Transcriptomic Response of the Diazotrophic Bacteria Gluconacetobacter diazotrophicus Strain PAL5 to Iron Limitation and Characterization of the fur Regulatory Network
Abstract
:1. Introduction
2. Results
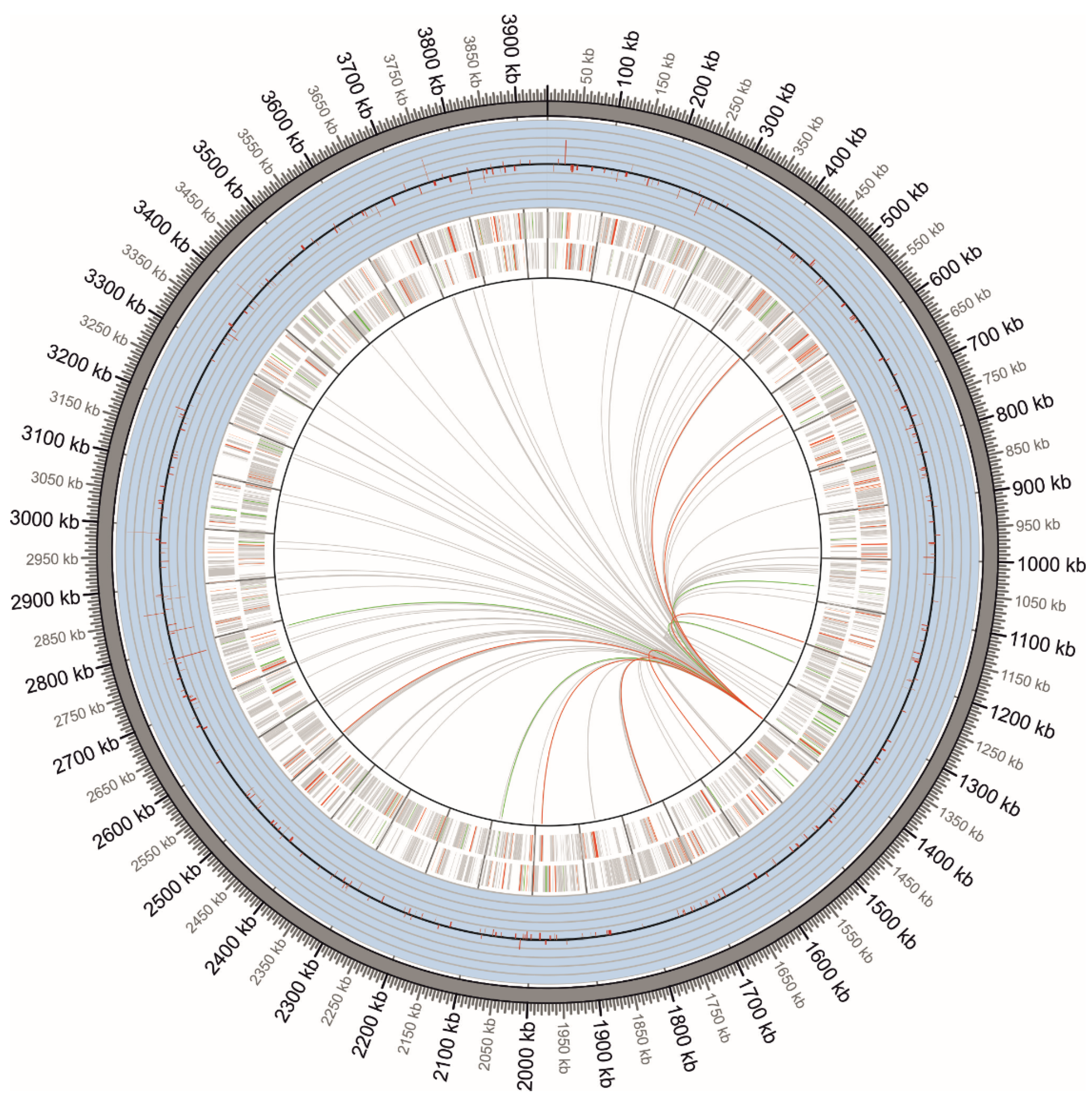
2.1. Differentially Expressed Gene Identification and Iron-Activated and Repressed Genes by Functional Category
2.2. Main Metabolic Changes in Bacterial Adaptation to Iron-Limiting Conditions
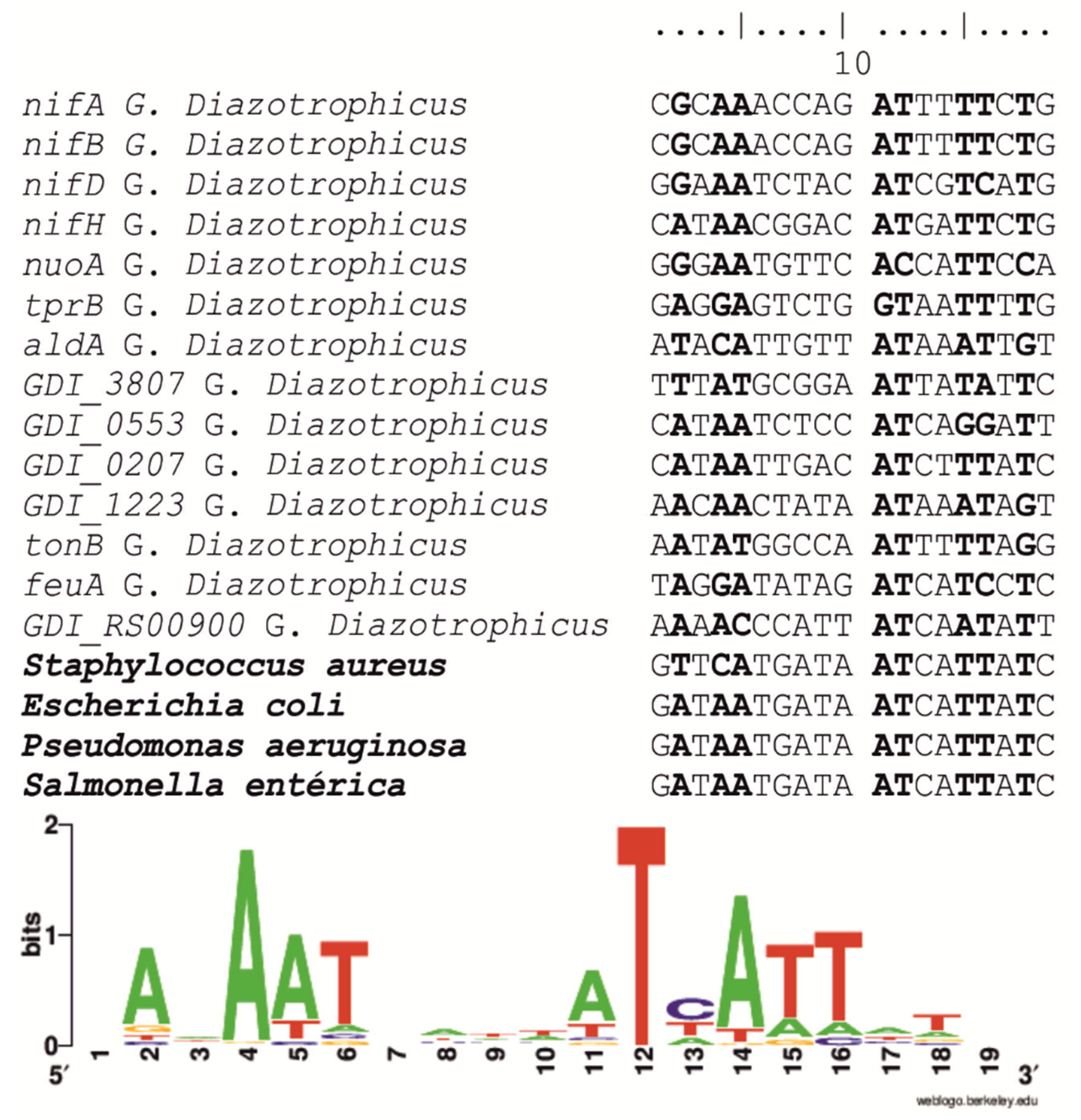
2.3. Identification of fur Box Sequences in the G. diazotrophicus Genome
2.4. Production of Siderophores in Response to Iron Limitations
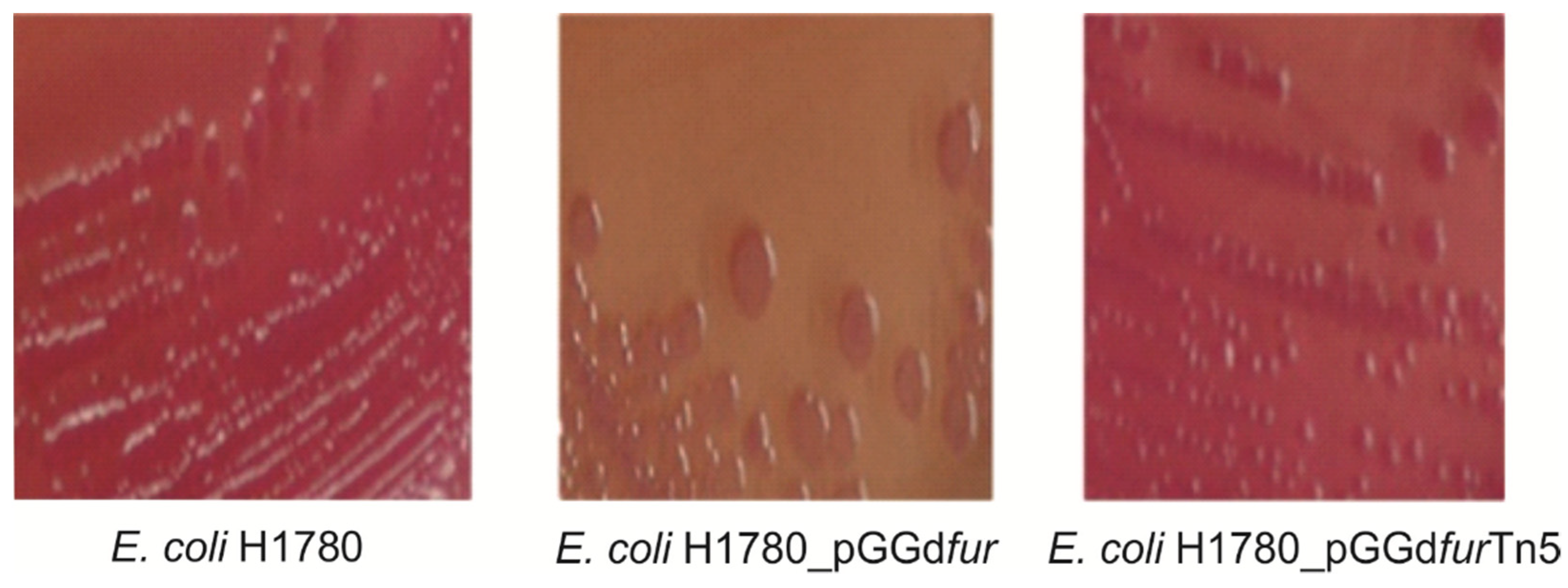
2.5. fur Insertional Mutants and Complementation of an E. coli fur Mutant by G. diazotrophicus fur Homologs
3. Discussion
3.1. Iron Availability Promotes Changes in the G. diazotrophicus Transcriptome
3.2. Metabolic Pathways and Iron Uptake
3.3. fur Regulatory Network
3.4. Molecular Characterization of fur from G. diazotrophicus
4. Materials and Methods
4.1. Bacterial Growth
4.2. RNA-Seq Profiling Experiment
4.3. Mapping, Clustering and Quality Control
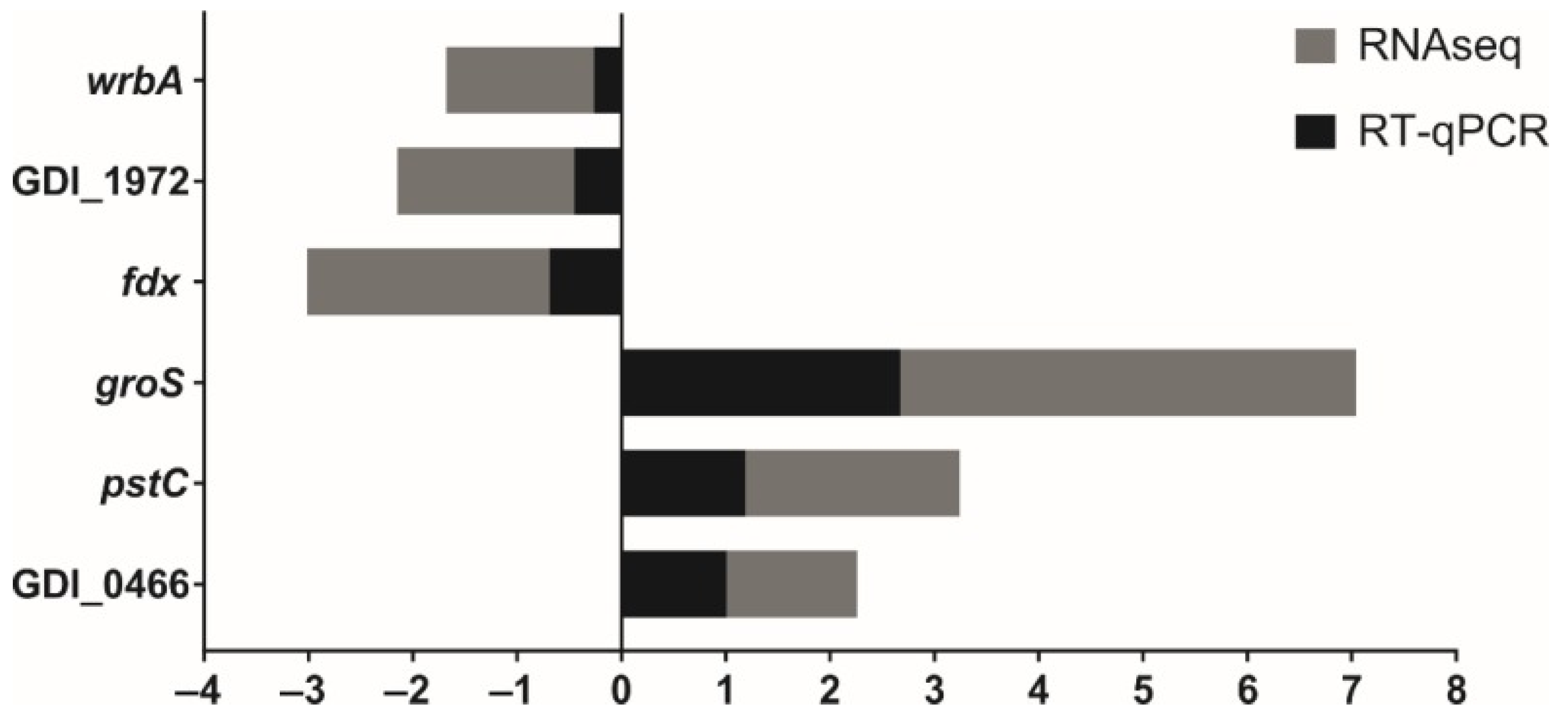
4.4. Real-Time Quantitative (RT-qPCR) Validation
4.5. In Silico Searching for fur Binding Sites (fur Boxes)
4.6. Generation of fur Insertional Mutants of G. diazotrophicus
4.7. Evaluation of the Production of Siderophores
4.8. Fur Titration Assays (FURTA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Posey, J.E.; Gherardini, F.C. Lack of a Role for Iron in the Lyme Disease Pathogen. Science 2000, 288, 1651–1653. [Google Scholar] [CrossRef]
- Lemanceau, P.; Expert, D.; Gaymard, F.; Bakker, P.A.H.M.; Briat, J.-F. Chapter 12 Role of iron in plant–microbe interactions. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2009; Volume 51, pp. 491–549. [Google Scholar]
- Stojiljkovic, I.; Cobeljic, M.; Hantke, K. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 1993, 108, 111–115. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Molina, M.A.; Godoy, P.; Ramos-González, M.I.; Muñoz, N.; Ramos, J.L.; Espinosa-Urgel, M. Role of iron and the TonB system in colonization of corn seeds and roots by Pseudomonas putida KT2440. Environ. Microbiol. 2005, 7, 443–449. [Google Scholar] [CrossRef]
- Khalid, S.; Asghar, H.N.; Akhtar, M.J.; Aslam, A.; Zahir, Z.A. Biofortification of iron in chickpea by plant growth promoting rhizobacteria. Pak. J. Bot. 2015, 47, 1191–1194. [Google Scholar]
