Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants
Abstract
:1. Introduction
2. Results
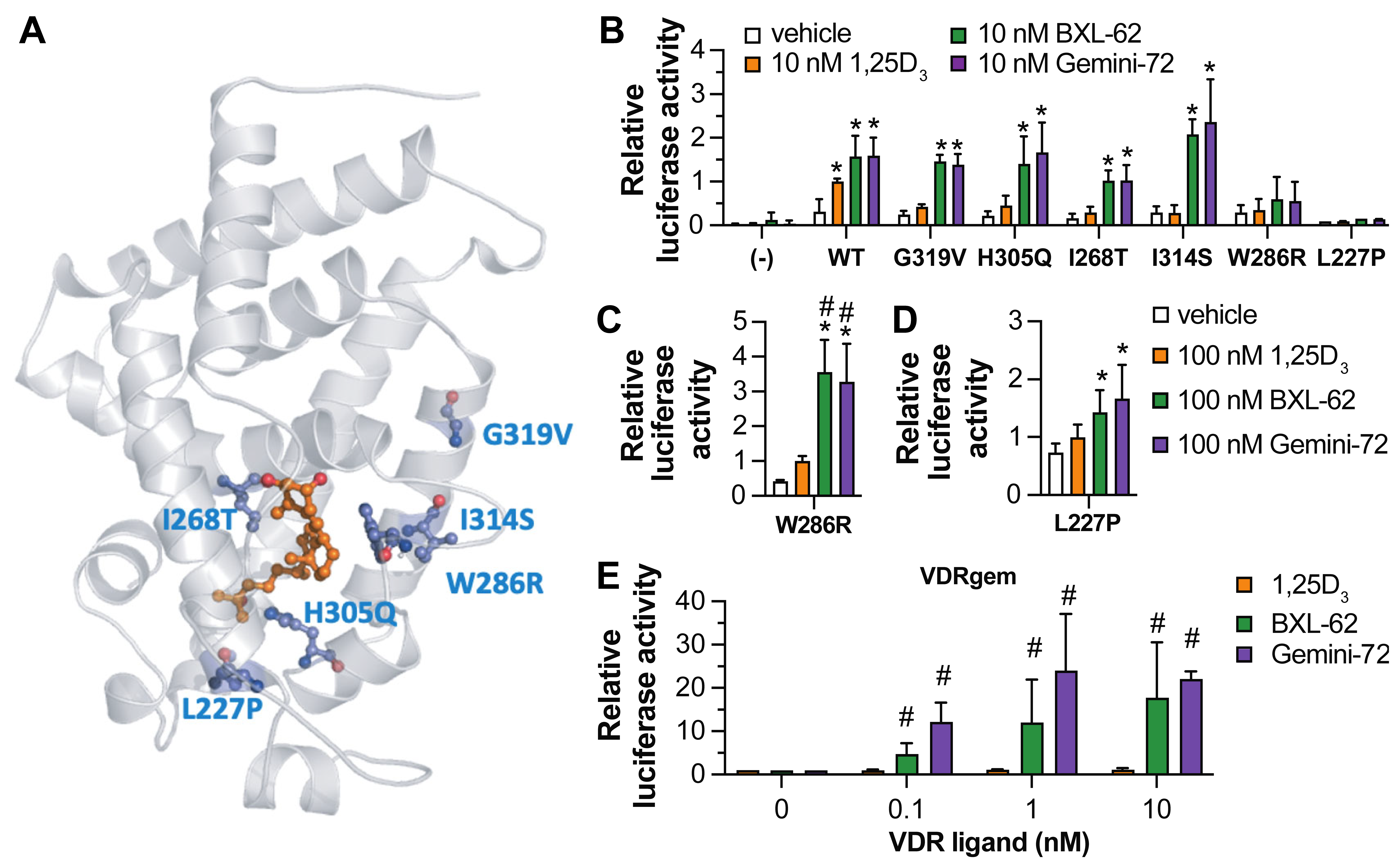
2.1. BXL-62 and Gemini-72 Induce the Transcriptional Activity of Point-Mutated VDR Unresponsive to 1,25D3
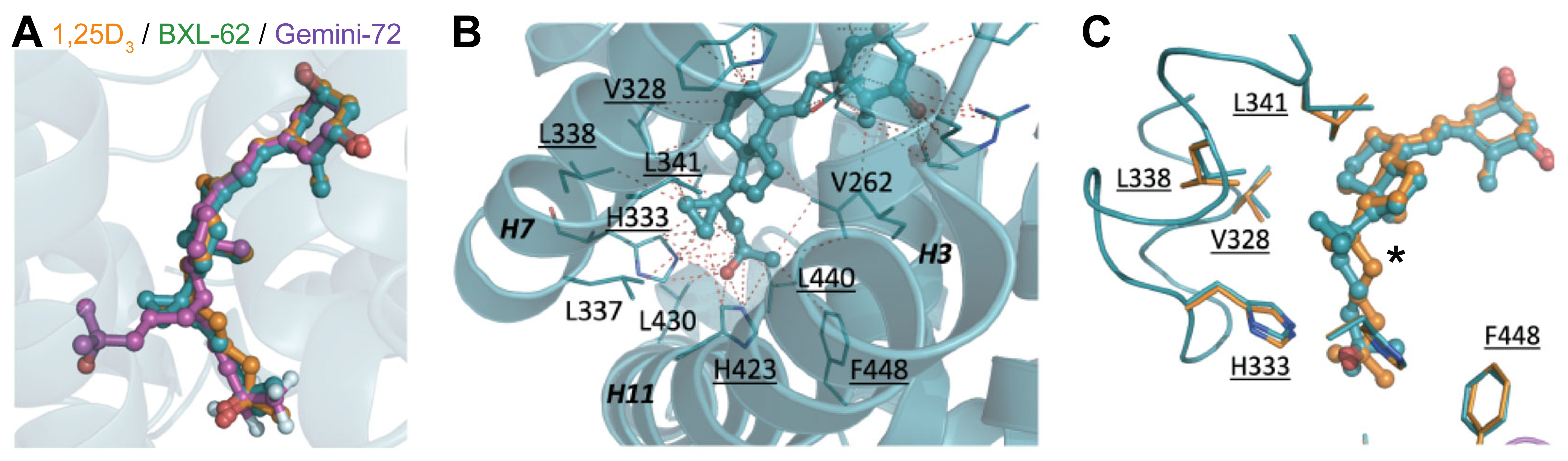
2.2. Binding Mode of BXL-62
2.3. BXL-62 Stabilizes VDRgem Active Conformation
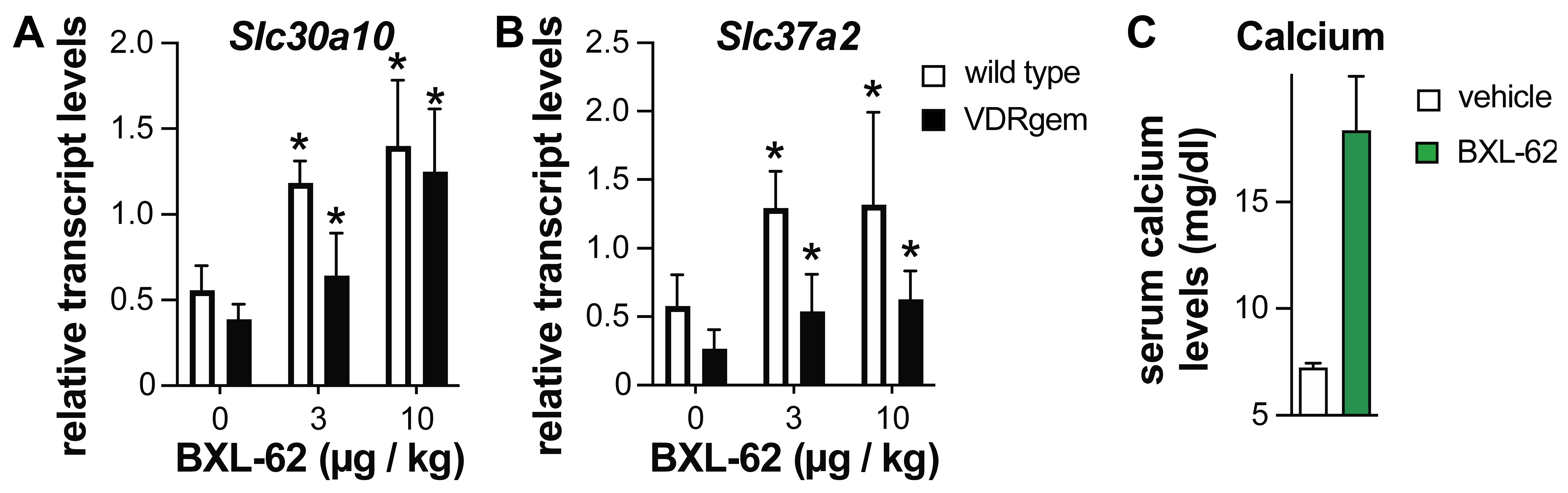
2.4. In Vivo Effect of BXL-62
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals
5.2. Cells
5.3. Transactivation Assays
5.4. Crystallization and Structure Determination
5.5. Molecular Dynamics
5.6. Mice
5.7. Analysis of Transcript Levels
- The set of primers were the following:
- Slc37a2, 5′-TAGGGCCAGACTAGAGCCA-3′ (sense) and
- 5′-ACATGCTCATCTCTGCCGAC-3′ (antisense);
- Slc30a10, 5′-GGTGATTCCCTGAACACCGA-3′ (sense) and
- 5′-ACGTGCAAAAGAACACCTCTG-3′ (antisense) and
- 18 S, 5′-AGCTCACTGGCATGGCCTTC-3′ (sense) and
- 5′-CGCCTGCTTCACCACCTTC-3′ (antisense).
5.8. Blood Sample Collection and Analysis
5.9. Data and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1,25D3 | 1α,25-dihydroxyvitamin D3 |
| VDR | vitamin D receptor |
| HVDRR | Hereditary Vitamin D-resistant rickets |
| LBD | ligand-binding domain |
| LBP | ligand-binding pocket |
| MD | molecular dynamics |
References
- Bikle, D.; Christakos, S. New aspects of vitamin D metabolism and action—Addressing the skin as source and target. Nat. Rev. Endocrinol. 2020, 16, 234–252. [Google Scholar] [CrossRef] [PubMed]
- Fleet, J.C. The role of vitamin D in the endocrinology controlling calcium homeostasis. Mol. Cell. Endocrinol. 