Virus-like Particles: Fundamentals and Biomedical Applications
Abstract
:1. Introduction
2. Brief Description of Viruses
3. Key Concepts about Virus-like Particles (VLPs)
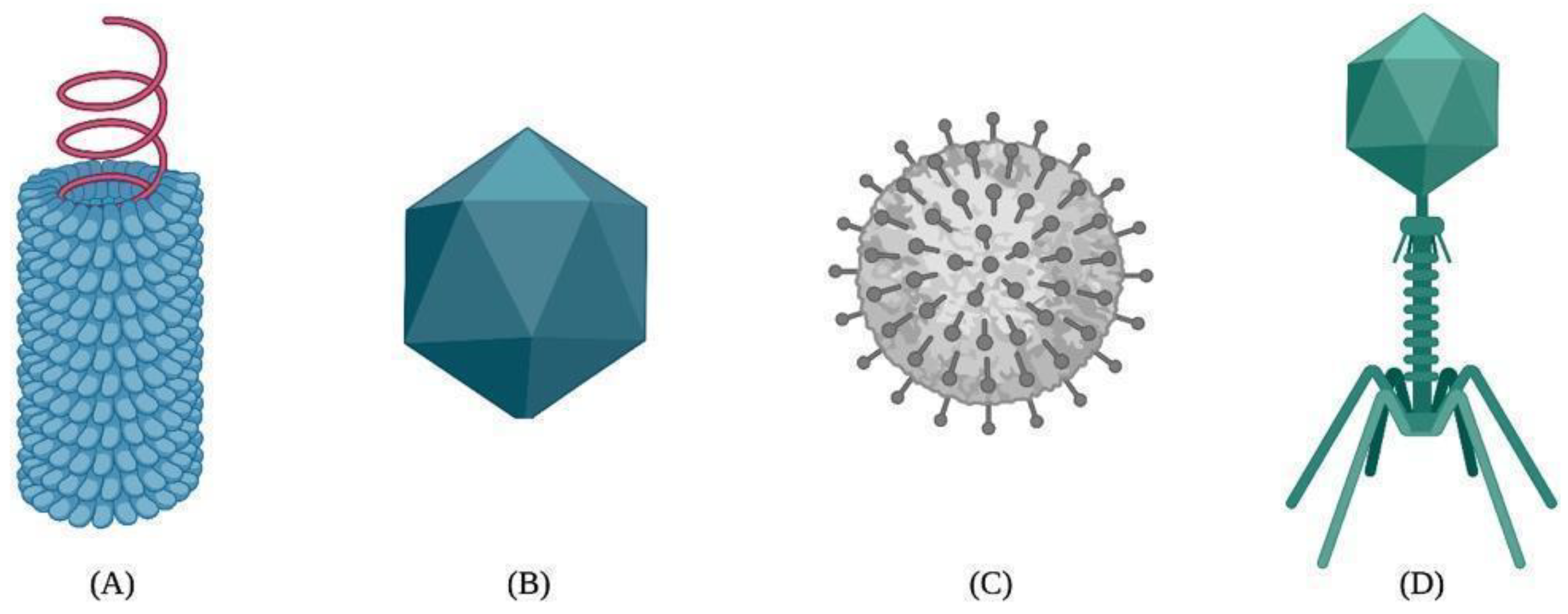
4. Structure Classification of VLPs
5. Expression of VLPs
6. Morphology and Manipulation of Viral Capsids
7. Functionalization of the VLPs
8. Characterization
9. Discussion
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murphy, C.J.; Vartanian, A.M.; Geiger, F.M.; Hamers, R.J.; Pedersen, J.; Cui, Q.; Haynes, C.L.; Carlson, E.E.; Hernandez, R.; Klaper, R.D.; et al. Biological Responses to Engineered Nanomaterials: Needs for the Next Decade. ACS Cent. Sci. 2015, 1, 117–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baig, N.; Kammakakam, I.; Falath, W. Nanomaterials: A Review of Synthesis Methods, Properties, Recent Progress, and Challenges. Mater. Adv. 2021, 2, 1821–1871. [Google Scholar] [CrossRef]
- DeFrates, K.G.; Moore, R.; Borgesi, J.; Lin, G.; Mulderig, T.; Beachley, V.; Hu, X. Protein-Based Fiber Materials in Medicine: A Review. Nanomaterials 2018, 8, 457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, D.; Nasrolahi Shirazi, A.; Parang, K. Self-Assembly of Peptides to Nanostructures. Org. Biomol. Chem. 2014, 12, 3544–3561. [Google Scholar] [CrossRef] [Green Version]
- Tariq, H.; Batool, S.; Asif, S.; Ali, M.; Abbasi, B.H. Virus-Like Particles: Revolutionary Platforms for Developing Vaccines Against Emerging Infectious Diseases. Front. Microbiol. 2022, 12, 790121. [Google Scholar] [CrossRef] [PubMed]
- Al-Halifa, S.; Gauthier, L.; Arpin, D.; Bourgault, S.; Archambault, D. Nanoparticle-Based Vaccines against Respiratory Viruses. Front. Immunol. 2019, 10, 22. [Google Scholar] [CrossRef] [Green Version]
- Jhaveri, J.; Raichura, Z.; Khan, T.; Momin, M.; Omri, A. Chitosan Nanoparticles-Insight into Properties, Functionalization and Applications in Drug Delivery and Theranostics. Molecules 2021, 26, 272. [Google Scholar] [CrossRef]
- Gama, P.; Cadena-Nava, R.D.; Juarez-Moreno, K.; Pérez-Robles, J.; Vazquez-Duhalt, R. Virus-Based Nanoreactors with GALT Activity for Classic Galactosemia Therapy. ChemMedChem 2021, 16, 1438–1445. [Google Scholar] [CrossRef]
- Wang, Y.; Douglas, T. Protein Nanocage Architectures for the Delivery of Therapeutic Proteins. Curr. Opin. Colloid Interface Sci. 2021, 51, 101395. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Speiser, D.E.; Knuth, A.; Bachmann, M.F. Virus-like Particles for Vaccination against Cancer. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1579. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.; Wang, X.; Nie, S.; Chen, Z.; Shin, D.M. Therapeutic Nanoparticles for Drug Delivery in Cancer. Clin. Cancer Res. 2008, 14, 1310–1316. [Google Scholar] [CrossRef] [Green Version]
