Bile Acid–Drug Interaction via Organic Anion-Transporting Polypeptide 4C1 Is a Potential Mechanism of Altered Pharmacokinetics of Renally Excreted Drugs
Abstract
:1. Introduction
2. Results
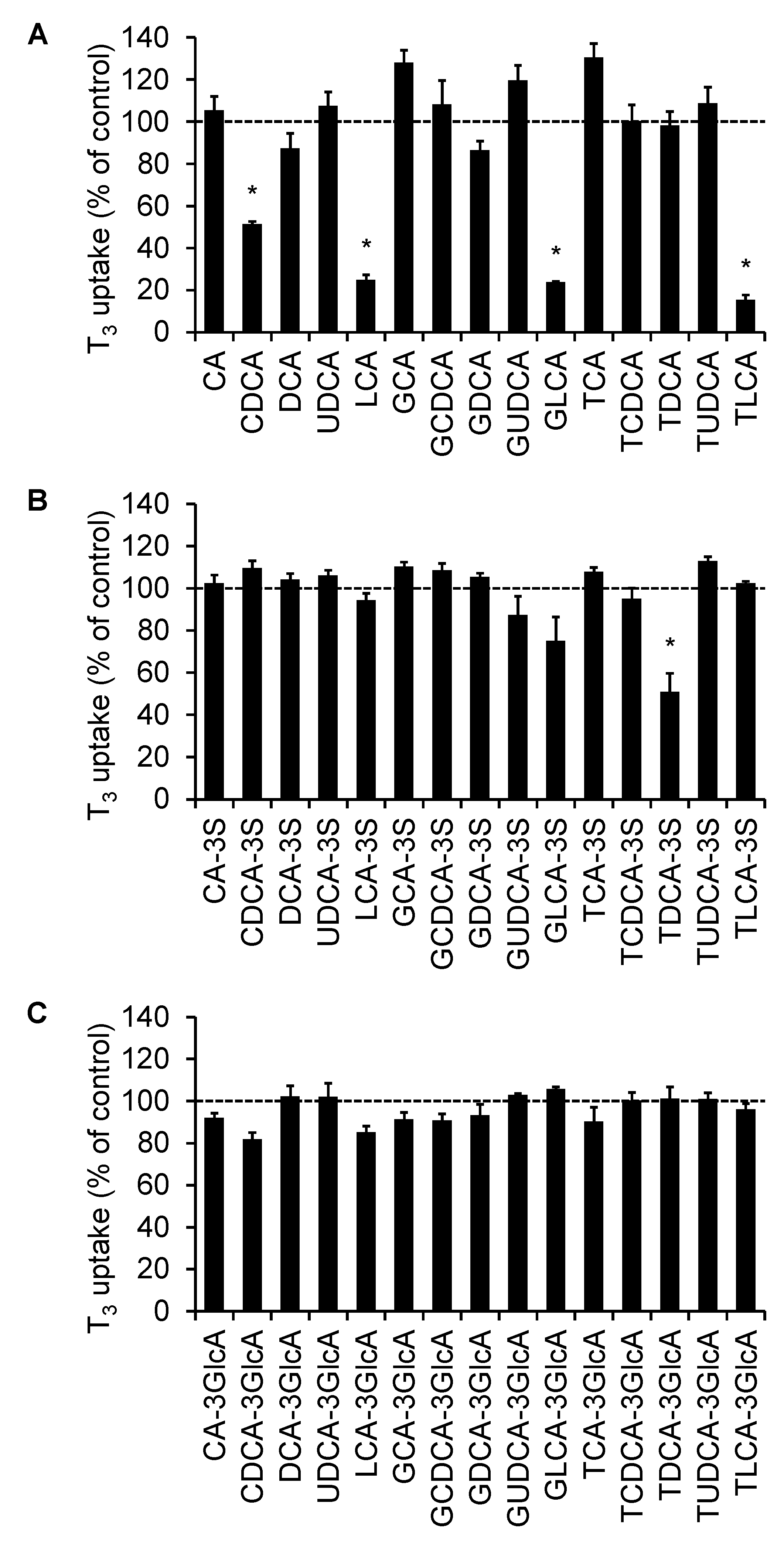
2.1. Screening of the Inhibitory Effect of Bile Acids on OATP4C1-Mediated Transport
2.2. The Concentration-Dependent Inhibitory Effect of Bile Acids
2.3. Calculation of the BDI Index
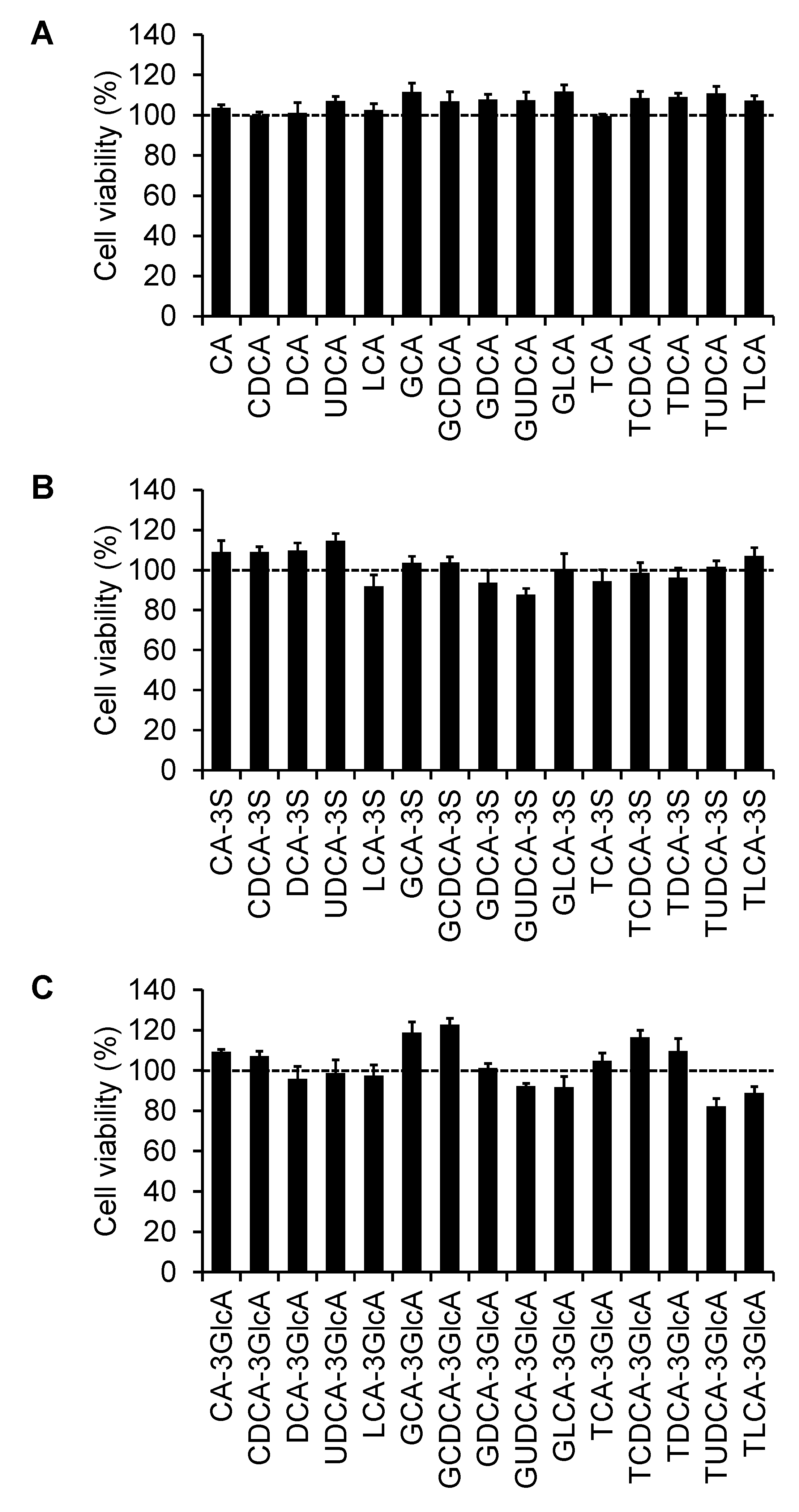
2.4. Effect of Bile Acids on Cell Viability
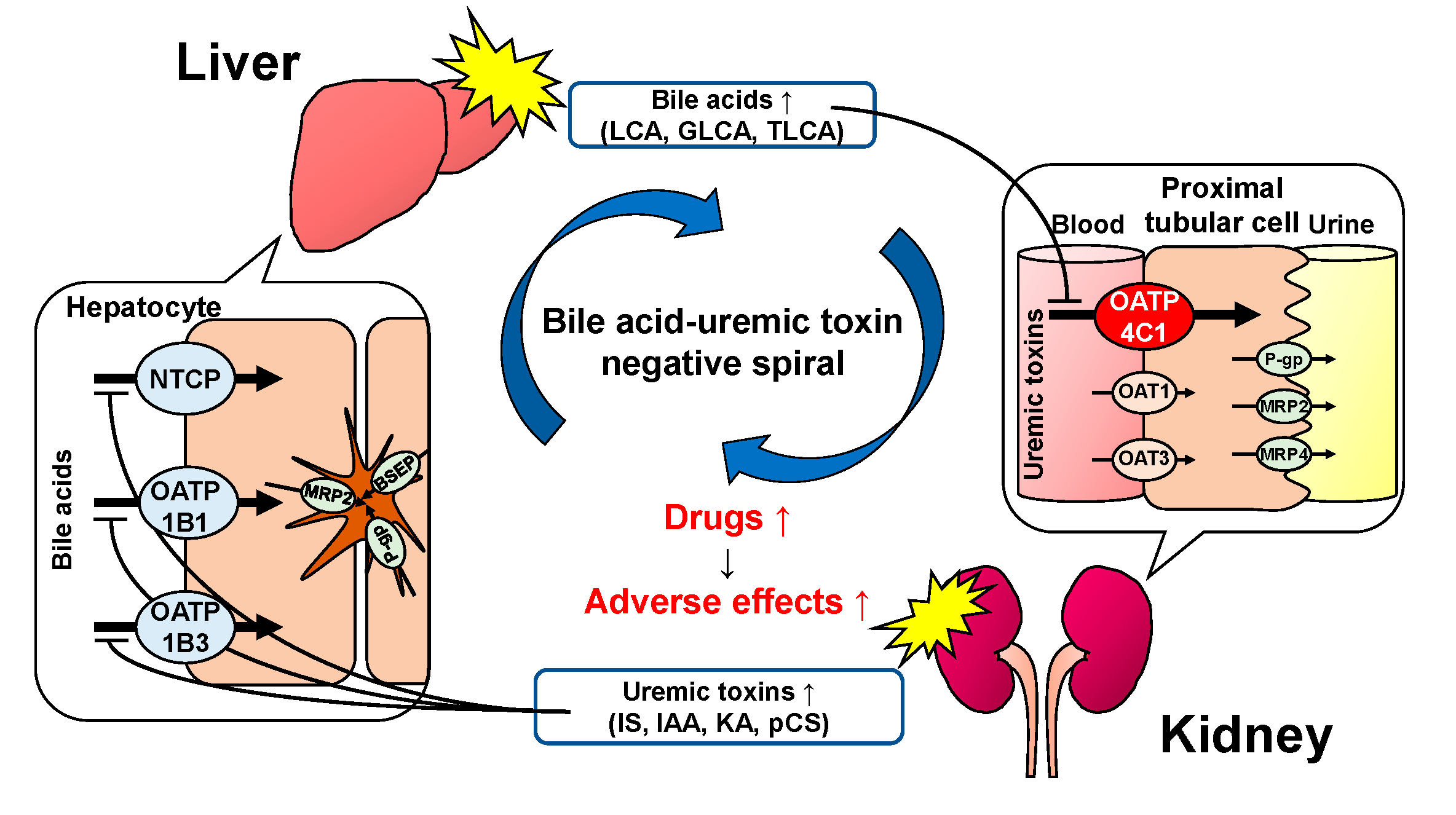
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Transport Study
4.4. Sample Preparation
4.5. Liquid Chromatography/Tandem Mass Spectrometry (LC/MS/MS) Condition
4.6. Inhibitory Effect of Bile Acids
4.7. BDI Index Prediction
4.8. Effect of Bile Acids on Cell Viability
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Narisue, M.; Sugimoto, Y.; Shibata, R.; Otsubo, T.; Hirata, S. High Incidence of Adverse Events in Patients with Impaired Renal Function Taking Pregabalin, Even When Dose Is Adjusted for Renal Function. Nihon Toseki Igakkai Zasshi 2015, 48, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; Sunakawa, Y.; Miwa, K.; Akiyama, Y.; Sugiyama, M.; Kawara, K.; Ishida, H.; Yamashita, K.; Mizuno, K.; Saji, S.; et al. Delayed Elimination of SN-38 in Cancer Patients with Severe Renal Failure. Drug Metab. Dispos. 2011, 39, 161–164. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.I.; Masuo, Y.; Okumura, H.; Watanabe, Y.; Suzuki, H.; Sunakawa, Y.; Shimada, K.; Kawara, K.; Akiyama, Y.; Kitamura, M.; et al. Increased Plasma Concentrations of Unbound SN-38, the Active Metabolite of Irinotecan, in Cancer Patients with Severe Renal Failure. Pharm. Res. 2016, 33, 269–282. [Google Scholar] [CrossRef]
- Yin, J.; Wang, J. Renal Drug Transporters and Their Significance in Drug-Drug Interactions. Acta Pharm. Sin. B 2016, 6, 363–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandoni, A.; Villar, S.R.; Picena, J.C.; Anzai, N.; Endou, H.; Torres, A.M. Expression of Rat Renal Cortical OAT1 and OAT3 in Response to Acute Biliary Obstruction. Hepatology 2006, 43, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Brandoni, A.; Anzai, N.; Kanai, Y.; Endou, H.; Torres, A.M. Renal