Basal Forebrain-Dorsal Hippocampus Cholinergic Circuit Regulates Olfactory Associative Learning
Abstract
:1. Introduction
2. Results
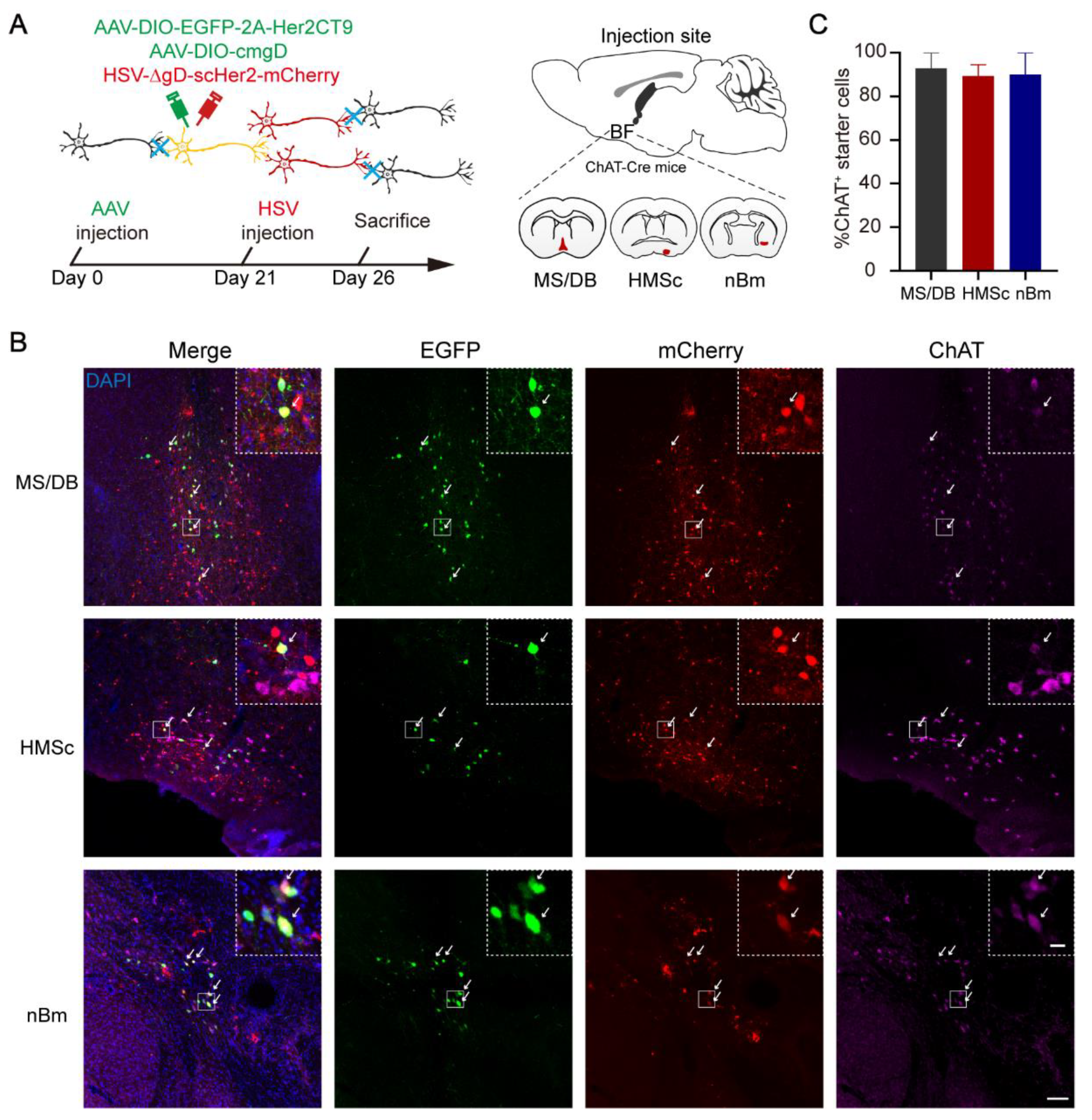
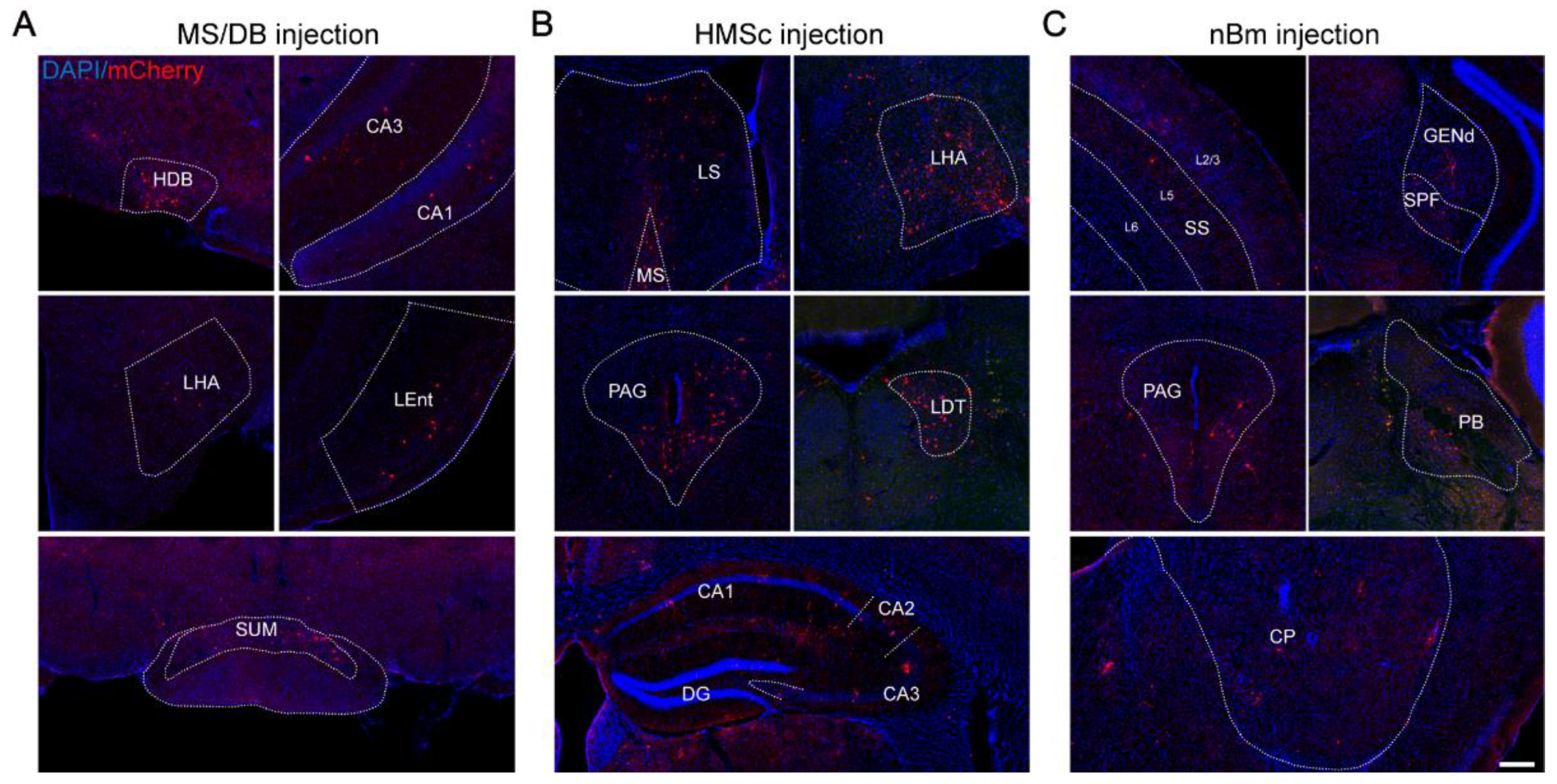
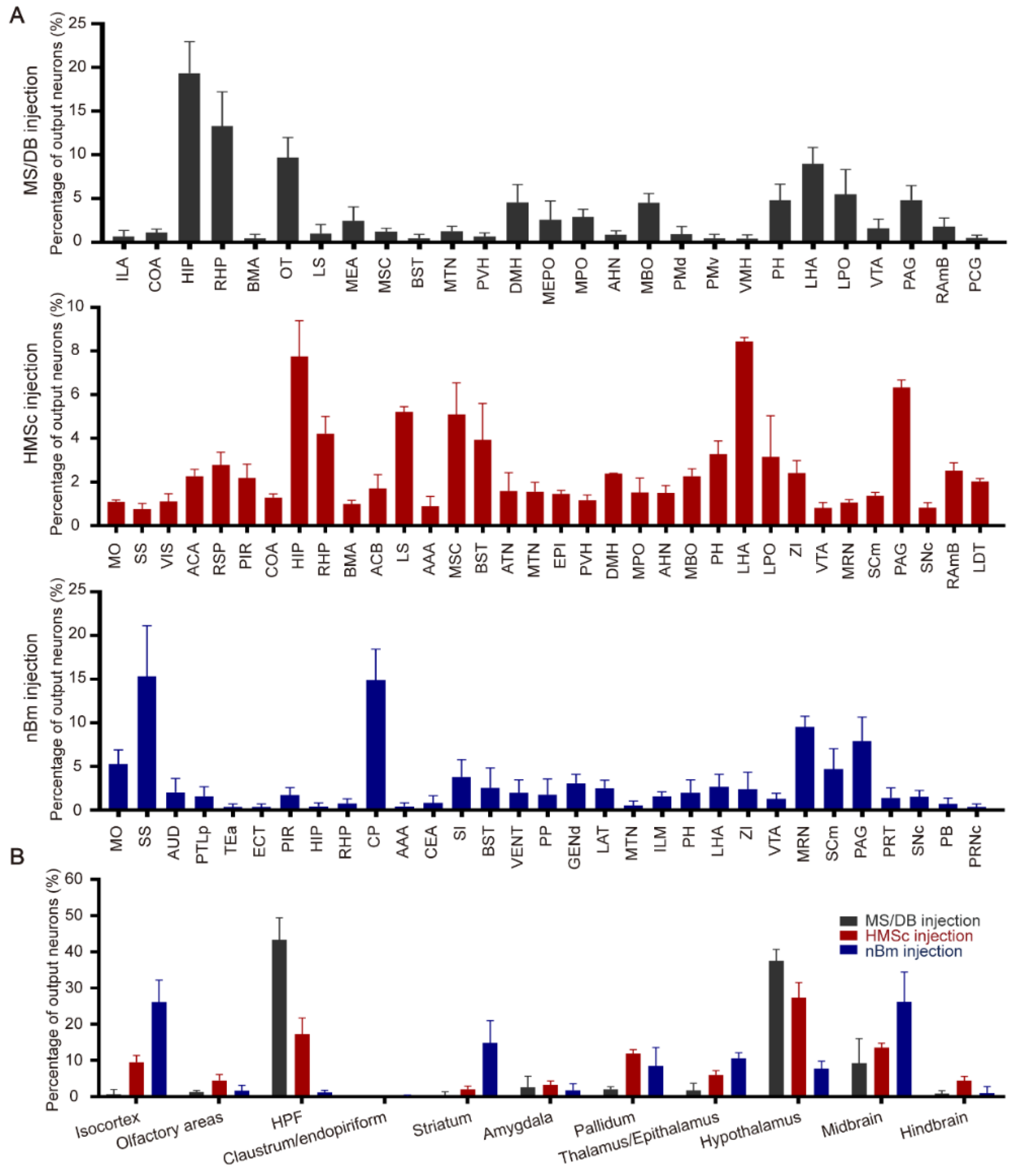
2.1. The Output Patterns of BFCNs in Different Subregions
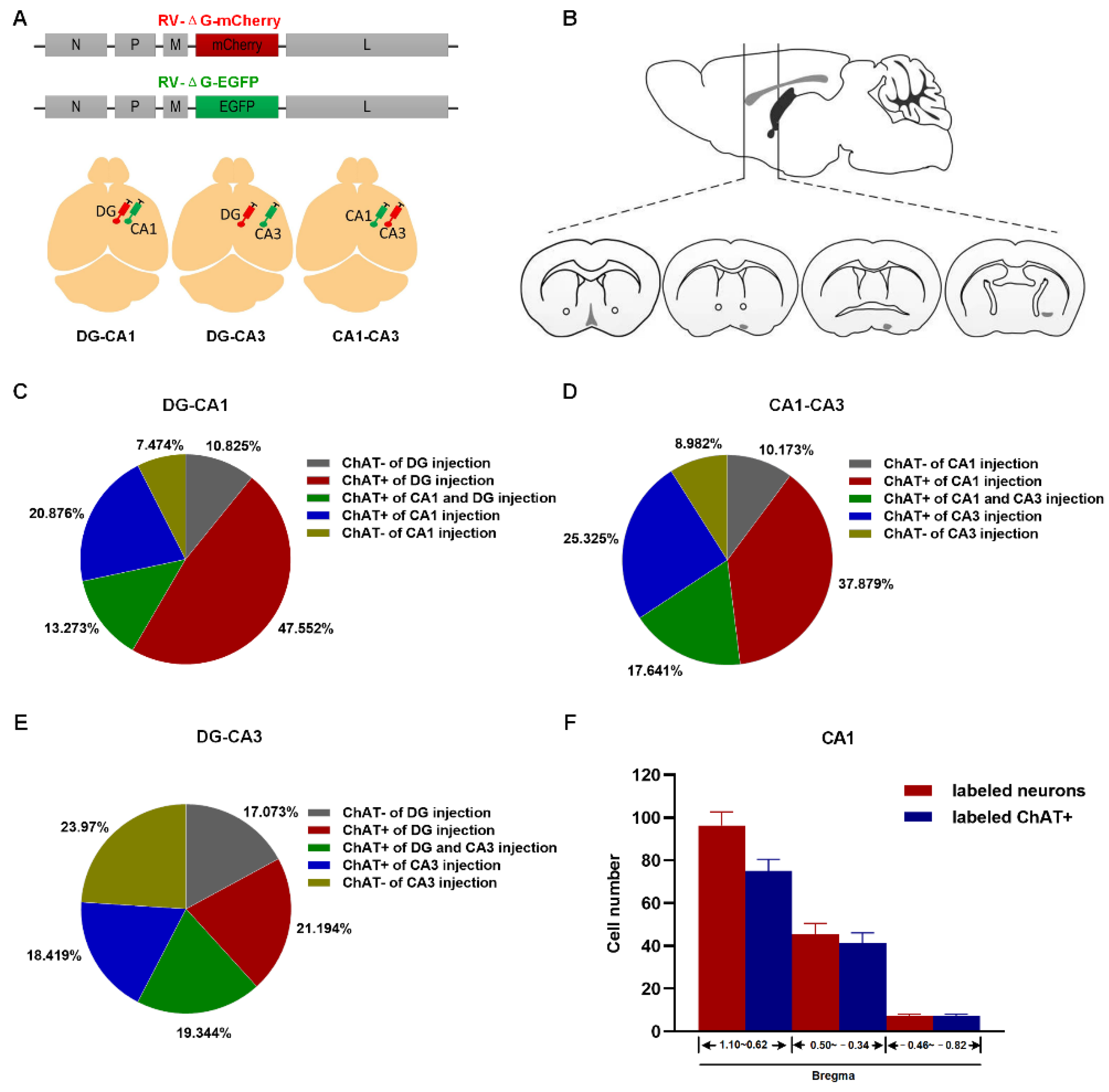
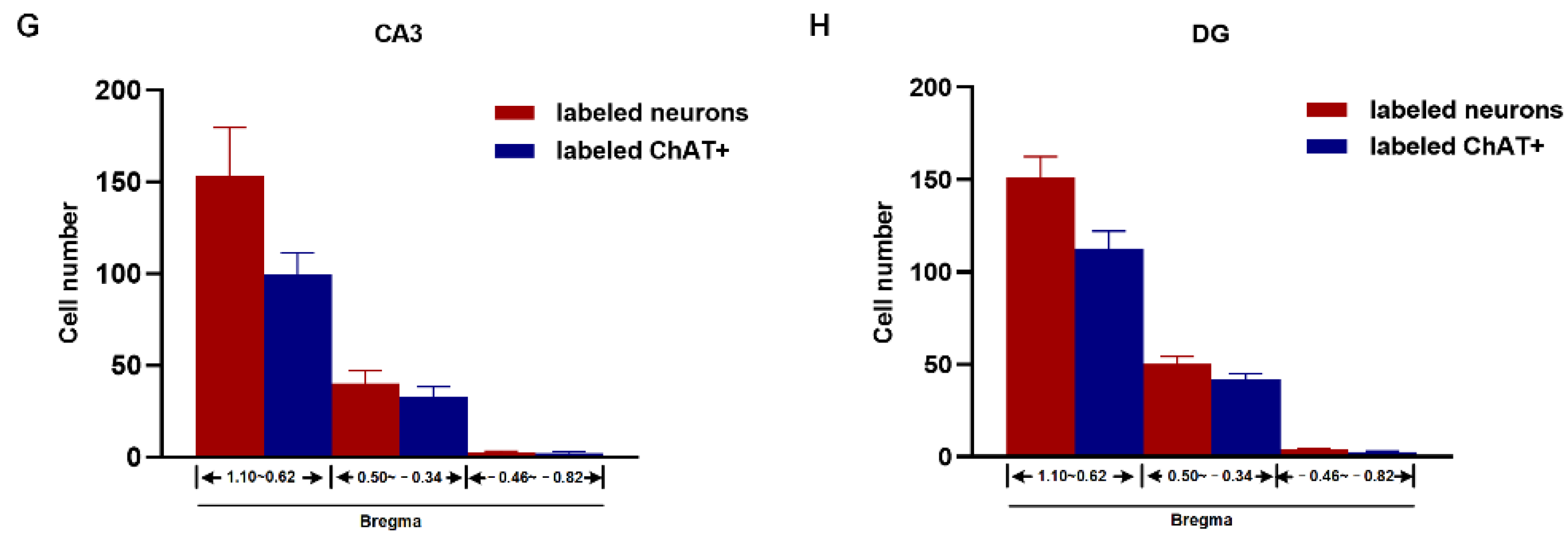
2.2. Topography of BFCN Projecting to Dorsal Hippocampus
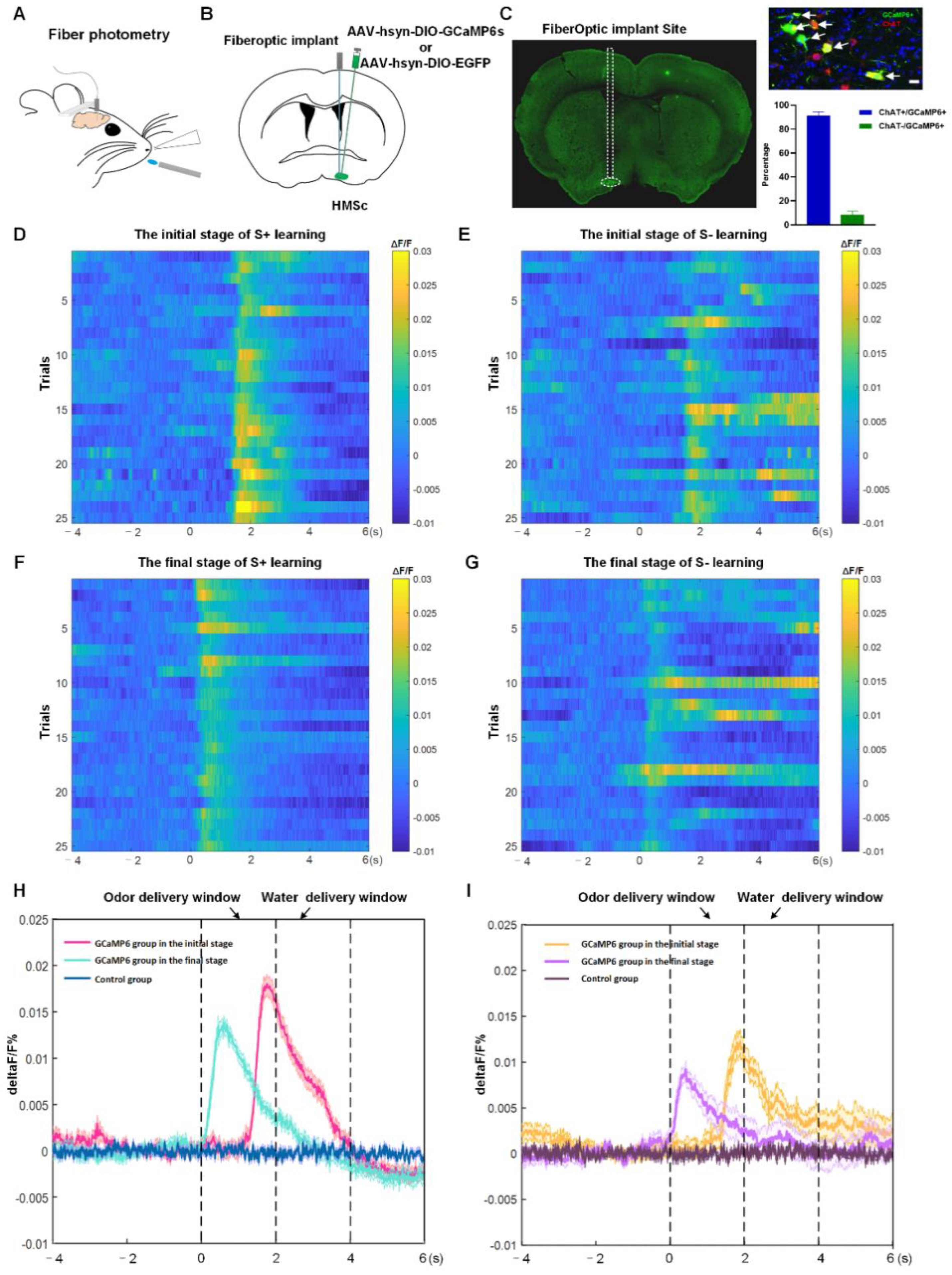
2.3. BFCNs in HMSc Were Activated in Odor-Cued Go/No-Go Discrimination Task
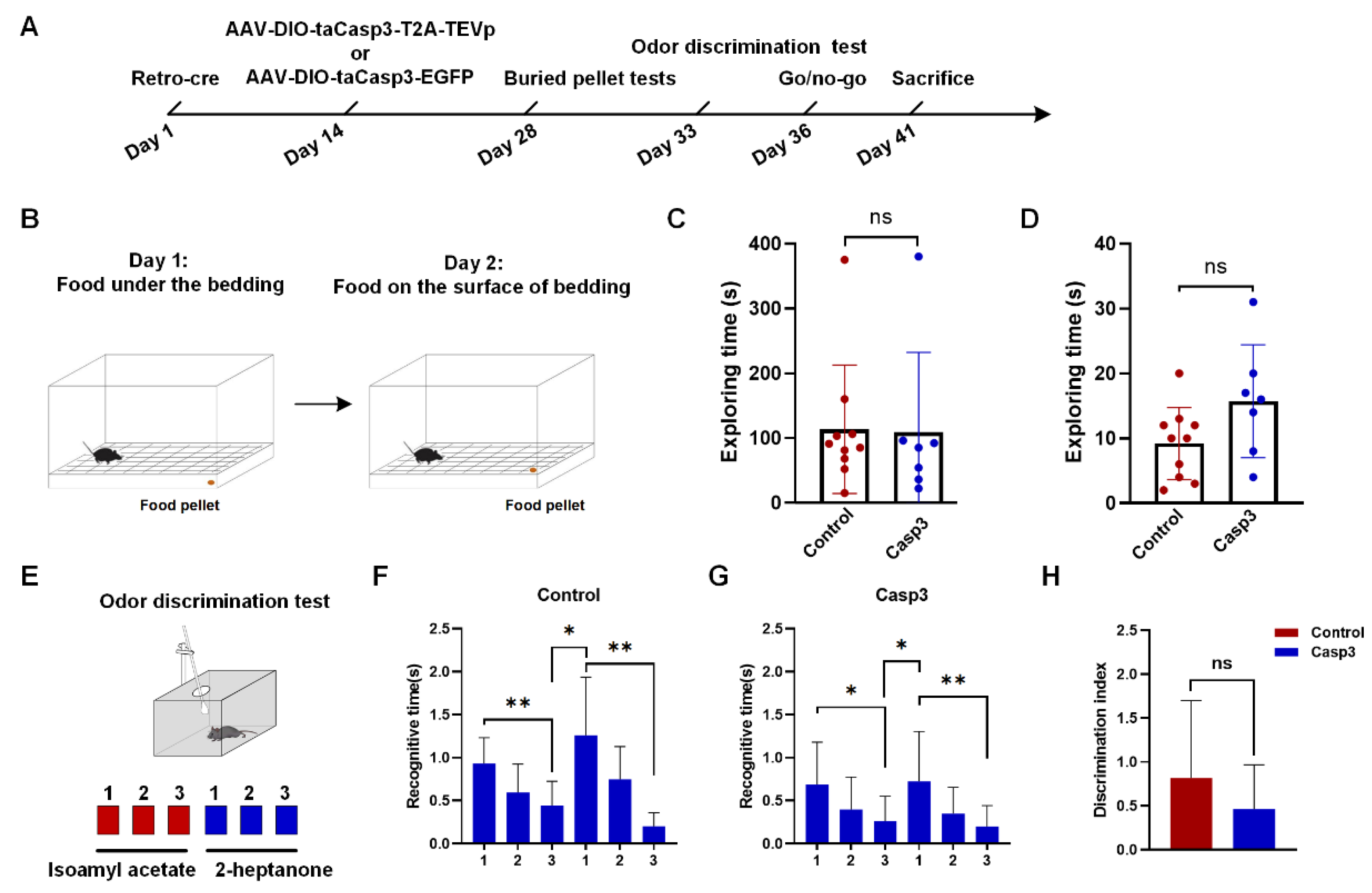
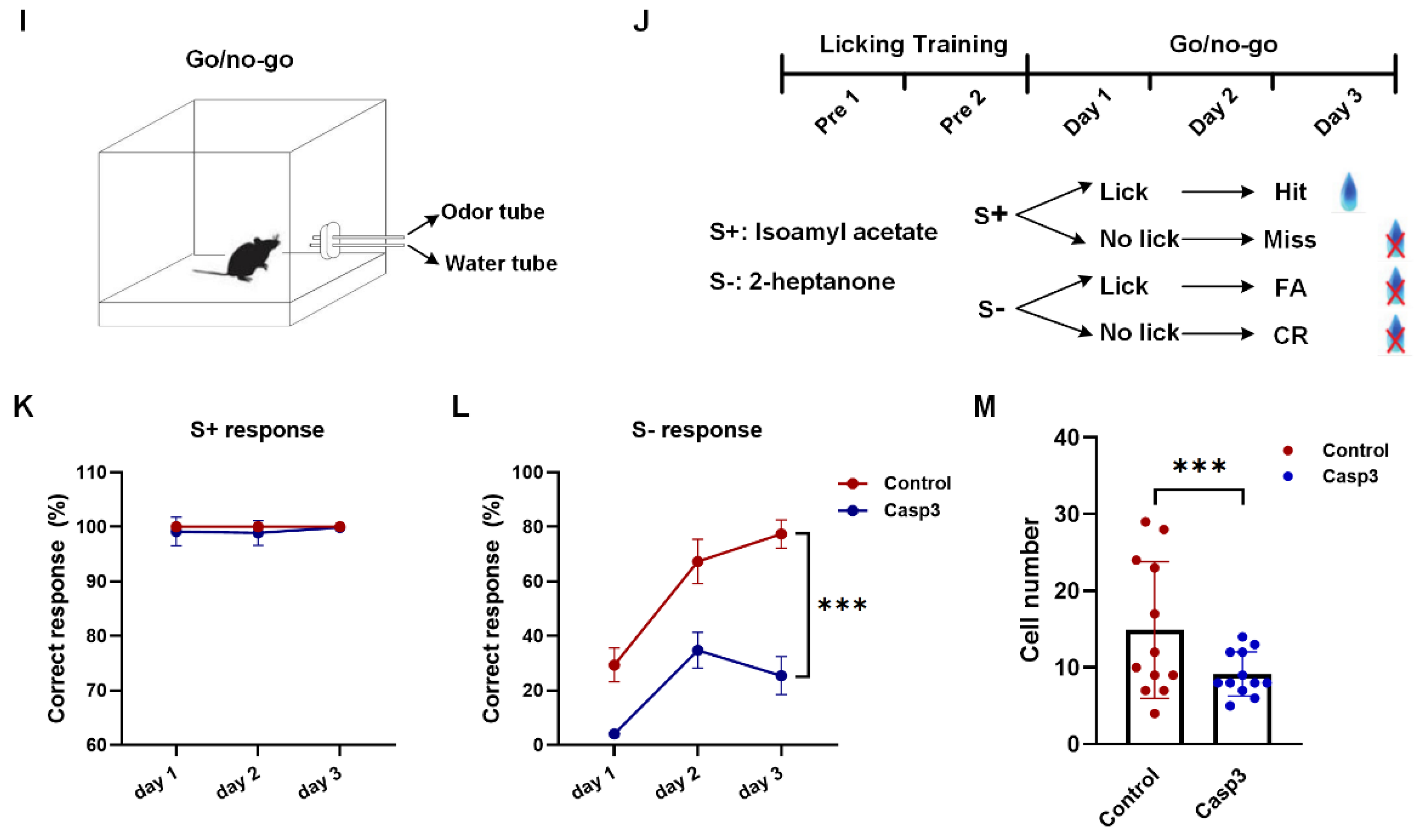
2.4. Selective Depletion of Cholinergic Neurons Innervating dHIP Impaired Correct Performance of Odor-Cued Go/No-Go Discrimination Task
3. Discussion
3.1. Projectomes of BFCNs
3.2. Anatomical Relationship between BFCNs and dHIP
3.3. Functions of HMSc-dHIP Circuit in Olfactory Associative Learning
4. Materials and Methods
4.1. Animal
4.2. Stereotaxic Injection of the Viruses
4.3. Histology and Immunofluorescence Staining
4.4. Fiber Implantation and Fiber Photometry
4.5. Behavioral Tests
4.5.1. Buried Pellet Tests
4.5.2. Odor Discrimination Test
4.5.3. Odor-Cued Go/No-Go Discrimination Task
4.6. Data Analysis
4.6.1. Cell Counting in Tracing Samples
4.6.2. Cell Counting in Go/No-Go Tasks
4.6.3. Analysis of Fiber Photometry Data
4.6.4. Analysis of Behavioral Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Definition |
| AAA | Anterior amygdalar area |
| ACA | Anterior cingulate area |
| ACB | Nucleus accumbens |
| AHN | Anterior hypothalamic nucleus |
| ATN | Anterior group of the dorsal thalamus |
| AUD | Auditory areas |
| BF | Basal forebrain |
| BFCNs | Basal forebrain cholinergic neurons |
| BMA | Basomedial amygdalar nucleus |
| BST | Bed nucleus of the stria terminalis |
| CA1 | Field CA1 |
| CA2 | Field CA2 |
| CA3 | Field CA3 |
| CEA | Central amygdalar nucleus |
| COA | Cortical amygdalar area |