- Cavalcante, V.A.; Dobereiner, J. A New acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil 1988, 108, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Salgado, T.; Fuentes-Ramirez, L.E.; Tapia-Hernandez, A.; Mascarua-Esparza, M.A.; Martinez-Romero, E.; Caballero-Mellado, J. Coffea arabica L., a new host plant for Acetobacter diazotrophicus, and isolation of other nitrogen-fixing Acetobacteria. Appl. Environ. Microbiol. 1997, 63, 3676–3683. [Google Scholar] [CrossRef] [Green Version]
- Tapia-Hernández, A.; Bustillos-Cristales, M.R.; Jiménez-Salgado, T.; Caballero-Mellado, J.; Fuentes-Ramírez, L.E. Natural endophytic occurrence of Acetobacter diazotrophicus in pineapple plants. Microb. Ecol. 2000, 39, 49–55. [Google Scholar] [CrossRef]
- Muthukumarasamy, R.; Kang, U.G.; Park, K.D.; Jeon, W.T.; Park, C.Y.; Cho, Y.S.; Kwon, S.W.; Song, J.; Roh, D.H.; Revathi, G. Enumeration, isolation and identification of diazotrophs from korean wetland rice varieties grown with long-term application of N and compost and their short-term inoculation effect on rice plants. J. Appl. Microbiol. 2007, 102, 981–991. [Google Scholar] [CrossRef]
- Saravanan, V.S.; Madhaiyan, M.; Osborne, J.; Thangaraju, M.; Sa, T.M. Ecological occurrence of Gluconacetobacter diazotrophicus and nitrogen-fixing Acetobacteraceae members: Their possible role in plant growth promotion. Microb. Ecol. 2008, 55, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Rouws, L.F.M.; Meneses, C.H.S.G.; Guedes, H.V.; Vidal, M.S.; Baldani, J.I.; Schwab, S. Monitoring the colonization of sugarcane and rice plants by the endophytic diazotrophic bacterium Gluconacetobacter diazotrophicus marked with Gfp and GusA reporter genes. Lett. Appl. Microbiol. 2010, 51, 325–330. [Google Scholar] [CrossRef] [PubMed]
- de Souza, A.L.S.R.; De Souza, S.A.; De Oliveira, M.V.V.; Ferraz, T.M.; Figueiredo, F.A.M.M.A.; Da Silva, N.D.; Rangel, P.L.; Panisset, C.R.S.; Olivares, F.L.; Campostrini, E.; et al. Endophytic colonization of Arabidopsis thaliana by Gluconacetobacter diazotrophicus and its effect on plant growth promotion, plant physiology, and activation of plant defense. Plant Soil 2015, 399, 257–270. [Google Scholar] [CrossRef]
- Yoon, V.; Tian, G.; Vessey, J.K.; Macfie, S.M.; Dangi, O.P.; Kumer, A.K.; Tian, L. Colonization efficiency of different sorghum genotypes by Gluconacetobacter diazotrophicus. Plant Soil 2016, 398, 243–256. [Google Scholar] [CrossRef]
- Bertalan, M.; Albano, R.; de Pádua, V.; Rouws, L.; Rojas, C.; Hemerly, A.; Teixeira, K.; Schwab, S.; Araujo, J.; Oliveira, A.; et al. Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genom. 2009, 10, 450. [Google Scholar] [CrossRef] [Green Version]
- Velayudhan, J.; Hughes, N.J.; McColm, A.A.; Bagshaw, J.; Clayton, C.L.; Andrews, S.C.; Kelly, D.J. Iron acquisition and virulence in Helicobacter pylori: A major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 2000, 37, 274–286. [Google Scholar] [CrossRef]