Elimination of P-Aminohippurate (PAH) in Response to Three Days of Biliary Obstruction in the Rat. The Role of OAT1 and OAT3. Biochim. Biophys. Acta Mol. Basis Dis. 2006, 1762, 673–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, Y.; Kobayashi, Y.; Gabazza, E.C.; Higuchi, K.; Kamisako, T.; Kuroda, M.; Takeuchi, K.; Iwasa, M.; Kaito, M.; Adachi, Y. Increased Renal Expression of Bilirubin Glucuronide Transporters in a Rat Model of Obstructive Jaundice. Am. J. Physiol. Liver Physiol. 2002, 282, G656–G662. [Google Scholar] [CrossRef] [Green Version]
- Denk, G.U.; Soroka, C.J.; Takeyama, Y.; Chen, W.S.; Schuetz, J.D.; Boyer, J.L. Multidrug Resistance-Associated Protein 4 Is up-Regulated in Liver but down-Regulated in Kidney in Obstructive Cholestasis in the Rat. J. Hepatol. 2004, 40, 585–591. [Google Scholar] [CrossRef] [PubMed]
- De Aguiar Vallim, T.Q.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.; Gioiello, A.; Noriega, L.; Strehle, A.; Oury, J.; Rizzo, G.; Macchiarulo, A.; Yamamoto, H.; Mataki, C.; Pruzanski, M.; et al. TGR5-Mediated Bile Acid Sensing Controls Glucose Homeostasis. Cell Metab. 2009, 10, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Makishima, M.; Okamoto, A.Y.; Repa, J.J.; Tu, H.; Learned, R.M.; Luk, A.; Hull, M.V.; Lustig, K.D.; Mangelsdorf, D.J.; Shan, B. Identification of a Nuclear Receptor for Bite Acids. Science 1999, 284, 1362–1365. [Google Scholar] [CrossRef]
- Parks, D.J.; Blanchard, S.G.; Bledsoe, R.K.; Chandra, G.; Consler, T.G.; Kliewer, S.A.; Stimmel, J.B.; Willson, T.M.; Zavacki, A.M.; Moore, D.D.; et al. Bile Acids: Natural Ligands for an Orphan Nuclear Receptor. Science 1999, 284, 1365–1368. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Hollister, K.; Sowers, L.C.; Forman, B.M. Endogenous Bile Acids Are Ligands for the Nuclear Receptor FXR/BAR. Mol. Cell 1999, 3, 543–553. [Google Scholar] [CrossRef]
- Staudinger, J.L.; Goodwin, B.; Jones, S.A.; Hawkins-Brown, D.; MacKenzie, K.I.; LaTour, A.; Liu, Y.; Klaassen, C.D.; Brown, K.K.; Reinhard, J.; et al. The Nuclear Receptor PXR Is a Lithocholic Acid Sensor That Protects against Liver Toxicity. Proc. Natl. Acad. Sci. USA 2001, 98, 3369–3374. [Google Scholar] [CrossRef] [Green Version]
