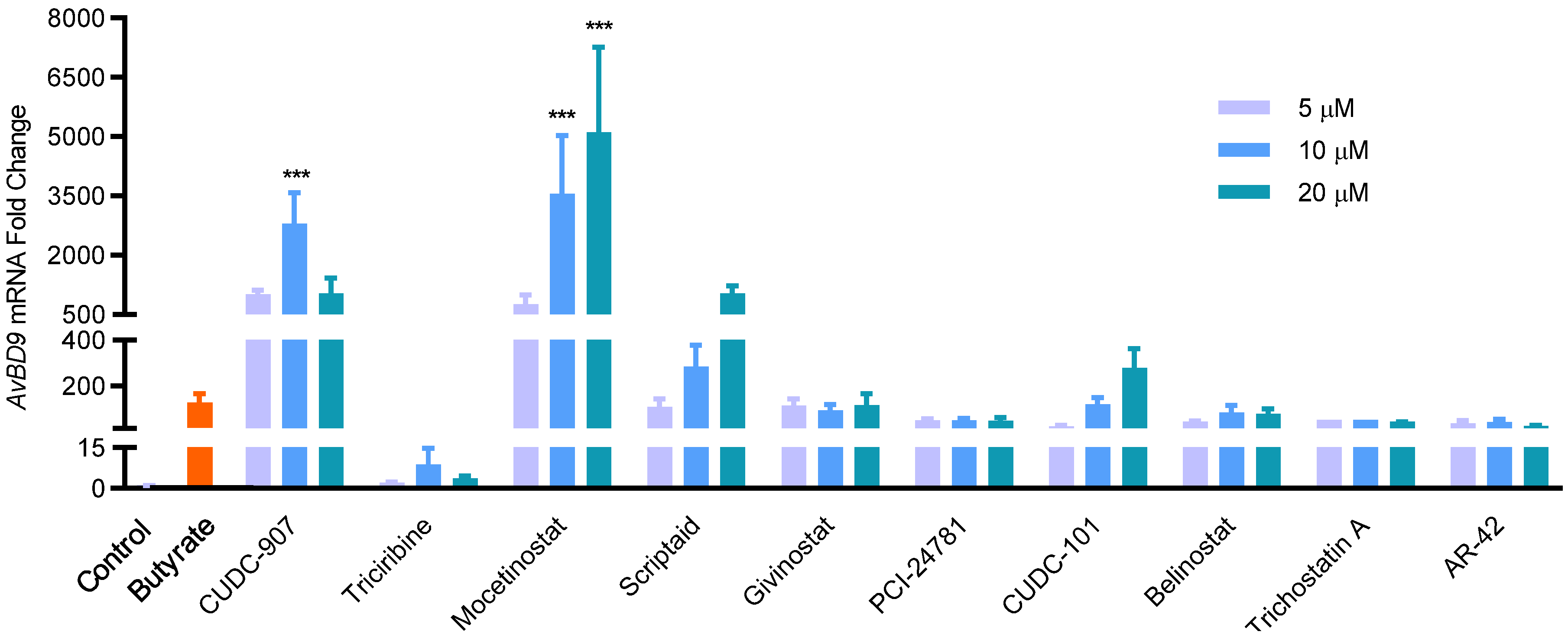

| Compound | Major Function | Z-Score | Fold Change | Cell Viability (%) | ||||
|---|---|---|---|---|---|---|---|---|
| 5 μM | 10 μM | 20 μM | 5 μM | 10 μM | 20 μM | |||
| CUDC-907 | HDACi/PI3Ki 2 | 9.97 | 137.04 | 482.67 | 264.88 | 88.1 | 69.1 | 51.2 |
| Triciribine | AKTi | 3.25 | 37.36 | 77.73 | 156.08 | 92.8 | 74.7 | 79.4 |
| Mocetinostat | HDACi | 3.45 | 21.36 | 41.72 | 59.69 | 96.7 | 96.0 | 74.4 |
| Scriptaid | HDACi | 4.33 | 6.17 | 25.71 | 50.42 | 100.3 | 92.4 | 80.7 |
| Givinostat | HDACi | 5.68 | 52.38 | 113.64 | 46.13 | 99.1 | 85.0 | 92.5 |
| PCI-24781 | HDACi | 2.85 | 77.51 | 82.80 | 37.43 | 98.3 | 87.9 | 80.9 |
| CUDC-101 | HDACi/EGFRi/HER2i | 2.54 | 4.69 | 12.52 | 34.17 | 105.0 | 66.5 | 70.3 |
| Belinostat | HDACi | 6.58 | 23.05 | 41.44 | 33.96 | 97.5 | 98.8 | 79.7 |
| Trichostatin A | HDACi | 17.00 | 68.08 | 47.64 | 27.62 | 96.1 | 106.2 | 68.4 |
| AR-42 | HDACi | 2.27 | 107.71 | 80.82 | 25.88 | 103.0 | 111.2 | 85.8 |
| Vorinostat | HDACi | 3.35 | 1.59 | 3.52 | 14.53 | 101.3 | 87.5 | 74.4 |
| Wortmannin | PI3Ki | 2.38 | 29.61 | 33.69 | 12.09 | 92.4 | 67.9 | 58.9 |
| Maprotiline hydrochloride | NRi | 5.61 | 1.53 | 3.85 | 7.01 | 102.3 | 97.7 | 64.1 |
| Desloratadine | HH1Ri | 3.00 | 1.73 | 2.47 | 6.83 | 97.6 | 93.8 | 76.6 |
| Doxorubicin | TOPIIi | 5.64 | 7.89 | 101.89 | 6.56 | 95.9 | 71.9 | 65.8 |
| Tetrandrine | CaChi | 2.17 | 1.22 | 1.97 | 6.42 | 102.3 | 89.6 | 69.2 |
| Quinacrine | NF-κBi/p53a/HMTi | 3.02 | 1.78 | 2.10 | 6.03 | 97.7 | 75.2 | 60.9 |
| Promazine | D2DRi | 2.82 | 0.64 | 3.24 | 5.39 | 100.4 | 74.2 | 58.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, W.; Mi, D.; Vinson, P.N.; Xiao, Y.; Zhang, G. Large-Scale Identification of Multiple Classes of Host Defense Peptide-Inducing Compounds for Antimicrobial Therapy. Int. J. Mol. Sci. 2022, 23, 8400. https://doi.org/10.3390/ijms23158400
Lyu W, Mi D, Vinson PN, Xiao Y, Zhang G. Large-Scale Identification of Multiple Classes of Host Defense Peptide-Inducing Compounds for Antimicrobial Therapy. International Journal of Molecular Sciences. 2022; 23(15):8400. https://doi.org/10.3390/ijms23158400
Chicago/Turabian StyleLyu, Wentao, Dehui Mi, Paige N. Vinson, Yingping Xiao, and Guolong Zhang. 2022. "Large-Scale Identification of Multiple Classes of Host Defense Peptide-Inducing Compounds for Antimicrobial Therapy" International Journal of Molecular Sciences 23, no. 15: 8400. https://doi.org/10.3390/ijms23158400
APA StyleLyu, W., Mi, D., Vinson, P. N., Xiao, Y., & Zhang, G. (2022). Large-Scale Identification of Multiple Classes of Host Defense Peptide-Inducing Compounds for Antimicrobial Therapy. International Journal of Molecular Sciences, 23(15), 8400. https://doi.org/10.3390/ijms23158400







