miR-199a: A Tumor Suppressor with Noncoding RNA Network and Therapeutic Candidate in Lung Cancer
Abstract
:1. Introduction
2. Materials and Methods
3. Result and Discussion
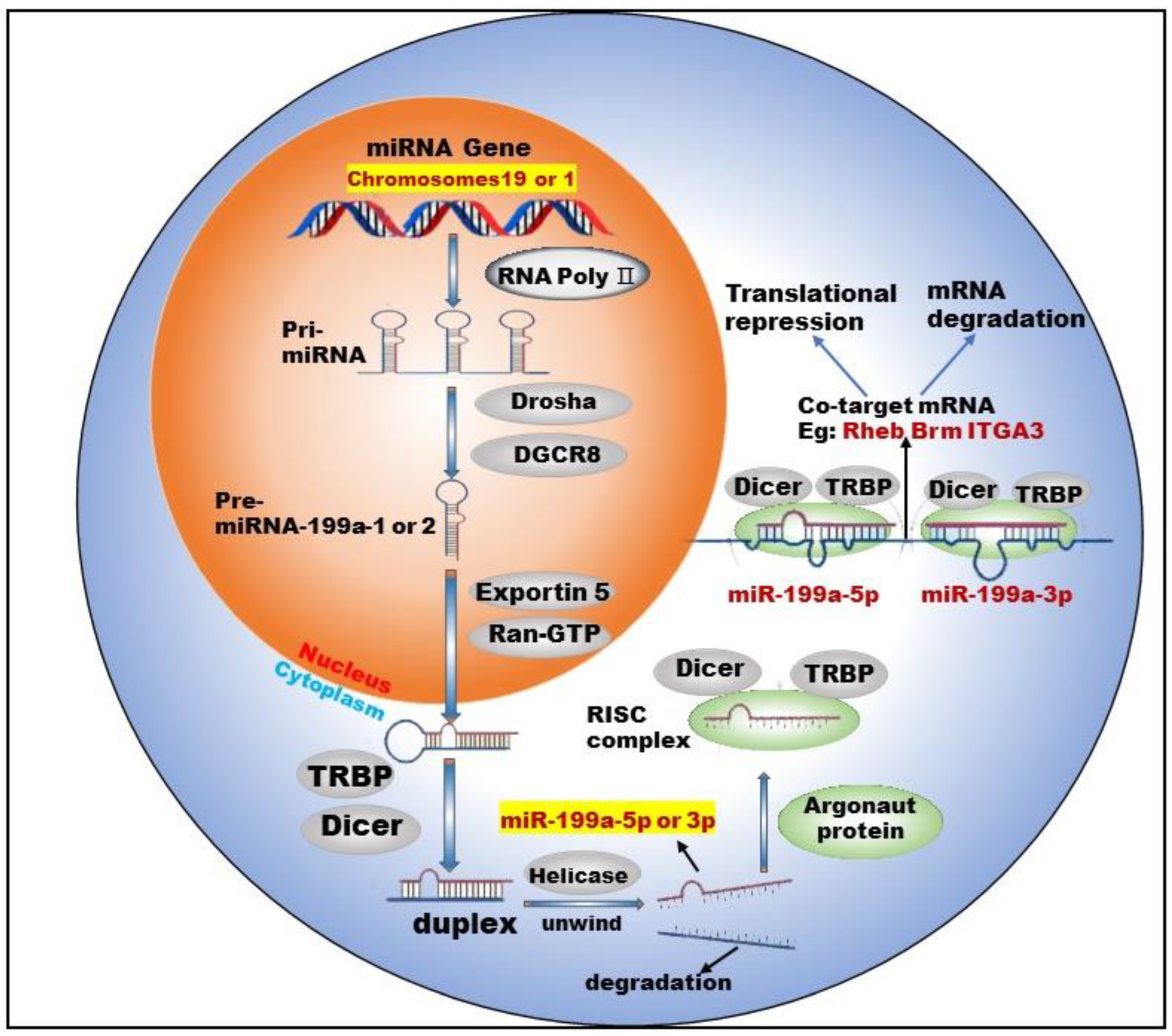
3.1. Biogenesis of miR-199a
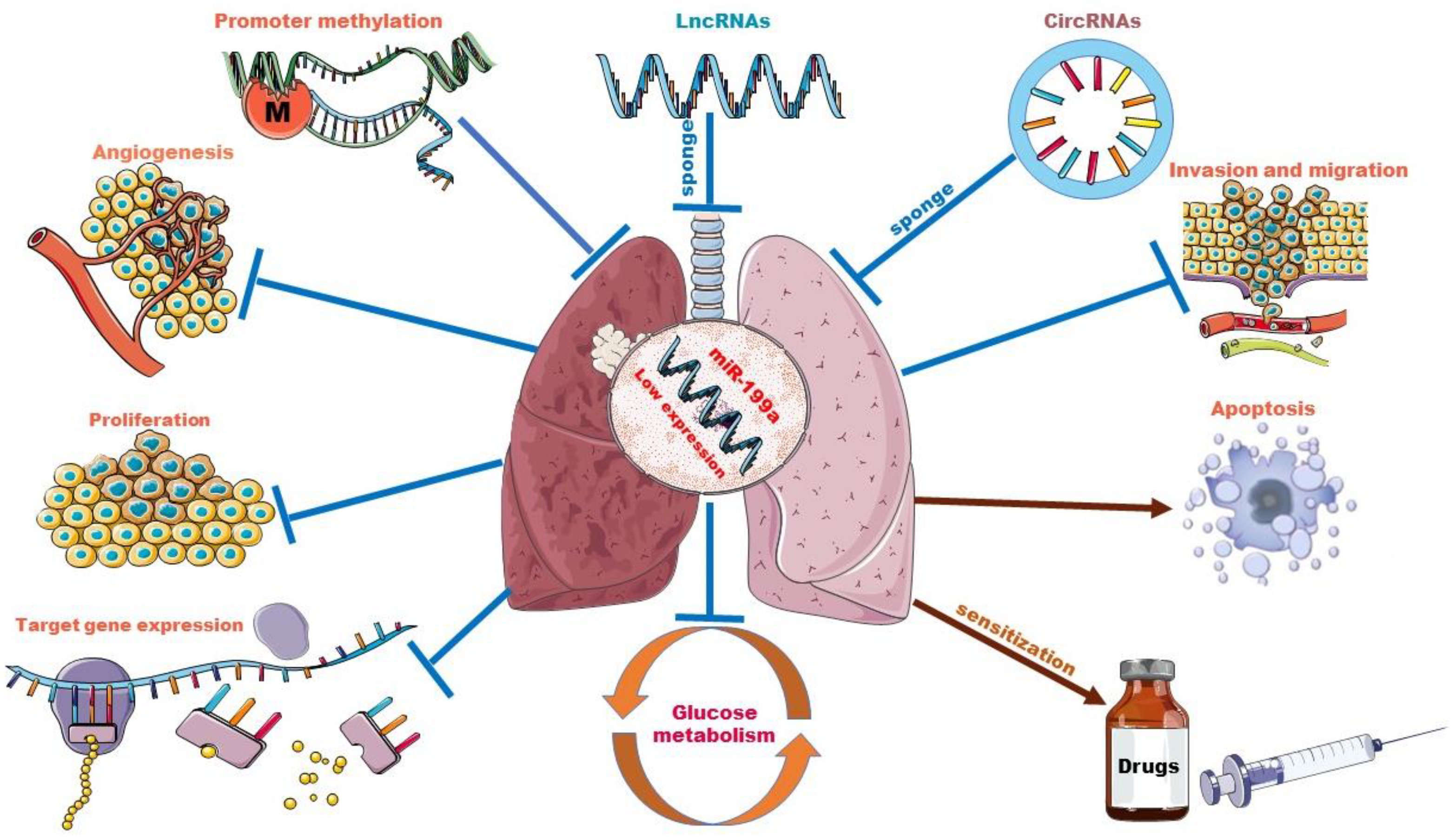
3.2. Biological Roles of miR-199a in Lung Cancer
3.2.1. Methylation and the Low Expression of miR-199a
3.2.2. miR-199a Regulates Cell Functions and Tumor Angiogenesis
3.2.3. miR-199a Suppresses the Progression of Lung Cancer via Mediating Signaling Pathway
3.2.4. miR-199a Improves the Accuracy of Lung Cancer Diagnosis
3.2.5. miR-199a Regulates Glucose Metabolism in Lung Cancer
3.2.6. miR-199a and Drug Resistance
3.3. Interplay between miR-199a and Other ncRNAs
3.3.1. miR-199a and lncRNAs
3.3.2. miR-199a and circRNAs
3.3.3. miR-199a and tRFs
3.4. Discussion and Perspectives
3.4.1. Recent Progress in Diagnosis and Treatment of Lung Cancer
3.4.2. Prospects of ncRNAs for Diagnosis and Treatment of Lung Cancer
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, R.; Zhang, S.; Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer incidence and mortality in China 2016. J. Natl. Cancer Cent. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.K.; Singh, D.P.; Choudhary, J. Lung cancer identification: A review on detection and classification. Cancer Metastasis Rev. 2020, 39, 989–998. [Google Scholar] [CrossRef]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef]
- Rami-Porta, R.; Bolejack, V.; Giroux, D.J.; Chansky, K.; Crowley, J.; Asamura, H.; Goldstraw, P. The IASLC Lung Cancer Staging Project: The New Database to Inform the Eighth Edition of the TNM Classification of Lung Cancer. J. Thorac. Oncol. 2014, 9, 1618–1624. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhu, S.; Meng, N.; He, Y.; Lu, R.; Yan, G.-R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol. Ther. 2019, 27, 1718–1725. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Anelli, L.; Zagaria, A.; Specchia, G.; Musto, P.; Albano, F. Dysregulation of miRNA in Leukemia: Exploiting miRNA Expression Profiles as Biomarkers. Int. J. Mol. Sci. 2021, 22, 7156. [Google Scholar] [CrossRef]
- Barwari, T.; Joshi, A.; Mayr, M. MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2577–2584. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Chan, W.-Y. Flexible and versatile as a chameleon-sophisticated functions of microRNA-199a. Int. J. Mol. Sci. 2012, 13, 8449–8466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Cao, Z.; Wang, W.; Jie, X.; Li, L. MicroRNA-199a-5p regulates FOXC2 to control human vascular smooth muscle cell phenotypic switch. Mol. Med. Rep. 2021, 24, 12266. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.-B.; Jia, B.; Wang, W.; Xu, G.-H.; Guo, J.-C.; Li, X.; Liu, J.-N. Role of miR-199a-5p in osteoblast differentiation by targeting TET2. Gene 2020, 726, 144193. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhang, H.; Huang, S.; Pei, L.; Feng, M.; Zhao, X.; Ouyang, Z.; Yao, S.; Jiang, R.; Wei, K. miR-199a-3p promotes cardiomyocyte proliferation by inhibiting Cd151 expression. Biochem. Biophys. Res. Commun. 2019, 516, 28–36. [Google Scholar] [CrossRef]
- Sakurai, K.; Furukawa, C.; Haraguchi, T.; Inada, K.-I.; Shiogama, K.; Tagawa, T.; Fujita, S.; Ueno, Y.; Ogata, A.; Ito, M.; et al. MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers. Cancer Res. 2011, 71, 1680–1689. [Google Scholar] [CrossRef] [Green Version]
- Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Arai, T.; Okato, A.; Kurozumi, A.; Kato, M.; Katada, K.; Okamoto, Y.; Seki, N. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017, 108, 1681–1692. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Yu, X.; Tang, Y.; Guo, Y.; Yuan, J.; Bai, J.; Yao, T.; Wu, X. MicroRNA-199a-3p inhibits ovarian cancer cell viability by targeting the oncogene YAP1. Mol. Med. Rep. 2021, 23, 11876. [Google Scholar] [CrossRef]
- Hou, G.; Wang, Y.; Zhang, M.; Hu, Y.; Zhao, Y.; Jia, A.; Wang, P.; Zhao, W.; Zhao, W.; Lu, Z. miR-199a-3p suppresses progression of esophageal squamous cell carcinoma through inhibiting mTOR/p70S6K pathway. Anticancer Drugs 2021, 32, 157–167. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, X.; Cai, Q.; Wang, X.; Yu, B.; Cai, Q.; Liu, B.; Zhu, Z.; Li, C. miR-199a-3p promotes gastric cancer progression by targeting ZHX1. FEBS Lett. 2014, 588, 4504–4512. [Google Scholar] [CrossRef] [Green Version]