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction between Virus-Like Particles (VLPs) and Pattern Recognition Receptors (PRRs) from Dendritic Cells (DCs): Toward Better Engineering of VLPs. Front. Immunol. 2020, 11, 1100. [Google Scholar] [CrossRef]
- Ding, X.; Liu, D.; Booth, G.; Gao, W.; Lu, Y. Virus-Like Particle Engineering: From Rational Design to Versatile Applications. Biotechnol. J. 2018, 13, 1700324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, G.; Duan, W.-T.; Sun, M.-X.; Wang, M.-H.; Wang, S.-H.; Cai, X.-H.; Tu, Y. Self-Assembly into Virus–like Particles of the Recombinant Capsid Protein of Porcine Circovirus Type 3 and Its Application on Antibodies Detection. AMB Expr. 2020, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- Wilkerson, J.W.; Yang, S.-O.; Funk, P.J.; Stanley, S.K.; Bundy, B.C. Nanoreactors: Strategies to Encapsulate Enzyme Biocatalysts in Virus-like Particles. New Biotechnol. 2018, 44, 59–63. [Google Scholar] [CrossRef]
- Donaldson, B.; Lateef, Z.; Walker, G.F.; Young, S.L.; Ward, V.K. Virus-like Particle Vaccines: Immunology and Formulation for Clinical Translation. Expert Rev. Vaccines 2018, 17, 833–849. [Google Scholar] [CrossRef]
- Qian, C.; Liu, X.; Xu, Q.; Wang, Z.; Chen, J.; Li, T.; Zheng, Q.; Yu, H.; Gu, Y.; Li, S.; et al. Recent Progress on the Versatility of Virus-Like Particles. Vaccines 2020, 8, 139. [Google Scholar] [CrossRef] [Green Version]
- Cervera, L.; Gòdia, F.; Tarrés-Freixas, F.; Aguilar-Gurrieri, C.; Carrillo, J.; Blanco, J.; Gutiérrez-Granados, S. Production of HIV-1-Based Virus-like Particles for Vaccination: Achievements and Limits. Appl. Microbiol. Biotechnol. 2019, 103, 7367–7384. [Google Scholar] [CrossRef]
- Carvalho, S.B.; Silva, R.J.S.; Moreira, A.S.; Cunha, B.; Clemente, J.J.; Alves, P.M.; Carrondo, M.J.T.; Xenopoulos, A.; Peixoto, C. Efficient Filtration Strategies for the Clarification of Influenza Virus-like Particles Derived from Insect Cells. Sep. Purif. Technol. 2019, 218, 81–88. [Google Scholar] [CrossRef]
- Lu, W.; Zhao, Z.; Huang, Y.-W.; Wang, B. Review: A Systematic Review of Virus-like Particles of Coronavirus: Assembly, Generation, Chimerism and Their Application in Basic Research and in the Clinic. Int. J. Biol. Macromol. 2022, 200, 487–497. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Wilusz, J. The Fundamentals of Human Virology. In Microbial Forensics; Academic Press: Cambridge, MA, USA, 2005; pp. 41–53. Available online: https://reader.elsevier.com/reader/sd/pii/B9780120884834500068?token=D05E2ABB6D7A53A0599AEC3B4DA2198639C8B92C5885FD1DAD03FD3B815E2AD22A5A5B7DA37908EDC614D5F34F18C331&originRegion=us-east-1&originCreation=20220418192715 (accessed on 18 April 2022).