2017, 453, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Goltzman, D. Functions of vitamin D in bone. Histochem. Cell Biol. 2018, 149, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Malloy, P.J. Mutations in the vitamin D receptor and hereditary vitamin D-resistant rickets. Bonekey Rep. 2014, 3, 510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malloy, P.J.; Xu, R.; Peng, L.; Peleg, S.; Al-Ashwal, A.; Feldman, D. Hereditary 1,25-dihydroxyvitamin D resistant rickets due to a mutation causing multiple defects in vitamin D receptor function. Endocrinology 2004, 145, 5106–5114. [Google Scholar] [CrossRef] [Green Version]
- Rochel, N.; Molnar, F. Structural aspects of Vitamin D endocrinology. Mol. Cell. Endocrinol. 2017, 453, 22–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leyssens, C.; Verlinden, L.; Verstuyf, A. The future of vitamin D analogs. Front. Physiol. 2014, 5, 122. [Google Scholar] [CrossRef] [Green Version]
- Belorusova, A.Y.; Rochel, N. Structural Studies of Vitamin D Nuclear Receptor Ligand-Binding Properties. Vitam. Horm. 2016, 100, 83–116. [Google Scholar] [PubMed]
- Maestro, M.A.; Molnar, F.; Carlberg, C. Vitamin D and Its Synthetic Analogs. J. Med. Chem. 2019, 62, 6854–6875. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K. Discovery of Nuclear Receptor Ligands and Elucidation of Their Mechanisms of Action. Chem. Pharm. Bull. 2019, 67, 609–619. [Google Scholar] [CrossRef] [Green Version]
- Peleg, S.; Sastry, M.; Collins, E.D.; Bishop, J.E.; Norman, A.W. Distinct conformational changes induced by 20-epi analogues of 1 alpha,25-dihydroxyvitamin D3 are associated with enhanced activation of the vitamin D receptor. J. Biol. Chem. 1995, 270, 10551–10558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uskokovic, M.R.; Manchand, P.; Marczak, S.; Maehr, H.; Jankowski, P.; Adorini, L.; Reddy, G.S. C-20 cyclopropyl vitamin D3 analogs. Curr. Top. Med. Chem. 2006, 6, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Tocchini-Valentini, G.; Rochel, N.; Wurtz, J.M.; Mitschler, A.; Moras, D. Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands. Proc. Natl. Acad. Sci. USA 2001, 98, 5491–5496. [Google Scholar] [CrossRef] [Green Version]