- Shatseva, T.; Lee, D.Y.; Deng, Z.; Yang, B.B. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J. Cell Sci. 2011, 124, 2826–2836. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.; Assen, F.P.; Leithner, A.; Abe, J.; Schachner, H.; Asfour, G.; Bago-Horvath, Z.; Stein, J.V.; Uhrin, P.; Sixt, M.; et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 2018, 359, 1408–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadi, A.; Khansarinejad, B.; Hosseinkhani, S.; Ghanei, M.; Mowla, S.J. miR-199a-5p and miR-495 target GRP78 within UPR pathway of lung cancer. Gene 2017, 620, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, D.; Li, X.; Shao, Y.; He, Y.; Yu, H.; Ma, Z.-L. miR-199a-5p suppresses non-small cell lung cancer via targeting MAP3K11. J. Cancer 2019, 10, 2472–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, N.; Liang, Y.; Zhu, T.; Long, Y.; Chen, Z.; Zhang, X.; Jiang, L. Epigenetic silencing of microRNA-199a-5p promotes the proliferation of non-small cell lung cancer cells by increasing AKAP1 expression. Oncol. Lett. 2021, 21, 434. [Google Scholar] [CrossRef]
- Mudduluru, G.; Ceppi, P.; Kumarswamy, R.; Scagliotti, G.V.; Papotti, M.; Allgayer, H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene 2011, 30, 2888–2899. [Google Scholar] [CrossRef]
- Wu, F.; Lin, X.; Shan, S.-K.; Li, F.; Xu, F.; Zhong, J.-Y.; Guo, B.; Zheng, M.-H.; Wang, Y.; Mo, Z.-H.; et al. The Suppression of miR-199a-3p by Promoter Methylation Contributes to Papillary Thyroid Carcinoma Aggressiveness by Targeting RAP2a and DNMT3a. Front. Cell Dev. Biol. 2020, 8, 594528. [Google Scholar] [CrossRef]
- Deng, Y.; Zhao, F.; Hui, L.; Li, X.; Zhang, D.; Lin, W.; Chen, Z.; Ning, Y. Suppressing miR-199a-3p by promoter methylation contributes to tumor aggressiveness and cisplatin resistance of ovarian cancer through promoting DDR1 expression. J. Ovarian Res. 2017, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Konno, M.; Koseki, J.; Asai, A.; Yamagata, A.; Shimamura, T.; Motooka, D.; Okuzaki, D.; Kawamoto, K.; Mizushima, T.; Eguchi, H.; et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019, 10, 3888. [Google Scholar] [CrossRef] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Xue, S.; Zhu, W.; Liu, D.; Su, Z.; Zhang, L.; Chang, Q.; Li, P. Circulating miR-26a-1, miR-146a and miR-199a-1 are potential candidate biomarkers for acute myocardial infarction. Mol. Med. 2019, 25, 18. [Google Scholar] [CrossRef]
- Villalobos, P.; Wistuba, I.I. Lung Cancer Biomarkers. Hematol. Oncol. Clin. N. Am. 2017, 31, 13–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, G.; Huang, G.; Liu, H.-D.; Liang, H.-X.; Ni, Y.-F.; Ding, Z.-H.; Ni, G.-Y.; Hua, H.-W. miR-199a suppresses the hypoxia-induced proliferation of non-small cell lung cancer cells through targeting HIF1α. Mol. Cell Biochem. 2013, 384, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zheng, Y.; Tan, J.; Tian, R.; Shen, P.; Cai, W.; Liao, H. miR-199a-5p-HIF-1α-STAT3 Positive Feedback Loop Contributes to the Progression of Non-Small Cell Lung Cancer. Front. Cell Dev. Biol. 2020, 8, 620615. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Jiao, W.-Y. Down-Regulation of ZEB1 by miR-199a-3p Overexpression Restrains Tumor Stem-Like Properties and Mitochondrial Function of Non-Small Cell Lung Cancer. Onco Targets Ther. 2020, 13, 4607–4616. [Google Scholar] [CrossRef]
- Dong, C.; Cao, H.; Liu, Z.; Xi, L.; Shi, Y.; Yang, R. CHML targeted by miR-199a-3p promotes non-small cell lung cancer cell growth via binding to Rab5A. Pathol. Res. Pract. 2021, 227, 153626. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.; Wang, Y.; Wu, D.; Zhang, H. miR-199a-3p plays an anti-tumorigenic role in lung adenocarcinoma by suppressing anterior gradient 2. Bioengineered 2021, 12, 7859–7871. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Chai, B.; Wu, Z.; Gu, Z.; Zou, H.; Zhang, H.; Li, Y.; Sun, Q.; Fang, W.; et al. miR-199a-3p/5p regulate tumorgenesis via targeting Rheb in non-small cell lung cancer. Int. J. Biol. Sci. 2022, 18, 4187–4202. [Google Scholar] [CrossRef]
- He, J.; Wang, M.; Jiang, Y.; Chen, Q.; Xu, S.; Xu, Q.; Jiang, B.-H.; Liu, L.-Z. Chronic arsenic exposure and angiogenesis in human bronchial epithelial cells via the ROS/miR-199a-5p/HIF-1α/COX-2 pathway. Environ. Health Perspect. 2014, 122, 255–261. [Google Scholar] [CrossRef]
- Zuo, Y.; Qu, C.; Tian, Y.; Wen, Y.; Xia, S.; Ma, M. The HIF-1/SNHG1/miR-199a-3p/TFAM axis explains tumor angiogenesis and metastasis under hypoxic conditions in breast cancer. Biofactors 2021, 47, 444–460. [Google Scholar] [CrossRef]