- Kawamata, Y.; Fujii, R.; Hosoya, M.; Harada, M.; Yoshida, H.; Miwa, M.; Fukusumi, S.; Habata, Y.; Itoh, T.; Shintani, Y.; et al. A G Protein-Coupled Receptor Responsive to Bile Acids. J. Biol. Chem. 2003, 278, 9435–9440. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.D.; Chen, W.D.; Yu, D.; Forman, B.M.; Huang, W. The G-Protein-Coupled Bile Acid Receptor, Gpbar1 (TGR5), Negatively Regulates Hepatic Inflammatory Response through Antagonizing Nuclear Factor Kappa Light-Chain Enhancer of Activated B Cells (NF-ΚB) in Mice. Hepatology 2011, 54, 1421–1432. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Houten, S.M.; Mataki, C.; Christoffolete, M.A.; Kim, B.W.; Sato, H.; Messaddeq, N.; Harney, J.W.; Ezaki, O.; Kodama, T.; et al. Bile Acids Induce Energy Expenditure by Promoting Intracellular Thyroid Hormone Activation. Nature 2006, 439, 484–489. [Google Scholar] [CrossRef]
- Li, T.; Chiang, J.Y.L. Bile Acid Signaling in Metabolic Disease and Drug Therapy. Pharmacol. Rev. 2014, 66, 948–983. [Google Scholar] [CrossRef] [Green Version]
- Hagenbuch, B.; Meier, P.J. Molecular Cloning, Chromosomal Localization, and Functional Characterization of a Human Liver Na+/Bile Acid Cotransporter. J. Clin. Investig. 1994, 93, 1326–1331. [Google Scholar] [CrossRef] [Green Version]
- Meier, P.J. Molecular Mechanisms of Hepatic Bile Salt Transport from Sinusoidal Blood into Bile. Am. J. Physiol. Liver Physiol. 1995, 269, G801–G812. [Google Scholar] [CrossRef]
- Meier, P.J.; Stieger, B. Bile Salt Transporters. Annu. Rev. Physiol. 2002, 64, 635–661. [Google Scholar] [CrossRef] [PubMed]
- Keppler, D.; König, J.; Büchler, M. The Canalicular Multidrug Resistance Protein, CMRP/MRP2, a Novel Conjugate Export Pump Expressed in the Apical Membrane of Hepatocytes. Adv. Enzyme Regul. 1997, 37, 321–333. [Google Scholar] [CrossRef]