| CP | Caudoputamen |
| DB | Diagonal band nuclei |
| DG | Dentate gyrus |
| dHIP | Dorsal hippocampus |
| DMH | Dorsomedial nucleus of the hypothalamus |
| ECT | Ectorhinal area |
| EPI | Epithalamus |
| GENd | Geniculate group dorsal thalamus |
| HDB | Nucleus of the horizontal limb of the diagonal band |
| HIP | Hippocampal region |
| HMSc | HMS complex |
| ILA | Infralimbic area |
| ILM | Intralaminar nuclei of the dorsal thalamus |
| LAT | Lateral group of the dorsal thalamus |
| LDT | Laterodorsal tegmental nucleus |
| LEnt | Lateral entorhinal cortex |
| LHA | Lateral hypothalamic area |
| LPO | Lateral preoptic area |
| LS | Lateral septal nucleus |
| MBO | Mammillary body |
| MCPO | Magnocellular preoptic nucleus |
| MEA | Medial amygdalar nucleus |
| MEPO | Median preoptic nucleus |
| MO | Somatomotor area |
| MPO | Medial preoptic area |
| MS | Medial septal nucleus |
| MSC | Medial septal complex |
| MRN | Midbrain reticular nucleus |
| MTN | Midline group of the dorsal thalamus |
| nBm | Nucleus basalis Meynert |
| OB | Olfactory bulb |
| OT | Olfactory tubercle |
| PAG | Periaqueductal gray |
| PB | Parabrachial nucleus |
| PCG | Pontine central gray |
| PH | Posterior hypothalamic nucleus |
| PIR | Piriform area |
| PMd | Dorsal premammillary nucleus |
| PMv | Ventral premammillary nucleus |
| PP | Peripeduncular nucleus |
| PRNc | Pontine reticular nucleus, caudal part |
| PRT | Pretectal region |
| PTLp | Posterior parietal association area |
| PVH | Paraventricular hypothalamic nucleus |
| RAmB | Midbrain raphe nuclei |
| RHP | Retrohippocampal region |
| RSP | Retrosplenial area |
| SCm | Superior colliculus, motor related |
| SI | Substantia innominata |
| SNc | Substantia nigra, compact part |
| SPF | Subparafascicular nucleus |
| SS | Somatosensory area |
| SUB | Subiculum |
| SUM | Supramammillary nucleus |
| TEa | Temporal association area |
| VDB | The vertical limb of the diagonal band |