- Song, X.; Guo, Y.; Song, P.; Duan, D.; Guo, W. Non-coding RNAs in Regulating Tumor Angiogenesis. Front. Cell Dev. Biol. 2021, 9, 751578. [Google Scholar] [CrossRef]
- Wang, L.-M.; Zhang, L.-L.; Wang, L.-W.; Zhu, L.; Ma, X.-X. Influence of miR-199a on rats with non-small cell lung cancer via regulating the HIF-1α/VEGF signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10363–10369. [Google Scholar] [PubMed]
- Qi, H.; Liu, Y.; Wang, N.; Xiao, C. Lentinan Attenuated the PM2.5 Exposure-Induced Inflammatory Response, Epithelial-Mesenchymal Transition and Migration by Inhibiting the PVT1/miR-199a-5p/caveolin1 Pathway in Lung Cancer. DNA Cell Biol. 2021, 40, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, H.; Wang, Z.; Gao, S.; Zhang, X.; Zhu, H.; Wang, N.; Li, H. The regulation of autophagy by the miR-199a-5p/p62 axis was a potential mechanism of small cell lung cancer cisplatin resistance. Cancer Cell Int. 2022, 22, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, T.; Chen, G.; Yan, G.; Zhang, X.; Wan, Y.; Li, Q.; Zhu, B.; Zhuo, W. Identification of a serum microRNA expression signature for detection of lung cancer, involving miR-23b, miR-221, miR-148b and miR-423-3p. Lung Cancer 2017, 114, 6–11. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ren, S.; Wang, Y.; Li, X.; Zhou, C.; Hirsch, F.R. Serum microRNAs improving the diagnostic accuracy in lung cancer presenting with pulmonary nodules. J. Thorac. Dis. 2018, 10, 5080–5085. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Posener, K.; Negelein, E. Über den Stoffwechsel der Carcinomzelle. Naturwissenschaften 1924, 12, 1131–1137. [Google Scholar] [CrossRef]
- Li, F.; Han, X.; Li, F.; Wang, R.; Wang, H.; Gao, Y.; Wang, X.; Fang, Z.; Zhang, W.; Yao, S.; et al. LKB1 Inactivation Elicits a Redox Imbalance to Modulate Non-small Cell Lung Cancer Plasticity and Therapeutic Response. Cancer Cell 2015, 27, 698–711. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Wu, Q.; Wang, J.; Yao, B.; Ma, L.; Yang, Z.; Li, J.; Liu, B. NOX4 supports glycolysis and promotes glutamine metabolism in non-small cell lung cancer cells. Free Radic. Biol. Med. 2016, 101, 236–248. [Google Scholar] [CrossRef]
- Zhou, L.; Li, M.; Yu, X.; Gao, F.; Li, W. Repression of Hexokinases II-Mediated Glycolysis Contributes to Piperlongumine-Induced Tumor Suppression in Non-Small Cell Lung Cancer Cells. Int. J. Biol. Sci. 2019, 15, 826–837. [Google Scholar] [CrossRef]
- Martinez, C.A.; Scafoglio, C. Heterogeneity of Glucose Transport in Lung Cancer. Biomolecules 2020, 10, 868. [Google Scholar] [CrossRef]
- Ghanem, N.; El-Baba, C.; Araji, K.; El-Khoury, R.; Usta, J.; Darwiche, N. The Pentose Phosphate Pathway in Cancer: Regulation and Therapeutic Opportunities. Chemotherapy 2021, 66, 179–191. [Google Scholar] [CrossRef]
- Liu, X.; Olszewski, K.; Zhang, Y.; Lim, E.W.; Shi, J.; Zhang, X.; Zhang, J.; Lee, H.; Koppula, P.; Lei, G.; et al. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 2020, 22, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Movahed, Z.G.; Yarani, R.; Mohammadi, P.; Mansouri, K. Sustained oxidative stress instigates differentiation of cancer stem cells into tumor endothelial cells: Pentose phosphate pathway, reactive oxygen species and autophagy crosstalk. Biomed. Pharmacother. 2021, 139, 111643. [Google Scholar] [CrossRef] [PubMed]
- Ghanavat, M.; Shahrouzian, M.; Zayeri, Z.D.; Banihashemi, S.; Kazemi, S.M.; Saki, N. Digging deeper through glucose metabolism and its regulators in cancer and metastasis. Life Sci. 2021, 264, 118603. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Feng, S.; Nie, Z.; Deng, Y.; Xuan, Y.; Chen, X.; Lu, Y.; Liang, L.; Chen, Y. TRAF6 Promoted Tumor Glycolysis in Non-Small-Cell Lung Cancer by Activating the Akt-HIFα Pathway. BioMed Res. Int. 2021, 2021, 3431245. [Google Scholar] [CrossRef]
- Hua, Q.; Wang, D.; Zhao, L.; Hong, Z.; Ni, K.; Shi, Y.; Liu, Z.; Mi, B. AL355338 acts as an oncogenic lncRNA by interacting with protein ENO1 to regulate EGFR/AKT pathway in NSCLC. Cancer Cell Int. 2021, 21, 525. [Google Scholar] [CrossRef]
- Cao, H.-J.; Zhou, W.; Xian, X.-L.; Sun, S.-J.; Ding, P.-J.; Tian, C.-Y.; Tian, F.-L.; Jiang, C.-H.; Fu, T.-T.; Zhao, S.; et al. A Mixture of Baicalein, Wogonin, and Oroxylin-A Inhibits EMT in the A549 Cell Line via the PI3K/AKT-TWIST1-Glycolysis Pathway. Front. Pharmacol. 2021, 12, 821485. [Google Scholar] [CrossRef]