- Urzúa, L.S.; Vázquez-Candanedo, A.P.; Sánchez-Espíndola, A.; Ramírez, C.Á.; Baca, B.E. Identification and characterization of an iron ABC transporter operon in Gluconacetobacter diazotrophicus Pal 5. Arch. Microbiol. 2013, 195, 431–438. [Google Scholar] [CrossRef] [PubMed]
- de Paula Soares, C.; Rodrigues, E.P.; de Paula Ferreira, J.; Simões Araújo, J.L.; Rouws, L.F.M.; Baldani, J.I.; Vidal, M.S. Tn5 insertion in the TonB gene promoter affects iron-related phenotypes and increases extracellular siderophore levels in Gluconacetobacter diazotrophicus. Arch. Microbiol. 2015, 197, 223–233. [Google Scholar] [CrossRef]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Pandey, S.S.; Patnana, P.K.; Lomada, S.K.; Tomar, A.; Chatterjee, S. Co-regulation of iron metabolism and virulence associated fuctions by iron and XibR, a novel iron binding transcription factor, in the plant pathogen Xanthomonas. PLoS Pathog. 2016, 30, 12. [Google Scholar]
- Cheng, Y.; Yang, R.; Lyu, M.; Wang, S.; Liu, X.; Wen, Y.; Song, Y.; Li, J.; Chen, Z. IdeR, a DtxR family iron response regulator, controls iron homeostasis, morphological differentiation, secondary metabolism, and the oxidative stress response in Streptomyces avermitilis. Appl. Environ. Microbiol. 2018, 84, e01503-18. [Google Scholar] [CrossRef] [Green Version]
- Rodionov, D.A.; Gelfand, M.S.; Todd, J.D.; Curson, A.R.J.; Johnston, A.W.B. Computational reconstruction of iron- and manganese-responsive transcriptional networks in α-Proteobacteria. PLoS Comput. Biol. 2006, 2, 1568–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trovero, M.F.; Scavone, P.; Platero, R.; de Souza, E.M.; Fabiano, E.; Rosconi, F. Herbaspirillum seropedicae differentially expressed genes in response to iron availability. Front. Microbiol. 2018, 9, 1430. [Google Scholar] [CrossRef] [PubMed]
- Koebnik, R. TonB-dependent trans-envelope signalling: The exception or the rule? Trends Microbiol. 2005, 13, 343–347. [Google Scholar] [CrossRef]
- Bouveret, E.; Journet, L.; Walburger, A.; Cascales, E.; Benedetti, H.; Lloubès, R. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 2002, 84, 413–421. [Google Scholar] [CrossRef]
- Llamas, M.A.; Rodríguez-Herva, J.J.; Hancock, R.E.W.; Bitter, W.; Tommassen, J.; Ramos, J.L. Role of Pseudomonas putida Tol-OprL gene products in uptake of solutes through the cytoplasmic membrane. J. Bacteriol. Res. 2003, 185, 4707–4716. [Google Scholar] [CrossRef] [Green Version]
- Piddock, L.J.V. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.; Worgall, S.; Quadri, L.E.N. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch. Microbiol. 2003, 180, 374–379. [Google Scholar] [CrossRef]