- Ciesielski, F.; Rochel, N.; Moras, D. Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: Remodelling the LBP with one side chain rotation. J. Steroid Biochem. Mol. Biol. 2007, 103, 235–242. [Google Scholar] [CrossRef]
- Huet, T.; Maehr, H.; Lee, H.J.; Uskokovic, M.R.; Suh, N.; Moras, D.; Rochel, N. Structure-function study of gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097. Med. Chem. Comm. 2011, 2, 424–429. [Google Scholar] [CrossRef] [Green Version]
- Abu El Maaty, M.A.; Grelet, E.; Keime, C.; Rerra, A.I.; Gantzer, J.; Emprou, C.; Terzic, J.; Lutzing, R.; Bornert, J.M.; Laverny, G.; et al. Single-cell analyses unravel cell type-specific responses to a vitamin D analog in prostatic precancerous lesions. Sci. Adv. 2021, 7, eabg5982. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Wislocki, A.; Goodman, C.; Ji, Y.; Ge, R.; Maehr, H.; Uskokovic, M.; Reiss, M.; Suh, N. A novel vitamin D derivative activates bone morphogenetic protein signaling in MCF10 breast epithelial cells. Mol. Pharm. 2006, 69, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Belorusova, A.Y.; Suh, N.; Lee, H.J.; So, J.Y.; Maehr, H.; Rochel, N. Structural analysis and biological activities of BXL0124, a gemini analog of vitamin D. J. Steroid Biochem. Mol. Biol. 2017, 173, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Maehr, H.; Rochel, N.; Lee, H.J.; Suh, N.; Uskokovic, M.R. Diastereotopic and deuterium effects in gemini. J. Med. Chem. 2013, 56, 3878–3888. [Google Scholar] [CrossRef] [PubMed]
- Huet, T.; Laverny, G.; Ciesielski, F.; Molnar, F.; Ramamoorthy, T.G.; Belorusova, A.Y.; Antony, P.; Potier, N.; Metzger, D.; Moras, D.; et al. A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects. Cell. Rep. 2015, 10, 516–526. [Google Scholar] [CrossRef] [Green Version]