- Xie, Z.; Li, H.; Zang, J. Knockdown of lysine (K)-specific demethylase 2B KDM2B inhibits glycolysis and induces autophagy in lung squamous cell carcinoma cells by regulating the phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin pathway. Bioengineered 2021, 12, 12227–12235. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Li, Y.; Huang, C.; Xiao, X.; Zhong, Z.; Tang, J.; Lu, H.; Tang, Y.; Yang, J. Dihydroartemisinin and artesunate inhibit aerobic glycolysis via suppressing c-Myc signaling in non-small cell lung cancer. Biochem. Pharmacol. 2022, 198, 114941. [Google Scholar] [CrossRef]
- Tian, W.; Yuan, X.; Song, Y.; Zhai, J.; Wei, H.; Wang, L.; Li, D.; Chen, Q. miR-218 inhibits glucose metabolism in non-small cell lung cancer via the NF-κB signaling pathway. Exp. Ther. Med. 2021, 21, 106. [Google Scholar] [CrossRef]
- Xia, M.; Li, X.; Diao, Y.; Du, B.; Li, Y. Targeted inhibition of glutamine metabolism enhances the antitumor effect of selumetinib in KRAS-mutant NSCLC. Transl. Oncol. 2021, 14, 100920. [Google Scholar] [CrossRef] [PubMed]
- Giatromanolaki, A.; Sivridis, E.; Arelaki, S.; Koukourakis, M.I. Expression of enzymes related to glucose metabolism in non-small cell lung cancer and prognosis. Exp. Lung Res. 2017, 43, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Ciscato, F.; Ferrone, L.; Masgras, I.; Laquatra, C.; Rasola, A. Hexokinase 2 in Cancer: A Prima Donna Playing Multiple Characters. Int. J. Mol. Sci. 2021, 22, 4716. [Google Scholar] [CrossRef]
- Shen, J.; Jin, Z.; Lv, H.; Jin, K.; Jonas, K.; Zhu, C.; Chen, B. PFKP is highly expressed in lung cancer and regulates glucose metabolism. Cell. Oncol. 2020, 43, 617–629. [Google Scholar] [CrossRef]
- Ding, M.; Li, F.; Wang, B.; Chi, G.; Liu, H. A comprehensive analysis of WGCNA and serum metabolomics manifests the lung cancer-associated disordered glucose metabolism. J. Cell. Biochem. 2019, 120, 10855–10863. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chai, B.; Wang, X.; Wu, Z.; Gu, Z.; Liu, X.; Zhao, Y.; Chen, T.; Ma, Z.; Sun, Q. miRNA-199a-5p/SLC2A1 axis regulates glucose metabolism in non-small cell lung cancer. J. Cancer 2022, 13, 2352–2361. [Google Scholar] [CrossRef] [PubMed]
- Brody, R.; Zhang, Y.; Ballas, M.; Siddiqui, M.K.; Gupta, P.; Barker, C.; Midha, A.; Walker, J. PD-L1 expression in advanced NSCLC: Insights into risk stratification and treatment selection from a systematic literature review. Lung Cancer 2017, 112, 200–215. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Zhan, Y.; Liu, S.; Lu, J.; Luo, J.; Feng, J.; Fan, S. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. J. Exp. Clin. Cancer Res. 2018, 37, 226. [Google Scholar] [CrossRef]
- Crucitta, S.; Cucchiara, F.; Mathijssen, R.; Mateo, J.; Jager, A.; Joosse, A.; Passaro, A.; Attili, I.; Petrini, I.; van Schaik, R.; et al. Treatment-driven tumour heterogeneity and drug resistance: Lessons from solid tumours. Cancer Treat. Rev. 2022, 104, 102340. [Google Scholar] [CrossRef]
- Wang, J.; Yang, M.; Li, Y.; Han, B. The Role of MicroRNAs in the Chemoresistance of Breast Cancer. J. Mod. Oncol. 2015, 76, 368–374. [Google Scholar] [CrossRef]
- Toscano-Garibay, J.D.; Aquino-Jarquin, G. Regulation exerted by miRNAs in the promoter and UTR sequences: MDR1/P-gp expression as a particular case. DNA Cell Biol. 2012, 31, 1358–1364. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Sun, J.; Zhang, N.; Zheng, Y.; Wang, X.; Lv, L.; Liu, J.; Xu, Y.; Shen, Y.; Yang, M. Noncoding RNAs in gastric cancer: Implications for drug resistance. Mol. Cancer 2020, 19, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, Y.; Qin, Z.; Cai, S.; Yu, L.; Hu, H.; Zeng, S. The role of non-coding RNAs in ABC transporters regulation and their clinical implications of multidrug resistance in cancer. Expert Opin. Drug Metab. Toxicol. 2021, 17, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, C.; Xu, F.; Zhang, A.; Jin, M.; Zhang, K.; Liu, L.; Hua, Q.; Zhao, J.; Liu, J.; et al. Cisplatin-resistant NSCLC cells induced by hypoxia transmit resistance to sensitive cells through exosomal PKM2. Theranostics 2021, 11, 2860–2875. [Google Scholar] [CrossRef]
- Xia, M.; Sheng, L.; Qu, W.; Xue, X.; Chen, H.; Zheng, G.; Chen, W. miR-194-5p enhances the sensitivity of nonsmall-cell lung cancer to doxorubicin through targeted inhibition of hypoxia-inducible factor-1. World J. Surg. Oncol. 2021, 19, 174. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Tóth, S.; Szepesi, Á.; Tran-Nguyen, V.-K.; Sarkadi, B.; Német, K.; Falson, P.; Di Pietro, A.; Szakács, G.; Boumendjel, A. Synthesis and Anticancer Cytotoxicity of Azaaurones Overcoming Multidrug Resistance. Molecules 2020, 25, 764. [Google Scholar] [CrossRef] [Green Version]