- Hentzer, M.; Eberl, L.; Givskov, M. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: Anaerobic respiration and iron limitation. Biofilms 2005, 2, 37–61. [Google Scholar] [CrossRef]
- Quail, M.A.; Jordan, P.; Grogan, J.M.; Butt, J.N.; Lutz, M.; Thomson, A.J.; Andrews, S.C.; Guest, J.R. Spectroscopic and voltametric characterisation of the bacterioferritin-associated ferredoxin of Escherichia coli. Biochem. Biophys. Res. Commun. 1996, 229, 635–642. [Google Scholar] [CrossRef]
- Weeratunga, S.K.; Gee, C.E.; Lovell, S.; Zeng, Y.; Woodin, C.L.; Rivera, M. Binding of Pseudomonas aeruginosa apobacterioferritin-associated ferredoxin to bacterioferritin B promotes heme mediation of electron delivery and mobilization of core mineral iron. Biochemistry 2009, 48, 7420–7431. [Google Scholar] [CrossRef] [Green Version]
- Imlay, J.A. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Harris, D.P.; Luo, F.; Xiong, W.; Joachimiak, M.; Wu, L.; Dehal, P.; Jacobsen, J.; Yang, Z.; Palumbo, A.V. Snapshot of iron response in Shewanella oneidensis by gene network reconstruction. BMC Genom. 2009, 10, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva Neto, J.F.; Lourenço, R.F.; Marques, M.V. Global transcriptional response of Caulobacter crescentus to iron availability. BMC Genom. 2013, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Machuca, A.; Martinez, V. Transcriptome analysis of the intracellular facultative pathogen Piscirickettsia salmonis: Expression of putative groups of genes associated with virulence and iron metabolism. PLoS ONE 2016, 11, e0168855. [Google Scholar] [CrossRef] [Green Version]
- McHugh, J.P.; Rodríguez-Quiñones, F.; Abdul-Tehrani, H.; Svistunenko, D.A.; Poole, R.K.; Cooper, C.E.; Andrews, S.C. Global iron-dependent gene regulation in Escherichia coli: A new mechanism for iron homeostasis. J. Biol. Chem. 2003, 278, 29478–29486. [Google Scholar] [CrossRef] [Green Version]
- Holmes, K.; Mulholland, F.; Pearson, B.M.; Pin, C.; McNicholl-Kennedy, J.; Ketley, J.M.; Wells, J.M. Campylobacter jejuni gene expression in response to iron limitation and the role of fur. Microbiology 2005, 151, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Mesleh, M.F.; Rajaratnam, P.; Conrad, M.; Chandrasekaran, V.; Liu, C.M.; Pandya, B.A.; Hwang, Y.S.; Rye, P.T.; Muldoon, C.; Becker, B.; et al. Targeting bacterial cell wall peptidoglycan synthesis by inhibition of glycosyltransferase activity. Chem. Biol. Drug Des. 2016, 87, 190–199. [Google Scholar] [CrossRef]
- Liu, M.F.; Huang, M.; Zhu, D.K.; Wang, M.S.; Jia, R.Y.; Chen, S.; Sun, K.F.; Yang, Q.; Wu, Y.; Biville, F.; et al. Identifying the genes responsible for iron-limited condition in Riemerella anatipestifer CH-1 through RNA-Seq-based analysis. Biomed. Res. Int. 2017, 201, 1–10. [Google Scholar] [CrossRef]