| VENT | Ventral group of the dorsal thalamus |
| vHIP | Ventral hippocampus |
| VP | Ventral pallidum |
| VIS | Visual area |
| VMH | Ventromedial hypothalamic nucleus |
| VTA | Ventral tegmental area |
| ZI | Zona incerta |
References
- Woolf, N.J. Cholinergic systems in mammalian brain and spinal cord. Prog. Neurobiol. 1991, 37, 475–524. [Google Scholar] [CrossRef]
- Zaborszky, L.; Csordas, A.; Mosca, K.; Kim, J.; Gielow, M.R.; Vadasz, C.; Nadasdy, Z. Neurons in the basal forebrain project to the cortex in a complex topographic organization that reflects corticocortical connectivity patterns: An experimental study based on retrograde tracing and 3D reconstruction. Cereb. Cortex 2015, 25, 118–137. [Google Scholar] [CrossRef] [Green Version]
- Gielow, M.R.; Zaborszky, L. The Input-Output Relationship of the Cholinergic Basal Forebrain. Cell Rep. 2017, 18, 1817–1830. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Feng, S.; Zhu, X.; Jiang, W.; Wen, P.; Ye, F.; Rao, X.; Jin, S.; He, X.; Xu, F. Different Subgroups of Cholinergic Neurons in the Basal Forebrain Are Distinctly Innervated by the Olfactory Regions and Activated Differentially in Olfactory Memory Retrieval. Front. Neural Circuits 2018, 12, 99. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Lv, F.; Yuan, Y.; Fan, C.; Li, J.; Sun, W.; Hu, J. Whole-Brain Mapping of Monosynaptic Afferent Inputs to Cortical CRH Neurons. Front. Neurosci. 2019, 13, 565. [Google Scholar] [CrossRef] [Green Version]
- Kondo, H.; Zaborszky, L. Topographic organization of the basal forebrain projections to the perirhinal, postrhinal, and entorhinal cortex in rats. J. Comp. Neurol. 2016, 524, 2503–2515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Yu, B.; Sun, Q.; Zhang, Y.; Ren, M.; Zhang, X.; Li, A.; Yuan, J.; Madisen, L.; Luo, Q.; et al. Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc. Natl. Acad. Sci. USA 2018, 115, 415–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Williams, J.; Nathans, J. Complete morphologies of basal forebrain cholinergic neurons in the mouse. eLife 2014, 3, e02444. [Google Scholar] [CrossRef] [PubMed]