- Sousa, D.; Matthiesen, R.; Lima, R.T.; Vasconcelos, M.H. Deep Sequencing Analysis Reveals Distinctive Non-Coding RNAs When Comparing Tumor Multidrug-Resistant Cells and Extracellular Vesicles with Drug-Sensitive Counterparts. Cancers 2020, 12, 200. [Google Scholar] [CrossRef] [Green Version]
- Zeng, T.; Xu, M.; Zhang, W.; Gu, X.; Zhao, F.; Liu, X.; Zhang, X. Autophagy inhibition and microRNA-199a-5p upregulation in paclitaxel-resistant A549/T lung cancer cells. Oncol. Rep. 2021, 46, 8100. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, H.; Zhu, Y.; Feng, H.; Wang, G.; Wang, S. miR-199a-5p is involved in doxorubicin resistance of non-small cell lung cancer (NSCLC) cells. Eur. J. Pharmacol. 2020, 878, 173105. [Google Scholar] [CrossRef]
- Lou, G.; Chen, L.; Xia, C.; Wang, W.; Qi, J.; Li, A.; Zhao, L.; Chen, Z.; Zheng, M.; Liu, Y. miR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J. Exp. Clin. Cancer Res. 2020, 39, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagkouni, D.; Karavangeli, A.; Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Characterizing miRNA–lncRNA Interplay, in Long Non-Coding RNAs: Methods and Protocols; Zhang, L., Hu, X., Eds.; Springer: New York, NY, USA, 2021; pp. 243–262. [Google Scholar]
- Peng, X.; Yan, J.; Cheng, F. LncRNA TMPO-AS1 up-regulates the expression of HIF-1α and promotes the malignant phenotypes of retinoblastoma cells via sponging miR-199a-5p. Pathol. Res. Pract. 2020, 216, 152853. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-Z.; Liu, G.-Y.; Pang, W.-W.; Zhang, H.; Zeng, Z.-J.; Wang, H.-J. LncRNA LUCAT1 promotes proliferation of ovarian cancer cells by regulating miR-199a-5p expression. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1682–1687. [Google Scholar] [PubMed]
- Yin, D.; Hu, Z.; Luo, C.; Wang, X.; Xin, H.; Sun, R.; Wang, P.; Li, J.; Fan, J.; Zhou, Z.; et al. LINC01133 promotes hepatocellular carcinoma progression by sponging miR-199a-5p and activating annexin A2. Clin. Transl. Med. 2021, 11, e409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cheng, Q.; Xia, M.; Huang, X.; He, X.; Liao, J. Hypoxia-Induced lncRNA-NEAT1 Sustains the Growth of Hepatocellular Carcinoma via Regulation of miR-199a-3p/UCK2. Front. Oncol. 2020, 10, 998. [Google Scholar] [CrossRef]
- Yang, B.; Jia, L.; Ren, H.; Jin, C.; Ren, Q.; Zhang, H.; Hu, D.; Zhang, H.; Hu, L.; Xie, T. LncRNA DLX6-AS1 increases the expression of HIF-1α and promotes the malignant phenotypes of nasopharyngeal carcinoma cells via targeting miR-199a-5p. Mol. Genet. Genom. Med. 2020, 8, e1017. [Google Scholar] [CrossRef]
- Li, H.; Huang, F.; Liu, X.Q.; Liu, H.C.; Dai, M.; Zeng, J. LncRNA TUG1 promotes Ewing’s sarcoma cell proliferation, migration, and invasion via the miR-199a-3p-MSI2 signaling pathway. Neoplasma 2021, 68, 590–601. [Google Scholar] [CrossRef]
- Li, C.; Zheng, X.; Liu, P.; Li, M. Clinical value of lncRNA TUG1 in temporal lobe epilepsy and its role in the proliferation of hippocampus neuron via sponging miR-199a-3p. Bioengineered 2021, 12, 10666–10673. [Google Scholar] [CrossRef]
- Xin, Y.; Zhang, W.; Mao, C.; Li, J.; Liu, X.; Zhao, J.; Xue, J.; Li, J.; Ren, Y. LncRNA LINC01140 Inhibits Glioma Cell Migration and Invasion via Modulation of miR-199a-3p/ZHX1 Axis. Onco Targets Ther. 2020, 13, 1833–1844. [Google Scholar] [CrossRef] [Green Version]
- Zhong, W.; Li, Y.-C.; Huang, Q.-Y.; Tang, X.-Q. lncRNA ANRIL Ameliorates Oxygen and Glucose Deprivation (OGD) Induced Injury in Neuron Cells via miR-199a-5p/CAV-1 Axis. Neurochem. Res. 2020, 45, 772–782. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, M.-M.; Liu, M.; Tan, Z.-G.; Qin, Q.-L.; Jiang, Y.G. LncRNA XIST sponges miR-199a-3p to modulate the Sp1/LRRK2 signal pathway to accelerate Parkinson’s disease progression. Aging 2021, 13, 4115–4137. [Google Scholar] [CrossRef] [PubMed]
- Hua, Q.; Jin, M.; Mi, B.; Xu, F.; Li, T.; Zhao, L.; Liu, J.; Huang, G. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J. Hematol. Oncol. 2019, 12, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Tang, G.; Zhou, S.; Niu, Y. LncRNA-miRNA interaction prediction through sequence-derived linear neighborhood propagation method with information combination. BMC Genom. 2019, 20 (Suppl. 11), 946. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, R.; Gong, Q.; Shi, D.; Song, L.; Song, Y. Circular RNA circVMA21 ameliorates lipopolysaccharide (LPS)-induced acute kidney injury by targeting the miR-199a-5p/NRP1 axis in sepsis. Biochem. Biophys. Res. Commun. 2021, 548, 174–181. [Google Scholar] [CrossRef]