- Laverny, G.; Penna, G.; Uskokovic, M.; Marczak, S.; Maehr, H.; Jankowski, P.; Ceailles, C.; Vouros, P.; Smith, B.; Robinson, M.; et al. Synthesis and anti-inflammatory properties of 1alpha,25-dihydroxy-16-ene-20-cyclopropyl-24-oxo-vitamin D3, a hypocalcemic, stable metabolite of 1alpha,25-dihydroxy-16-ene-20-cyclopropyl-vitamin D3. J. Med. Chem. 2009, 52, 2204–2213. [Google Scholar] [CrossRef] [PubMed]
- Laverny, G.; Penna, G.; Vetrano, S.; Correale, C.; Nebuloni, M.; Danese, S.; Adorini, L. Efficacy of a potent and safe vitamin D receptor agonist for the treatment of inflammatory bowel disease. Immunol. Lett. 2010, 131, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.J.; Lee, Y.Y.; Pakkala, S.; de Vos, S.; Elstner, E.; Norman, A.W.; Green, J.; Uskokovic, M.; Koeffler, H.P. 1,25(OH)2-16ene-vitamin D3 is a potent antileukemic agent with low potential to cause hypercalcemia. Leuk. Res. 1994, 18, 453–463. [Google Scholar] [CrossRef]
- Nakagawa, K.; Sowa, Y.; Kurobe, M.; Ozono, K.; Siu-Caldera, M.L.; Reddy, G.S.; Uskokovic, M.R.; Okano, T. Differential activities of 1alpha,25-dihydroxy-16-ene-vitamin D(3) analogs and their 3-epimers on human promyelocytic leukemia (HL-60) cell differentiation and apoptosis. Steroids 2001, 66, 327–337. [Google Scholar] [CrossRef]
- Yamada, S.; Yamamoto, K.; Masuno, H.; Ohta, M. Conformation-function relationship of vitamin D: Conformational analysis predicts potential side-chain structure. J. Med. Chem. 1998, 41, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Futawaka, K.; Tagami, T.; Fukuda, Y.; Koyama, R.; Nushida, A.; Nezu, S.; Yamamoto, H.; Imamoto, M.; Kasahara, M.; Moriyama, K. Transcriptional activation of the wild-type and mutant vitamin D receptors by vitamin D3 analogs. J. Mol. Endocrinol. 