- Vale-Martínez, A.; Baxter, M.G.; Eichenbaum, H. Selective lesions of basal forebrain cholinergic neurons produce anterograde and retrograde deficits in a social transmission of food preference task in rats. Eur. J. Neurosci. 2002, 16, 983–998. [Google Scholar] [CrossRef] [PubMed]
- Miasnikov, A.A.; Chen, J.C.; Weinberger, N.M. Rapid induction of specific associative behavioral memory by stimulation of the nucleus basalis in the rat. Neurobiol. Learn. Mem. 2006, 86, 47–65. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Shi, Y.F.; Xi, W.; Zhou, R.; Tan, Z.B.; Wang, H.; Li, X.M.; Chen, Z.; Feng, G.; Luo, M.; et al. Selective activation of cholinergic basal forebrain neurons induces immediate sleep-wake transitions. Curr. Biol. CB 2014, 24, 693–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hangya, B.; Ranade, S.P.; Lorenc, M.; Kepecs, A. Central Cholinergic Neurons Are Rapidly Recruited by Reinforcement Feedback. Cell 2015, 162, 1155–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, M.; Luo, M. Optogenetic activation of basal forebrain cholinergic neurons modulates neuronal excitability and sensory responses in the main olfactory bulb. J. Neurosci. Off. J. Soc. Neurosci. 2012, 32, 10105–10116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunez-Parra, A.; Cea-Del Rio, C.A.; Huntsman, M.M.; Restrepo, D. The Basal Forebrain Modulates Neuronal Response in an Active Olfactory Discrimination Task. Front. Cell. Neurosci. 2020, 14, 141. [Google Scholar] [CrossRef] [PubMed]
- Devore, S.; de Almeida, L.; Linster, C. Distinct roles of bulbar muscarinic and nicotinic receptors in olfactory discrimination learning. J. Neurosci. Off. J. Soc. Neurosci. 2014, 34, 11244–11260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roman, F.S.; Simonetto, I.; Soumireu-Mourat, B. Learning and memory of odor-reward association: Selective impairment following horizontal diagonal band lesions. Behav. Neurosci. 1993, 107, 72–81. [Google Scholar] [CrossRef]