- Han, W.; Tao, X.; Weng, T.; Chen, L. Circular RNA PVT1 inhibits tendon stem/progenitor cell senescence by sponging microRNA-199a-5p. Toxicol. Vitr. 2022, 79, 105297. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Zhuang, Y.; Tang, M.; Qian, Q.; Chen, J.-P. CircRNA UBAP2 facilitates the progression of colorectal cancer by regulating miR-199a/VEGFA pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7963–7971. [Google Scholar]
- Gan, X.; Zhu, H.; Jiang, X.; Obiegbusi, S.C.; Yong, M.; Long, X.; Hu, J. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer 2020, 19, 45. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Han, X.; Zhang, C.; Sun, C.; Huang, S.; Xiao, W.; Gao, Y.; Liang, Q.; Luo, F.; Lu, W.; et al. Hsa_circ_0060450 Negatively Regulates Type I Interferon-Induced Inflammation by Serving as miR-199a-5p Sponge in Type 1 Diabetes Mellitus. Front. Immunol. 2020, 11, 576903. [Google Scholar] [CrossRef]
- Song, R.; Li, Y.; Hao, W.; Yang, L.; Chen, B.; Zhao, Y.; Sun, B.; Xu, F. Circular RNA MTO1 inhibits gastric cancer progression by elevating PAWR via sponging miR-199a-3p. Cell Cycle 2020, 19, 3127–3139. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Zheng, R.; Wu, P.; Sun, Z.; Chen, J.; Zhang, L.; Zhang, C.; Qian, H.; Jiang, J.; et al. Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle 2021, 20, 522–536. [Google Scholar] [CrossRef]
- Meng, Y.; Hao, D.; Huang, Y.; Jia, S.; Zhang, J.; He, X.; Liu, D.; Sun, L. Circular RNA circNRIP1 plays oncogenic roles in the progression of osteosarcoma. Mamm. Genome 2021, 32, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.; Raghavan, S.; DasGupta, R.; Palakodeti, D. tRNA-derived fragments (tRFs): Establishing their turf in post-transcriptional gene regulation. Cell Mol. Life Sci. 2021, 78, 2607–2619. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, T.; Suresh, P.S.; Tsutsumi, R. tRFs: miRNAs in disguise. Gene 2016, 579, 133–138. [Google Scholar] [CrossRef]
- Hussain, S.A.; Deepak, K.V.; Nanjappa, D.P.; Sherigar, V.; Nandan, N.; Suresh, P.S.; Venkatesh, T. Comparative expression analysis of tRF-3001a and tRF-1003 with corresponding miRNAs (miR-1260a and miR-4521) and their network analysis with breast cancer biomarkers. Mol. Biol. Rep. 2021, 48, 7313–7324. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, H.; Chen, Z.; Wang, Y.; Gao, B. Effects of tRNA-derived fragments and microRNAs regulatory network on pancreatic acinar intracellular trypsinogen activation. Bioengineered 2022, 13, 3207–3220. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Model. Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef]
- Wang, Q.; Ye, B.; Wang, P.; Yao, F.; Zhang, C.; Yu, G. Overview of microRNA-199a Regulation in Cancer. Cancer Manag. Res. 2019, 11, 10327–10335. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Kramer, T.; Annema, J.T. Advanced bronchoscopic techniques for the diagnosis and treatment of peripheral lung cancer. Lung Cancer 2021, 161, 152–162. [Google Scholar] [CrossRef]
- Zeng, H.; Tian, P.; Li, W. Progress of Liquid Biopsy in the Diagnosis and Treatment of Lung Cancer with Malignant Pleural Effusion. Zhongguo Fei Ai Za Zhi 2021, 24, 653–659. [Google Scholar]
- Snowsill, T.; Yang, H.; Griffin, E.; Long, L.; Varley-Campbell, J.; Coelho, H.; Robinson, S.; Hyde, C. Low-dose computed tomography for lung cancer screening in high-risk populations: A systematic review and economic evaluation. Health Technol. Assess. 2018, 22, 1–276. [Google Scholar] [CrossRef] [PubMed]
- Vinod, S.K.; Hau, E. Radiotherapy treatment for lung cancer: Current status and future directions. Respirology 2020, 25 (Suppl. 2), 61–71. [Google Scholar] [CrossRef] [PubMed]
- El-Hussein, A.; Manoto, S.L.; Ombinda-Lemboumba, S.; Alrowaili, Z.A.; Mthunzi-Kufa, P. A Review of Chemotherapy and Photodynamic Therapy for Lung Cancer Treatment. Anticancer Agents Med. Chem. 2021, 21, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Imyanitov, E.N.; Iyevleva, A.G.; Levchenko, E.V. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit. Rev. Oncol. Hematol. 2021, 157, 103194. [Google Scholar] [CrossRef] [PubMed]