- Gerloff, T.; Stieger, B.; Hagenbuch, B.; Madon, J.; Landmann, L.; Roth, J.; Hofmann, A.F.; Meier, P.J. The Sister of P-Glycoprotein Represents the Canalicular Bile Salt Export Pump of Mammalian Liver. J. Biol. Chem. 1998, 273, 10046–10050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chignard, N.; Mergey, M.; Veissière, D.; Parc, R.; Capeau, J.; Poupon, R.; Paul, A.; Housset, C. Bile Acid Transport and Regulating Functions in the Human Biliary Epithelium. Hepatology 2001, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Shneider, B.L. Intestinal Bile Acid Transport: Biology, Physiology, and Pathophysiology. J. Pediatr. Gastroenterol. Nutr. 2001, 32, 407–417. [Google Scholar] [CrossRef]
- Seward, D.J.; Koh, A.S.; Boyer, J.L.; Ballatori, N. Functional Complementation between a Novel Mammalian Polygenic Transport Complex and an Evolutionarily Ancient Organic Solute Transporter, OSTα-OSTβ. J. Biol. Chem. 2003, 278, 27473–27482. [Google Scholar] [CrossRef] [Green Version]
- Dawson, P.A.; Hubbert, M.; Haywood, J.; Craddock, A.L.; Zerangue, N.; Christian, W.V.; Ballatori, N. The Heteromeric Organic Solute Transporter α-β, Ostα-Ostβ, Is an Ileal Basolateral Bile Acid Transporter. J. Biol. Chem. 2005, 280, 6960–6968. [Google Scholar] [CrossRef] [Green Version]
- Takehara, I.; Terashima, H.; Nakayama, T.; Yoshikado, T.; Yoshida, M.; Furihata, K.; Watanabe, N.; Maeda, K.; Ando, O.; Sugiyama, Y.; et al. Investigation of Glycochenodeoxycholate Sulfate and Chenodeoxycholate Glucuronide as Surrogate Endogenous Probes for Drug Interaction Studies of OATP1B1 and OATP1B3 in Healthy Japanese Volunteers. Pharm. Res. 2017, 34, 1601–1614. [Google Scholar] [CrossRef]
- Mikkaichi, T.; Suzuki, T.; Onogawa, T.; Tanemoto, M.; Mizutamari, H.; Okada, M.; Chaki, T.; Masuda, S.; Tokui, T.; Eto, N.; et al. Isolation and Characterization of a Digoxin Transporter and Its Rat Homologue Expressed in the Kidney. Proc. Natl. Acad. Sci. USA 2004, 101, 3569–3574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toyohara, T.; Suzuki, T.; Morimoto, R.; Akiyama, Y.; Souma, T.; Shiwaku, H.O.; Takeuchi, Y.; Mishima, E.; Abe, M.; Tanemoto, M.; et al. SLCO4C1 Transporter Eliminates Uremic Toxins and Attenuates Hypertension and Renal Inflammation. J. Am. Soc. Nephrol. 2009, 20, 2546–2555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, K.; Saigusa, D.; Kanemitsu, Y.; Matsumoto, Y.; Thanai, P.; Suzuki, N.; Mise, K.; Yamaguchi, H.; Nakamura, T.; Asaji, K.; et al. Gut Microbiome-Derived Phenyl Sulfate Contributes to Albuminuria in Diabetic Kidney Disease. Nat. Commun. 2019, 10, 1835. [Google Scholar] [CrossRef] [PubMed]