- Hanson, E.; Brandel-Ankrapp, K.L.; Arenkiel, B.R. Dynamic Cholinergic Tone in the Basal Forebrain Reflects Reward-Seeking and Reinforcement During Olfactory Behavior. Front. Cell. Neurosci. 2021, 15, 635837. [Google Scholar] [CrossRef] [PubMed]
- McDonald, R.J.; Balog, R.J.; Lee, J.Q.; Stuart, E.E.; Carrels, B.B.; Hong, N.S. Rats with ventral hippocampal damage are impaired at various forms of learning including conditioned inhibition, spatial navigation, and discriminative fear conditioning to similar contexts. Behav. Brain Res. 2018, 351, 138–151. [Google Scholar] [CrossRef]
- Lisman, J.E.; Otmakhova, N.A. Storage, recall, and novelty detection of sequences by the hippocampus: Elaborating on the SOCRATIC model to account for normal and aberrant effects of dopamine. Hippocampus 2001, 11, 551–568. [Google Scholar] [CrossRef]
- Kaifosh, P.; Losonczy, A. Mnemonic Functions for Nonlinear Dendritic Integration in Hippocampal Pyramidal Circuits. Neuron 2016, 90, 622–634. [Google Scholar] [CrossRef] [Green Version]
- Igarashi, K.M.; Ito, H.T.; Moser, E.I.; Moser, M.B. Functional diversity along the transverse axis of hippocampal area CA1. FEBS Lett. 2014, 588, 2470–2476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Castro, L.; Wasserman, E.A.; Freeman, J.H. Dorsal hippocampus is necessary for visual categorization in rats. Hippocampus 2018, 28, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Woods, N.I.; Stefanini, F.; Apodaca-Montano, D.L.; Tan, I.M.C.; Biane, J.S.; Kheirbek, M.A. The Dentate Gyrus Classifies Cortical Representations of Learned Stimuli. Neuron 2020, 107, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Chaillan, F.A.; Devigne, C.; Diabira, D.; Khrestchatisky, M.; Roman, F.S.; Ben-Ari, Y.; Soumireu-Mourat, B. Neonatal gamma-ray irradiation impairs learning and memory of an olfactory associative task in adult rats. Eur. J. Neurosci. 