- Berges, M.; Michel, A.M.; Lassek, C.; Nuss, A.M.; Beckstette, M.; Dersch, P.; Riedel, K.; Sievers, S.; Becher, D.; Otto, A.; et al. Iron regulation in Clostridioides difficile. Front. Microbiol. 2018, 9, 3183. [Google Scholar] [CrossRef] [Green Version]
- Nwugo, C.C.; Gaddy, J.A.; Zimbler, D.L.; Actis, L.A. Deciphering the iron response in Acinetobacter baumannii: A proteomics approach. J. Proteom. 2011, 74, 44–58. [Google Scholar] [CrossRef] [Green Version]
- Butcher, J.; Stintzi, A. The transcriptional landscape of Campylobacter jejuni under iron replete and iron limited growth conditions. PLoS ONE 2013, 8, e79475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouws, L.F.M.; Simões-Araújo, J.L.; Hemerly, A.S.; Baldani, J.I. Validation of a Tn5 transposon mutagenesis system for Gluconacetobacter diazotrophicus through characterization of a flagellar mutant. Arch. Microbiol. 2008, 189, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.; Buhrmester, J.; Hansmeier, N.; Pu, A.; Weidner, S. Role of the regulatory gene. Society 2005, 71, 5969–5982. [Google Scholar]
- Eijkelkamp, B.A.; Hassan, K.A.; Paulsen, I.T.; Brown, M.H. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 2011, 12, 126. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.K.; Hassan, K.A.; Tetu, S.G.; Loper, J.E.; Paulsen, I.T. The Effect of iron limitation on the transcriptome and proteome of Pseudomonas fluorescens Pf-5. PLoS ONE 2012, 7, e39139. [Google Scholar] [CrossRef] [Green Version]
- Thode, S.K.; Bækkedal, C.; Söderberg, J.J.; Hjerde, E.; Hansen, H.; Haugen, P. Construction of a fur null mutant and RNA-sequencing provide deeper global understanding of the Aliivibrio salmonicida fur regulon. PeerJ 2017, 2017, 3461. [Google Scholar] [CrossRef] [Green Version]
- Drepper, T.; Raabe, K.; Giaourakis, D.; Gendrullis, M.; Masepohl, B.; Klipp, W. The Hfq-like protein NrfA of the phototrophic purple bacterium Rhodobacter capsulatus controls nitrogen fixation via regulation of NifA and AnfA expression. FEMS Microbiol. Lett. 2002, 215, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Klassen, G.; De Oliveira Pedrosa, F.; De Souza, E.M.; Yates, M.G.; Rigo, L.U. Nitrogenase activity of Herbaspirillum seropedicae grown under low iron levels requires the products of NifXorf1 genes. FEMS Microbiol. Lett. 2003, 224, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Rosconi, F.; Souza, E.M.; Pedrosa, F.O.; Platero, R.A.; González, C.; González, M.; Batista, S.; Gill, P.R.; Fabiano, E.R. Iron depletion affects nitrogenase activity and expression of NifH and NifA genes in Herbaspirillum seropedicae. FEMS Microbiol. Lett. 2006, 258, 214–219. [Google Scholar] [CrossRef]