2016, 57, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Shen, Q.; Malloy, P.J.; Soliman, E.; Peng, X.; Kim, S.; Pike, J.W.; Feldman, D.; Christakos, S. Enhanced coactivator binding and transcriptional activation of mutant vitamin D receptors from patients with hereditary 1,25-dihydroxyvitamin D-resistant rickets by phosphorylation and vitamin D analogs. J. Bone Min. Res. 2005, 20, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Mano, H.; Nishikawa, M.; Yasuda, K.; Ikushiro, S.; Saito, N.; Sawada, D.; Honzawa, S.; Takano, M.; Kittaka, A.; Sakaki, T. Novel screening system for high-affinity ligand of heredity vitamin D-resistant rickets-associated vitamin D receptor mutant R274L using bioluminescent sensor. J. Steroid Biochem. Mol. Biol. 2017, 167, 61–66. [Google Scholar] [CrossRef]
- Huang, K.; Malloy, P.; Feldman, D.; Pitukcheewanont, P. Enteral calcium infusion used successfully as treatment for a patient with hereditary vitamin D resistant rickets (HVDRR) without alopecia: A novel mutation. Gene 2013, 512, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Adiceam, P.; Kottler, M.L.; Guillozo, H.; Rizk-Rabin, M.; Brouillard, F.; Lagier, P.; Palix, C.; Garnier, J.M.; Garabedian, M. Tryptophan missense mutation in the ligand-binding domain of the vitamin D receptor causes severe resistance to 1,25-dihydroxyvitamin D. J. Bone Min. Res. 2002, 17, 1728–1737. [Google Scholar] [CrossRef]
- Maehr, H.; Lee, H.J.; Perry, B.; Suh, N.; Uskokovic, M.R. Calcitriol derivatives with two different side chains at C-20. V. Potent inhibitors of mammary carcinogenesis and inducers of leukemia differentiation. J. Med. Chem. 2009, 52, 5505–5519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W. Xds. Acta Cryst. D Biol. Cryst. 2010, 66 Pt 2, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Cryst. D Biol. Cryst. 2006, 62 Pt 1, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Adams, P.D. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Cryst. D Biol. Cryst. 2005, 61 Pt 7, 850–855. [Google Scholar] [CrossRef] [PubMed]