- Koshikawa, K.; Terada, J.; Abe, M.; Iwasawa, S.; Sakayori, M.; Yoshioka, K.; Hirasawa, Y.; Kasai, H.; Kawasaki, Y.; Tsushima, K.; et al. Clinical characteristics and risk factors of drug-induced lung injury by ALK tyrosine kinase inhibitors: A single center retrospective analysis. Thorac. Cancer 2020, 11, 1495–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moro-Sibilot, D.; Cozic, N.; Pérol, M.; Mazières, J.; Otto, J.; Souquet, P.; Bahleda, R.; Wislez, M.; Zalcman, G.; Guibert, S.; et al. Crizotinib in c-MET- or ROS1-positive NSCLC: Results of the AcSé phase II trial. Ann. Oncol. 2019, 30, 1985–1991. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Chu, J.; Miao, S.; Wang, K.; Zhang, Q.; Wang, Y.; Xiao, Y.; Wu, L.; Liu, Y.; et al. Novel anilino quinazoline-based EGFR tyrosine kinase inhibitors for treatment of non-small cell lung cancer. Biomater. Sci. 2021, 9, 443–455. [Google Scholar] [CrossRef]
- Yoneda, K.; Imanishi, N.; Ichiki, Y.; Tanaka, F. Treatment of Non-small Cell Lung Cancer with EGFR-mutations. J. UOEH 2019, 41, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Le, P.; Romano, G.; Nana-Sinkam, P.; Acunzo, M. Non-Coding RNAs in Cancer Diagnosis and Therapy: Focus on Lung Cancer. Cancers 2021, 13, 1372. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Patel, S.K.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446–451. [Google Scholar] [CrossRef]
- Kwok, Z.; Ni, K.; Jin, Y. Extracellular Vesicle Associated Non-Coding RNAs in Lung Infections and Injury. Cells 2021, 10, 965. [Google Scholar] [CrossRef]
- O’Farrell, H.E.; Bowman, R.V.; Fong, K.M.; Yang, I.A. Plasma Extracellular Vesicle miRNAs Can Identify Lung Cancer, Current Smoking Status, and Stable COPD. Int. J. Mol. Sci. 2021, 22, 5803. [Google Scholar] [CrossRef]
- Messner, C.J.; Schmidt, S.; Özkul, D.; Gaiser, C.; Terracciano, L.; Krähenbühl, S.; Suter-Dick, L. Identification of miR-199a-5p, miR-214-3p and miR-99b-5p as Fibrosis-Specific Extracellular Biomarkers and Promoters of HSC Activation. Int. J. Mol. Sci. 2021, 22, 9799. [Google Scholar] [CrossRef]
- Ganju, A.; Khan, S.; Hafeez, B.B.; Behrman, S.W.; Yallapu, M.M.; Chauhan, S.C.; Jaggi, M. miRNA nanotherapeutics for cancer. Drug Discov. Today 2017, 22, 424–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [Green Version]
- Jing, Z.; Han, W.; Sui, X.; Xie, J.; Pan, H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015, 356, 332–338. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Q.; Zhong, Q.; Zhang, J.; Yang, M.; Li, C.; Zheng, P.; Bi, L.-J.; Ge, F. Identification of novel miR-21 target proteins in multiple myeloma cells by quantitative proteomics. J. Proteome Res. 2012, 11, 2078–2090. [Google Scholar] [CrossRef]
- Goodwin, D.; Rathi, V.; Conron, M.; Wright, G.M. Genomic and Clinical Significance of Multiple Primary Lung Cancers as Determined by Next-Generation Sequencing. J. Thorac. Oncol. 2021, 16, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Ruiz, S.; Urióstegui-Arcos, M.; Zurita, M. The transcriptional stress response and its implications in cancer treatment. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188620. [Google Scholar] [CrossRef] [PubMed]
| Biological Functions | Target | Ref | |
|---|---|---|---|
| miR-199a-5p | Playing a tumor suppressor role in NSCLC | MAP3K11 | [23] |
| miR-199a-5p | Inhibiting the proliferation, migration, and invasion of NSCLC cells and increasing cell apoptosis | HIF-1α | [33] |
| miR-199a-5p | Inhibiting proliferation and invasion of NSCLC cells | AKAP1 | [24] |
| miR-199a-5p | Inhibiting the generation of tumor blood vessels | HIF-1α | [38] |
| miR-199a-3p | Inhibiting the growth of NSCLC cells in vivo and promoting mitochondria-mediated cell apoptosis | ZEB1 | [34] |
| miR-199a-3p | Suppressing the viability and proliferation of NSCLC cells | CHML | [35] |
| miR-199a-3p | Inhibiting the growth of LUAD cells in vitro and in vivo and increasing cell apoptosis | AGR2 | [36] |
| miR-199a-5p/3p | Suppressing cell proliferation, migration ability and promote apoptosis | Rheb | [37] |
| Signaling Pathways Involved | Biological Functions | Ref | |
|---|---|---|---|
| miR-199a-5p | STAT signaling pathway | Inhibiting the progression of NSCLC | [33] |
| miR-199a-5p | MAPK signaling pathway | Suppressing the proliferation of lung cancer cells | [23] |
| miR-199a-5p | UPR signaling pathway | Having an effect on ER stress, as well as a causative role in lung tumorigenesis | [22] |
| miR-199a-5p | PI3K/Akt/mTOR signaling pathway | Inhibiting the expression of autophagy-related proteins | [43] |
| miR-199a-5p/3p | mTOR signaling pathway | Acting as cancer suppressor genes | [37] |
| Biological Functions | Diseases/Cell Lines | Ref | |
|---|---|---|---|
| miR-199a-5p | Increasing the formation of P62 | In SCLC lines (H446 and H69PR) | [43] |
| miR-199a-5p | Decreasing the expression of P62 | In multidrug-resistant SLCL cell lines (H446/EP) | [43] |