1997, 9, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.M.; Weeden, C.S.; Churchwell, J.C.; Kesner, R.P. The role of the dentate gyrus in the formation of contextual representations. Hippocampus 2013, 23, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Wang, Y.; Peng, J.; Zhao, Y.; He, X.; Yu, X.; Liu, Q.; Jin, S.; Xu, F. Whole-Brain Mapping the Direct Inputs of Dorsal and Ventral CA1 Projection Neurons. Front. Neural Circuits 2021, 15, 643230. [Google Scholar] [CrossRef]
- Amaral, D.G.; Kurz, J. An analysis of the origins of the cholinergic and noncholinergic septal projections to the hippocampal formation of the rat. J. Comp. Neurol. 1985, 240, 37–59. [Google Scholar] [CrossRef]
- Nyakas, C.; Luiten, P.G.; Spencer, D.G.; Traber, J. Detailed projection patterns of septal and diagonal band efferents to the hippocampus in the rat with emphasis on innervation of CA1 and dentate gyrus. Brain Res. Bull. 1987, 18, 533–545. [Google Scholar] [CrossRef] [Green Version]
- Matthews, D.A.; Salvaterra, P.M.; Crawford, G.D.; Houser, C.R.; Vaughn, J.E. An immunocytochemical study of choline acetyltransferase-containing neurons and axon terminals in normal and partially deafferented hippocampal formation. Brain Res. 1987, 402, 30–43. [Google Scholar] [CrossRef]
- Su, P.; Ying, M.; Xia, J.; Li, Y.; Wu, Y.; Wang, H.; Xu, F. Rigorous anterograde trans-monosynaptic tracing of genetic defined neurons with retargeted HSV1 H129. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mesulam, M.M.; Mufson, E.J.; Wainer, B.H.; Levey, A.I. Central cholinergic pathways in the rat: An overview based on an alternative nomenclature (Ch1-Ch6). Neuroscience 1983, 10, 1185–1201. [Google Scholar] [CrossRef]
- Paul, S.; Jeon, W.K.; Bizon, J.L.; Han, J.S. Interaction of basal forebrain cholinergic neurons with the glucocorticoid system in stress regulation and cognitive impairment. Front. Aging Neurosci. 2015, 7, 43. [Google Scholar] [CrossRef] [Green Version]