- Bricogne, G.; Blanc, E.; Brandl, M.; Flensburg, C.; Keller, P.; Paciorek, W.; Roversi, P.; Sharff, A.; Smart, O.S.; Vonrhein, C.; et al. BUSTER Version 2.11.2; Global Phasing Ltd.: Cambridge, UK, 2011. [Google Scholar]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Cryst. D Biol. Cryst. 2004, 60 Pt 12 Pt 1, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]

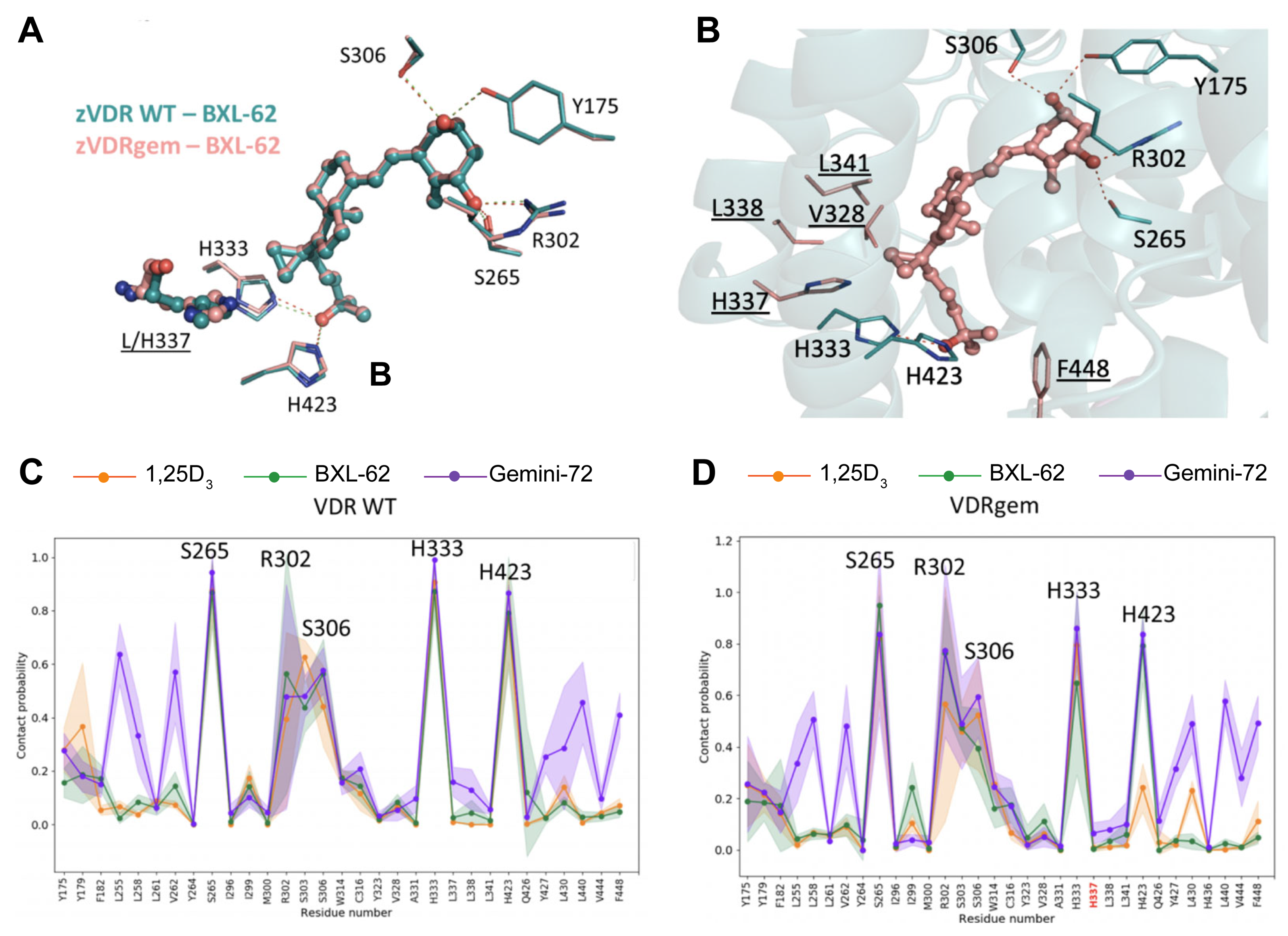
| His333-Lig | His423-Lig | His333-His423 | His423-His337 | ||
|---|---|---|---|---|---|
| zVDR WT | 1,25D3 | 0.91 | 0.77 | 0.17 | 0 |
| BXL-62 | 0.87 | 0.79 | 0.15 | 0 | |
| zVDRgem | 1,25D3 | 0.80 | 0.24 | 0.06 | 0.53 |
| BXL-62 | 0.65 | 0.79 | 0.21 | 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belorusova, A.Y.; Rovito, D.; Chebaro, Y.; Doms, S.; Verlinden, L.; Verstuyf, A.; Metzger, D.; Rochel, N.; Laverny, G. Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants. Int. J. Mol. Sci. 2022, 23, 8445. https://doi.org/10.3390/ijms23158445
Belorusova AY, Rovito D, Chebaro Y, Doms S, Verlinden L, Verstuyf A, Metzger D, Rochel N, Laverny G. Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants. International Journal of Molecular Sciences. 2022; 23(15):8445. https://doi.org/10.3390/ijms23158445
Chicago/Turabian StyleBelorusova, Anna Y., Daniela Rovito, Yassmine Chebaro, Stefanie Doms, Lieve Verlinden, Annemieke Verstuyf, Daniel Metzger, Natacha Rochel, and Gilles Laverny. 2022. "Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants" International Journal of Molecular Sciences 23, no. 15: 8445. https://doi.org/10.3390/ijms23158445