| miR-199a-5p | Inhibited autophagy and desensitized cells to a variety of chemotherapy drugs | In paclitaxel-resistant lung cancer cell lines (A549 and H1299) | [79] |
| miR-199a-5p | Increasing the sensitivity of lung cancer cells to doxorubicin | In doxorubicin-resistant lung cancer cell lines | [80] |
| miR-199a-5p | Increasing the sensitivity of lung cancer cells to bevacizumab | In NSCLC | [33] |
| miR-199a-5p/3p | Enhancing the sensitivity of gefitinib to EGFR-T790M | In NSCLC | [37] |
| LncRNAs | Mechanism | Biological Effects | Diseases | Ref | |
|---|---|---|---|---|---|
| miR-199a-5p | LncRNA TPPO-AS1 (TMPO antisense RNA 1) | LncRNA Tppo-AS1 has a binding site for miR-199a-5p | Negatively regulating the expression of miR-199a-5p | Retinoblastoma | [83] |
| miR-199a-5p | LncRNA LUCAT1 (lung cancer-associated transcription 1) | LncRNA LUCAT1 contains a highly conserved miR-199a-5p binding site in 3′-UTR | Driving the malignant development of OC | Ovarian cancer | [84] |
| miR-199a-5p | LncRNA 01133 | LncRNA 01133 sponges miR-199a-5p | Inducing EMT | Hepatocellular carcinoma | [85] |
| miR-199a-5p | LncRNA DLX6-AS1 (long-chain noncoding growth stasis specific protein 6 antisense RNA1) | LncRNA DLX6-AS1 could target miR-199a-5p | DLX6-AS1 promoted the malignant phenotype of cells a via targeting miR-199a-5p | Nasopharyngeal carcinoma | [87] |
| miR-199a-5p | LncRNA ANRIL | LncRNA ANRIL sponges miR-199a-5p | Activating the MEK/ERK pathway | Ischemic injury | [91] |
| miR-199a-5p | LncRNA 01123 | LncRNA 01123 sponges miR-199a-5p | Promoting NSCLC cell proliferation and aerobic glycolysis | NSCLC | [93] |
| miR-199a-3p | LncRNA NEAT1 (nuclear paraspeckle assembly transcript1) | LncRNA NEAT1 is a ceRNA of miR-199a-3p | Inhibiting the growth of HCC cells and increasing cell apoptosis | Hepatocellular carcinoma | [86] |
| miR-199a-3p | LncRNA TUG1 (taurine up-regulated gene 1) | LncRNA TUG1 sponges miR-199a-3p | Promoting the malignant phenotype of cells | Ewing’s sarcoma | [88] |
| miR-199a-3p | LncRNA TUG1 | miR-199a-3p was a target of LncRNA TUG1 | Enhancing cell activity and inhibiting cell apoptosis | Temporal lobe epilepsy | [89] |
| miR-199a-3p | LINC01140 | LINC01140 could target miR-199a-3p | Increasing the expression of ZHX1 | Gliomas | [90] |
| miR-199a-3p | LncRNA XIST (X-inactive specific transcript) | lncRNA XIST sponges miR-199a-3p | Accelerating the progression of Parkinson’s disease | Parkinson | [92] |
| CircRNAs | Mechanism | Biological Effects | Diseases/Cell Lines | Ref | |
|---|---|---|---|---|---|
| miR-199a-5p | CircVMA21 (circular RNAs vacuolar ATPase assembly factor) | CircVMA21 sponges miR-199a-5p | Inhibiting cell viability, apoptosis, and inflammation | LPS-mediated THP-1 | [95] |
| miR-199a-5p | CircPVT1 | CircPVT1 sponges miR-199a-5p | Suppressing the aging process of TSPC | TSPC (tendon stem/progenitor cell) | [96] |
| miR-199a-5p | CircMUC16 | CircMUC16 sponges miR-199a-5p | Promoting autophagy, invasion, and metastasis of cells | EOC (epithelial ovarian cancer) | [98] |
| miR-199a-5p | Circ0060450 | Circ0060450 could act as a sponge for miR-199a-5p | Inhibiting macrophage-mediated inflammation | Type 1 Diabetes Mellitus | [99] |
| miR-199a-5p | CircITCH | CircITCH could sponge miR-199a-5p | Inhibiting the metastasis of gastric cancer | Gastric cancer | [101] |
| miR-199a-3p | CircMTO1 | CircMTO1 could sponge miR-199a-3p | inhibiting the occurrence and development of gastric cancer | Gastric cancer | [100] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, W.; Li, Y.; Chai, B.; Liu, X.; Ma, Z. miR-199a: A Tumor Suppressor with Noncoding RNA Network and Therapeutic Candidate in Lung Cancer. Int. J. Mol. Sci. 2022, 23, 8518. https://doi.org/10.3390/ijms23158518
Meng W, Li Y, Chai B, Liu X, Ma Z. miR-199a: A Tumor Suppressor with Noncoding RNA Network and Therapeutic Candidate in Lung Cancer. International Journal of Molecular Sciences. 2022; 23(15):8518. https://doi.org/10.3390/ijms23158518
Chicago/Turabian StyleMeng, Wei, Yanli Li, Binshu Chai, Xiaomin Liu, and Zhongliang Ma. 2022. "miR-199a: A Tumor Suppressor with Noncoding RNA Network and Therapeutic Candidate in Lung Cancer" International Journal of Molecular Sciences 23, no. 15: 8518. https://doi.org/10.3390/ijms23158518
APA StyleMeng, W., Li, Y., Chai, B., Liu, X., & Ma, Z. (2022). miR-199a: A Tumor Suppressor with Noncoding RNA Network and Therapeutic Candidate in Lung Cancer. International Journal of Molecular Sciences, 23(15), 8518. https://doi.org/10.3390/ijms23158518