- López-Gomollón, S.; Hernández, J.A.; Pellicer, S.; Angarica, V.E.; Peleato, M.L.; Fillat, M.F. Cross-talk between iron and nitrogen regulatory networks in Anabaena (Nostoc) Sp. PCC 7120: Identification of overlapping genes in furA and NtcA regulons. J. Mol. Biol. 2007, 374, 267–281. [Google Scholar] [CrossRef]
- Mayer, S.M.; Lawson, D.M.; Gormal, C.A.; Roe, S.M.; Smith, B.E. New insights into structure-function relationships in nitrogenase: A 1.6 Å resolution x-ray crystallographic study of Klebsiella pneumoniae MoFe-protein. J. Mol. Biol. 1999, 292, 871–891. [Google Scholar] [CrossRef] [PubMed]
- Lawley, T.D.; Klimke, W.A.; Gubbins, M.J.; Frost, L.S. F factor conjugation is a true type iv secretion system. FEMS Microbiol. Lett. 2003, 224, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zaini, P.A.; Fogaça, A.C.; Lupo, F.G.N.; Nakaya, H.I.; Vêncio, R.Z.N.; Da Silva, A.M. The iron stimulon of Xylella fastidiosa includes genes for type IV pilus and colicin V-like bacteriocins. J. Bacteriol. 2008, 190, 2368–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janga, S.C.; Pérez-Rueda, E. Plasticity of transcriptional machinery in bacteria is increased by the repertoire of regulatory families. Comput. Biol. Chem. 2009, 33, 261–268. [Google Scholar] [CrossRef]
- Yu, C.; McClure, R.; Nudela, K.; Daou, N.; Genco, C.A. Characterization of the Neisseria gonorrhoeae iron and fur regulatory network. J. Bacteriol. 2016, 198, 2180–2191. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Quatrini, R.; Lefimil, C.; Veloso, F.A.; Pedroso, I.; Holmes, D.S.; Jedlicki, E. Bioinformatic Prediction and experimental verification of fur-regulated genes in the extreme acidophile Acidithiobacillus ferrooxidans. Nucleic Acids Res. 2007, 35, 2153–2166. [Google Scholar] [CrossRef]
- Da Silva Neto, J.F.; Braz, V.S.; Italiani, V.C.S.; Marques, M.V. fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res. 2009, 37, 4812–4825. [Google Scholar] [CrossRef]
- Yua, C.; Genco, C.A. fur-mediated activation of gene transcription in the human pathogen Neisseria gonorrhoeae. J. Bacteriol. 2012, 194, 1730–1742. [Google Scholar] [CrossRef] [Green Version]
- Grifantini, R.; Sebastian, S.; Frigimelica, E.; Draghi, M.; Bartolini, E.; Muzzi, A.; Rappuoli, R.; Grandi, G.; Genco, C.A. Identification of iron-activated and -repressed fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 2003, 100, 9542–9547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagg, A.; Neilands, J.B. Mapping of a mutation affecting regulation of iron uptake systems in Escherichia coli K-12. J. Bacteriol. 1985, 161, 450–453. [Google Scholar] [CrossRef] [Green Version]