- Okada, K.; Nishizawa, K.; Kobayashi, T.; Sakata, S.; Kobayashi, K. Distinct roles of basal forebrain cholinergic neurons in spatial and object recognition memory. Sci. Rep. 2015, 5, 13158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knox, D.; Keller, S.M. Cholinergic neuronal lesions in the medial septum and vertical limb of the diagonal bands of Broca induce contextual fear memory generalization and impair acquisition of fear extinction. Hippocampus 2016, 26, 718–726. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Kundu, S.; Lederman, J.D.; López-Hernández, G.Y.; Ballinger, E.C.; Wang, S.; Talmage, D.A.; Role, L.W. Cholinergic Signaling Controls Conditioned Fear Behaviors and Enhances Plasticity of Cortical-Amygdala Circuits. Neuron 2016, 90, 1057–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarter, M.; Bruno, J.P. Cortical cholinergic inputs mediating arousal, attentional processing and dreaming: Differential afferent regulation of the basal forebrain by telencephalic and brainstem afferents. Neuroscience 2000, 95, 933–952. [Google Scholar] [CrossRef]
- Risbrough, V.; Bontempi, B.; Menzaghi, F. Selective immunolesioning of the basal forebrain cholinergic neurons in rats: Effect on attention using the 5-choice serial reaction time task. Psychopharmacology 2002, 164, 71–81. [Google Scholar] [CrossRef]
- Wang, D.; Liu, P.; Mao, X.; Zhou, Z.; Cao, T.; Xu, J.; Sun, C.; Li, A. Task-Demand-Dependent Neural Representation of Odor Information in the Olfactory Bulb and Posterior Piriform Cortex. J. Neurosci. Off. J. Soc. Neurosci. 2019, 39, 10002–10018. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Tang, K.; Wu, J.; Xu, H.; Zhang, W.; Cao, T.; Zhou, Y.; Yu, T.; Li, A. Leptin modulates olfactory discrimination and neural activity in the olfactory bulb. Acta Physiol. 2019, 227, e13319. [Google Scholar] [CrossRef] [PubMed]
| Virus Name | Company | Cat. No. | Titers |