- Taghikhani, E.; Maas, R.; Fromm, M.F.; König, J. The Renal Transport Protein OATP4C1 Mediates Uptake of the Uremic Toxin Asymmetric Dimethylarginine (ADMA) and Efflux of Cardioprotective L-Homoarginine. PLoS ONE 2019, 14, e0213747. [Google Scholar] [CrossRef] [PubMed]
- Taghikhani, E.; Maas, R.; Taudte, R.V.; Gessner, A.; Fromm, M.F.; König, J. Vectorial Transport of the Arginine Derivatives Asymmetric Dimethylarginine (ADMA) and l-Homoarginine by OATP4C1 and P-Glycoprotein Studied in Double-Transfected MDCK Cells. Amino Acids 2020, 52, 975–985. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Miyamori, K.; Sato, T.; Ogura, J.; Kobayashi, M.; Yamada, T.; Mano, N.; Iseki, K. Quantification of Intracellular and Extracellular Digoxin and Ouabain by Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry. J. Chromatogr. B 2014, 972, 73–80. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Mano, N. Analysis of Membrane Transport Mechanisms of Endogenous Substrates Using Chromatographic Techniques. Biomed. Chromatogr. 2019, 33, e4495. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.Y.; Bleasby, K.; Yabut, J.; Cai, X.; Chan, G.H.; Hafey, M.J.; Xu, S.; Bergman, A.J.; Braun, M.P.; Dean, D.C.; et al. Transport of the Dipeptidyl Peptidase-4 Inhibitor Sitagliptin by Human Organic Anion Transporter 3, Organic Anion Transporting Polypeptide 4C1, and Multidrug Resistance P-Glycoprotein. J. Pharmacol. Exp. Ther. 2007, 321, 673–683. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Mishima, E.; Mano, N.; Abe, T.; Yamaguchi, H. Potential Drug Interactions Mediated by Renal Organic Anion Transporter OATP4C1. J. Pharmacol. Exp. Ther. 2017, 362, 271–277. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Maekawa, M.; Mano, N.; Abe, T.; Yamaguchi, H. Role of Oatp4c1 in Renal Handling of Remdesivir and Its Nucleoside Analog Gs-441524: The First Approved Drug for Patients with COVID-19. J. Pharm. Pharm. Sci. 2021, 24, 227–236. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Sugie, M.; Okada, M.; Mikkaichi, T.; Toyohara, T.; Abe, T.; Goto, J.; Hishinuma, T.; Shimada, M.; Mano, N. Transport of Estrone 3-Sulfate Mediated by Organic Anion Transporter OATP4C1: Estrone 3-Sulfate Binds to the Different Recognition Site for Digoxin in OATP4C1. Drug Metab. Pharmacokinet. 2010, 25, 314–317. [Google Scholar] [CrossRef]
- Brites, D.; Rodrigues, C.M.P.; Oliveira, N.; Cardoso, M.D.C.; Graça, L.M. Correction of Maternal Serum Bile Acid Profile during Ursodeoxycholic Acid Therapy in Cholestasis of Pregnancy. J. Hepatol. 1998, 28, 91–98. [Google Scholar] [CrossRef]
- Sang, C.; Wang, X.; Zhou, K.; Sun, T.; Bian, H.; Gao, X.; Wang, Y.; Zhang, H.; Jia, W.; Liu, P.; et al. Bile Acid Profiles Are Distinct among Patients with Different Etiologies of Chronic Liver Disease. J. Proteome Res. 2021, 20, 2340–2351. [Google Scholar] [CrossRef]
- Greco, A.V.; Mingrone, G. Serum Bile Acid Concentrations in Mild Liver Cirrhosis. Clin. Chim. Acta 1993, 221, 183–189. [Google Scholar] [CrossRef]
- Perez, M.J.; Britz, O. Bile-Acid-Induced Cell Injury and Protection. World J. Gastroenterol. 2009, 15, 1677–1689. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, L.; Chen, E.; Lu, J.; Xiao, L.; Liu, Q.; Zhu, D.; Zhang, F.; Xu, X.; Li, L. Targeted Metabolomics Analysis of Bile Acids in Patients with Idiosyncratic Drug-Induced Liver Injury. Metabolites 2021, 11, 852. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Z.; Shi, Z.; Zhu, Z.; Li, C.; Du, Z.; Zhang, Y.; Wang, Z.; Jiao, Z.; Tian, X.; et al. Analysis of Bile Acid Profile in Plasma to Differentiate Cholangiocarcinoma from Benign Biliary Diseases and Healthy Controls. J. Steroid Biochem. Mol. Biol. 2021, 205, 105775. [Google Scholar] [CrossRef]
- Malavolti, M.; Fromm, H.; Ceryak, S.; Shehan, K.L. Interaction of Potentially Toxic Bile Acids with Human Plasma Proteins: Binding of Lithocholic (3α-Hydroxy-5β-Cholan-24-Oic) Acid to Lipoproteins and Albumin. Lipids 1989, 24, 673–676. [Google Scholar] [CrossRef]
- Ceryak, S.; Bouscarel, B.; Fromm, H. Comparative Binding of Bile Acids to Serum Lipoproteins and Albumin. J. Lipid Res. 1993, 34, 1661–1674. [Google Scholar] [CrossRef]