- Hantke, K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol. Gen. Genet. 1987, 210, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.S.; Litwin, C.M.; Carroll, P.A.; Calderwood, S.B. Vibrio cholerae fur mutations associated with loss of repressor activity: Implications for the structural-functional relationships of fur. J. Bacteriol. 1994, 176, 5108–5115. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, M.P.; Payne, S.M. Genetic Analysis of the enterobactin gene cluster in Shigella flexneri. J. Bacteriol. 1991, 173, 816–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delany, I.; Ieva, R.; Alaimo, C.; Rappuoli, R.; Scarlato, V. The iron-responsive regulator fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J. Bacteriol. 2003, 185, 6032–6041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolmasky, M.E.; Wertheimer, A.M.; Actis, L.A.; Crosa, J.H. Characterization of the Vibrio anguillarum fur gene: Role in regulation of expression of the FatA outer membrane protein and catechols. J. Bacteriol. 1994, 176, 213–220. [Google Scholar] [CrossRef] [Green Version]
- Venturi, V.; Weisbeek, P.; Koster, M. gene regulation of siderophore-mediated iron acquisition in Pseudomonas: Not only the fur repressor. Mol. Microbiol. 1995, 17, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Frederick Sparling, P. Isolation and analysis of a fur mutant of Neisseria gonorrhoeae. J. Bacteriol. 1996, 178, 4224–4232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vajrala, N.; Sayavedra-Soto, L.A.; Bottomley, P.J.; Arp, D.J. Role of a fur homolog in iron metabolism in Nitrosomonas europaea. BMC Microbiol. 2011, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Hamza, I.; Hassett, R.; O’Brian, M.R. identification of a functional fur gene in Bradyrhizobium japonicum. J. Bacteriol. 1999, 181, 5843–5846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosconi, F.; de Vries, S.P.; Baig, A.; Fabiano, E.; Grant, A.J. Essential genes for in vitro growth of the endophyte Herbaspirillum seropedicae SmR1 as revealed by transposon insertion site sequencing. Appl. Environ. Microbiol. 2016, 27, 6664–6671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [Green Version]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar]
- Galisa, P.S.; da Silva, H.A.P.; Macedo, A.V.M.; Reis, V.M.; Vidal, M.S.; Baldani, J.I.; Simes-Araújo, J.L. Identification and validation of reference genes to study the gene expression in Gluconacetobacter diazotrophicus grown in different carbon sources using RT-qPCR. J. Microbiol. Methods 2012, 91, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2008, 8, 1–14. [Google Scholar]