|---|---|---|---|
| rAAV-hSyn-DIO-EGFP-T2A-Her2CT9-pA | — — | —— | 1.3 × 1013 VG/mL |
| rAAV-UL26.5p-DIO-cmgD-WPRE-pA | — — | — — | 1 × 1013 VG/mL |
| HSV-∆gD-hUbc-mcherry-2A-scHer2::gD-WPRE-pA | — — | — — | 1 × 108 pfu/mL |
| RV-dG-DsRed | BrainVTA | R02001 | 2 × 108 pfu/mL |
| RV-dG-GFP | BrainVTA | R02002 | 2 × 108 pfu/mL |
| rAAV-EF1α-DIO-GCaMP6s-WPRE-hGH-pA | BrainVTA | PT-0071 | 2 × 1012 VG/mL |
| rAAV-EF1α-DIO-EGFP-WPRE-hGH-pA | BrainVTA | PT-0795 | 2 × 1012 VG/mL |
| rAAV-Retro-hSyn-SV40-NLS-Cre-WPRE-hGH-pA | Brain Case | BC-0159 | 5.93 × 1012 VG/mL |
| rAAV-EF1α-DIO-taCasp3-T2A-TEVp-WPRE-hGH-pA | Brain Case | BC-0130 | 2.55 × 1012 VG/mL |
| rAAV-EF1α-DIO-taCasp3-EGFP-WPRE-hGH-pA | Brain Case | BC-0846 | 3.33 × 1012 VG/mL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Tao, S.; Liu, Y.; Liu, J.; Sun, L.; Zheng, Y.; Tian, Y.; Su, P.; Zhu, X.; Xu, F. Basal Forebrain-Dorsal Hippocampus Cholinergic Circuit Regulates Olfactory Associative Learning. Int. J. Mol. Sci. 2022, 23, 8472. https://doi.org/10.3390/ijms23158472
Zheng Y, Tao S, Liu Y, Liu J, Sun L, Zheng Y, Tian Y, Su P, Zhu X, Xu F. Basal Forebrain-Dorsal Hippocampus Cholinergic Circuit Regulates Olfactory Associative Learning. International Journal of Molecular Sciences. 2022; 23(15):8472. https://doi.org/10.3390/ijms23158472
Chicago/Turabian StyleZheng, Yingwei, Sijue Tao, Yue Liu, Jingjing Liu, Liping Sun, Yawen Zheng, Yu Tian, Peng Su, Xutao Zhu, and Fuqiang Xu. 2022. "Basal Forebrain-Dorsal Hippocampus Cholinergic Circuit Regulates Olfactory Associative Learning" International Journal of Molecular Sciences 23, no. 15: 8472. https://doi.org/10.3390/ijms23158472
APA StyleZheng, Y., Tao, S., Liu, Y., Liu, J., Sun, L., Zheng, Y., Tian, Y., Su, P., Zhu, X., & Xu, F. (2022). Basal Forebrain-Dorsal Hippocampus Cholinergic Circuit Regulates Olfactory Associative Learning. International Journal of Molecular Sciences, 23(15), 8472. https://doi.org/10.3390/ijms23158472