- Ceryak, S.; Bouscarel, B.; Malavolti, M.; Fromm, H. Extrahepatic Deposition and Cytotoxicity of Lithocholic Acid: Studies in Two Hamster Models of Hepatic Failure and in Cultured Human Fibroblasts. Hepatology 1998, 27, 546–556. [Google Scholar] [CrossRef] [Green Version]
- Fanali, G.; Di Masi, A.; Trezza, V.; Marino, M.; Fasano, M.; Ascenzi, P. Human Serum Albumin: From Bench to Bedside. Mol. Aspects Med. 2012, 33, 209–290. [Google Scholar] [CrossRef]
- Domenicali, M.; Baldassarre, M.; Giannone, F.A.; Naldi, M.; Mastroroberto, M.; Biselli, M.; Laggetta, M.; Patrono, D.; Bertucci, C.; Bernardi, M.; et al. Posttranscriptional Changes of Serum Albumin: Clinical and Prognostic Significance in Hospitalized Patients with Cirrhosis. Hepatology 2014, 60, 1851–1860. [Google Scholar] [CrossRef]
- Baldassarre, M.; Naldi, M.; Zaccherini, G.; Bartoletti, M.; Antognoli, A.; Laggetta, M.; Gagliardi, M.; Tufoni, M.; Domenicali, M.; Waterstradt, K.; et al. Determination of Effective Albumin in Patients with Decompensated Cirrhosis: Clinical and Prognostic Implications. Hepatology 2021, 74, 2058–2073. [Google Scholar] [CrossRef]
- Wasan, K.M.; Cassidy, S.M. Role of Plasma Lipoproteins in Modifying the Biological Activity of Hydrophobic Drugs. J. Pharm. Sci. 1998, 87, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Komazawa, H.; Yamaguchi, H.; Hidaka, K.; Ogura, J.; Kobayashi, M.; Iseki, K. Renal Uptake of Substrates for Organic Anion Transporters Oat1 and Oat3 and Organic Cation Transporters Oct1 and Oct2 Is Altered in Rats with Adenine-Induced Chronic Renal Failure. J. Pharm. Sci. 2013, 102, 1086–1094. [Google Scholar] [CrossRef]
- Sato, T.; Yamaguchi, H.; Kogawa, T.; Abe, T.; Mano, N. Organic Anion Transporting Polypeptides 1B1 and 1B3 Play an Important Role in Uremic Toxin Handling and Drug-Uremic Toxin Interactions in the Liver. J. Pharm. Pharm. Sci. 2014, 17, 475–484. [Google Scholar] [CrossRef]
- Weigand, K.M.; Schirris, T.J.J.; Houweling, M.; van den Heuvel, J.J.M.W.; Koenderink, J.B.; Dankers, A.C.A.; Russel, F.G.M.; Greupink, R. Uremic Solutes Modulate Hepatic Bile Acid Handling and Induce Mitochondrial Toxicity. Toxicol. Vitr. 2019, 56, 52–61. [Google Scholar] [CrossRef]