- Münch, R.; Hiller, K.; Barg, H.; Heldt, D.; Linz, S.; Wingender, E.; Jahn, D. PRODORIC: Prokaryotic database of gene regulation. Nucleic Acids Res. 2003, 31, 266–269. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.; Hon, G.; Chandonia, J.; Brenner, S. NCBI GenBank FTP site\nWebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Meneses, C.H.S.G.; Rouws, L.F.M.; Simões-Araújo, J.L.; Vidal, M.S.; Baldani, J.I. Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus. Mol. Plant-Microbe Interact. 2011, 24, 1448–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alquéres, S.; Meneses, C.; Rouws, L.; Rothballer, M.; Baldani, I.; Schmid, M.; Hartmann, A. The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus Pal5. Mol. Plant-Microbe Interact. 2013, 26, 937–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, C.d.P.; Trada-Sfeir, M.Z.; Terra, L.A.; Ferreira, J.d.P.; Dos-Santos, C.M.; Oliveira, I.G.B.d.; Araújo, J.L.S.; Meneses, C.H.S.G.; de Souza, E.M.; Baldani, J.I.; et al. Transcriptomic Response of the Diazotrophic Bacteria Gluconacetobacter diazotrophicus Strain PAL5 to Iron Limitation and Characterization of the fur Regulatory Network. Int. J. Mol. Sci. 2022, 23, 8533. https://doi.org/10.3390/ijms23158533
Soares CdP, Trada-Sfeir MZ, Terra LA, Ferreira JdP, Dos-Santos CM, Oliveira IGBd, Araújo JLS, Meneses CHSG, de Souza EM, Baldani JI, et al. Transcriptomic Response of the Diazotrophic Bacteria Gluconacetobacter diazotrophicus Strain PAL5 to Iron Limitation and Characterization of the fur Regulatory Network. International Journal of Molecular Sciences. 2022; 23(15):8533. https://doi.org/10.3390/ijms23158533
Chicago/Turabian StyleSoares, Cleiton de Paula, Michelle Zibetti Trada-Sfeir, Leonardo Araújo Terra, Jéssica de Paula Ferreira, Carlos Magno Dos-Santos, Izamara Gesiele Bezerra de Oliveira, Jean Luiz Simões Araújo, Carlos Henrique Salvino Gadelha Meneses, Emanuel Maltempi de Souza, José Ivo Baldani, and et al. 2022. "Transcriptomic Response of the Diazotrophic Bacteria Gluconacetobacter diazotrophicus Strain PAL5 to Iron Limitation and Characterization of the fur Regulatory Network" International Journal of Molecular Sciences 23, no. 15: 8533. https://doi.org/10.3390/ijms23158533
APA StyleSoares, C. d. P., Trada-Sfeir, M. Z., Terra, L. A., Ferreira, J. d. P., Dos-Santos, C. M., Oliveira, I. G. B. d., Araújo, J. L. S., Meneses, C. H. S. G., de Souza, E. M., Baldani, J. I., & Vidal, M. S. (2022). Transcriptomic Response of the Diazotrophic Bacteria Gluconacetobacter diazotrophicus Strain PAL5 to Iron Limitation and Characterization of the fur Regulatory Network. International Journal of Molecular Sciences, 23(15), 8533. https://doi.org/10.3390/ijms23158533