- Goto, J.; Miura, H.; Inada, M.; Nambara, T.; Nagakura, T.; Suzuki, H. Studies on Steroids. J. Chromatogr. A 1988, 452, 119–129. [Google Scholar] [CrossRef]
- Brouwer, K.L.R.; Keppler, D.; Hoffmaster, K.A.; Bow, D.A.J.; Cheng, Y.; Lai, Y.; Palm, J.E.; Stieger, B.; Evers, R. In Vitro Methods to Support Transporter Evaluation in Drug Discovery and Development. Clin. Pharmacol. Ther. 2013, 94, 95–112. [Google Scholar] [CrossRef] [Green Version]


| Bile Acid | IC50 (µM) | Ki (µM) |
|---|---|---|
| LCA | 6.12 ± 1.21 | 5.23 ± 1.03 |
| GLCA | 9.90 ± 0.475 | 8.46 ± 0.406 |
| TLCA | 12.3 ± 0.891 | 10.5 ± 0.762 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamauchi, M.; Sato, T.; Otake, A.; Kumondai, M.; Sato, Y.; Kikuchi, M.; Maekawa, M.; Yamaguchi, H.; Abe, T.; Mano, N. Bile Acid–Drug Interaction via Organic Anion-Transporting Polypeptide 4C1 Is a Potential Mechanism of Altered Pharmacokinetics of Renally Excreted Drugs. Int. J. Mol. Sci. 2022, 23, 8508. https://doi.org/10.3390/ijms23158508
Yamauchi M, Sato T, Otake A, Kumondai M, Sato Y, Kikuchi M, Maekawa M, Yamaguchi H, Abe T, Mano N. Bile Acid–Drug Interaction via Organic Anion-Transporting Polypeptide 4C1 Is a Potential Mechanism of Altered Pharmacokinetics of Renally Excreted Drugs. International Journal of Molecular Sciences. 2022; 23(15):8508. https://doi.org/10.3390/ijms23158508
Chicago/Turabian StyleYamauchi, Minami, Toshihiro Sato, Ayana Otake, Masaki Kumondai, Yu Sato, Masafumi Kikuchi, Masamitsu Maekawa, Hiroaki Yamaguchi, Takaaki Abe, and Nariyasu Mano. 2022. "Bile Acid–Drug Interaction via Organic Anion-Transporting Polypeptide 4C1 Is a Potential Mechanism of Altered Pharmacokinetics of Renally Excreted Drugs" International Journal of Molecular Sciences 23, no. 15: 8508. https://doi.org/10.3390/ijms23158508
APA StyleYamauchi, M., Sato, T., Otake, A., Kumondai, M., Sato, Y., Kikuchi, M., Maekawa, M., Yamaguchi, H., Abe, T., & Mano, N. (2022). Bile Acid–Drug Interaction via Organic Anion-Transporting Polypeptide 4C1 Is a Potential Mechanism of Altered Pharmacokinetics of Renally Excreted Drugs. International Journal of Molecular Sciences, 23(15), 8508. https://doi.org/10.3390/ijms23158508