- Louten, J. Chapter 2—Virus Structure and Classification. In Essential Human Virology; Academic Press: Cambridge, MA, USA, 2016; pp. 19–29. Available online: https://reader.elsevier.com/reader/sd/pii/B9780128009475000028?token=7BE372B98B22B5FE56B2BC5017996E8FFCD5C89A6B07CDF8E31722FCCE498BAAB6E0E964180573473995CB6AC8418EE3&originRegion=us-east-1&originCreation=20220418192735 (accessed on 18 April 2022).
- Lamarre, B.; Ryadnov, M.G. Self-Assembling Viral Mimetics: One Long Journey with Short Steps. Macromol. Biosci. 2011, 11, 503–513. [Google Scholar] [CrossRef]
- Pellett, P.E.; Mitra, S.; Holland, T.C. Basics of Virology. In Handbook of Clinical Neurology; Academic Press: Cambridge, MA, USA, 2014; pp. 45–66. Available online: https://reader.elsevier.com/reader/sd/pii/B978044453488000002X?token=8867B3E02B07251123A161859F00BE6AD3C8B28CADBE87515D622735E64D2280DD3A91ACEE30FBEA7FDDAA869EC19B6C&originRegion=us-east-1&originCreation=20220418192824 (accessed on 18 April 2022).
- Hellen, C.U.T.; Wimmer, E. The Role of Proteolytic Processing in the Morphogenesis of Virus Particles. Experientia 1992, 48, 201–215. [Google Scholar] [CrossRef]
- Wisskirchen, K.; Lucifora, J.; Michler, T.; Protzer, U. New Pharmacological Strategies to Fight Enveloped Viruses. Trends Pharmacol. Sci. 2014, 35, 470–478. [Google Scholar] [CrossRef]
- Lan, K.; Luo, M.-H. Herpesviruses: Epidemiology, Pathogenesis, and Interventions. Virol. Sin. 2017, 32, 347–348. [Google Scholar] [CrossRef] [Green Version]
- Fenner, F.; Bachmann, P.A.; Gibbs, E.P.J.; Murphy, F.A.; Studdert, M.J.; White, O.D. Structure and Composition of Viruses. In Veterinary Virology; Academic Press: Cambridge, MA, USA, 1987; pp. 3–19. Available online: https://reader.elsevier.com/reader/sd/pii/B9780122530555500050?token=839CB2DC605C5E4B8DDE945EEDC256BFFC9A0F47195CEEFD3E738A406FE0AD93976DB1DFC83C9863DE3C92ABD4B031B7&originRegion=us-east-1&originCreation=20220418192957 (accessed on 18 April 2022).
- Schoeman, D.; Fielding, B.C. Coronavirus Envelope Protein: Current Knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [Green Version]
- Cosset, F.-L.; Lavillette, D. 4—Cell Entry of Enveloped Viruses. In Advances in Genetics; Academic Press: Cambridge, MA, USA, 2011; pp. 121–183. Available online: https://reader.elsevier.com/reader/sd/pii/B9780123808608000045?token=5B7798F78728BD7418D95FAAFA97024C551D30E63F8BA3CA8B48A665E51F5D514C184E7B0F81D10B46C8A2A3A21D35B7&originRegion=us-east-1&originCreation=20220418193042 (accessed on 18 April 2022).
- López-Macías, C. Virus-like Particle (VLP)-Based Vaccines for Pandemic Influenza: Performance of a VLP Vaccine during the 2009 Influenza Pandemic. Hum. Vaccines Immunother. 2012, 8, 411–414. [Google Scholar] [CrossRef]
- Cui, Z.; Gorzelnik, K.V.; Chang, J.-Y.; Langlais, C.; Jakana, J.; Young, R.; Zhang, J. Structures of Qβ Virions, Virus-like Particles, and the Qβ–MurA Complex Reveal Internal Coat Proteins and the Mechanism of Host Lysis. Proc. Natl. Acad. Sci. USA 2017, 114, 11697–11702. [Google Scholar] [CrossRef] [Green Version]
- Golmohammadi, R.; Valegård, K.; Fridborg, K.; Liljas, L. The Refined Structure of Bacteriophage MS2 at 2·8 Å Resolution. J. Mol. Biol. 1993, 234, 620–639. [Google Scholar] [CrossRef]
- Herbert, F.C.; Brohlin, O.R.; Galbraith, T.; Benjamin, C.; Reyes, C.A.; Luzuriaga, M.A.; Shahrivarkevishahi, A.; Gassensmith, J.J. Supramolecular Encapsulation of Small-Ultrared Fluorescent Proteins in Virus-Like Nanoparticles for Noninvasive In Vivo Imaging Agents. Bioconj. Chem. 2020, 31, 1529–1536. [Google Scholar] [CrossRef]
- Rohovie, M.J.; Nagasawa, M.; Swartz, J.R. Virus-like Particles: Next-Generation Nanoparticles for Targeted Therapeutic Delivery. Bioeng. Transl. Med. 2017, 2, 43–57. [Google Scholar] [CrossRef]
- Syomin, B.V.; Ilyin, Y.V. Virus-Like Particles as an Instrument of Vaccine Production. Mol. Biol. 2019, 53, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Bárcena, J.; Blanco, E. Design of Novel Vaccines Based on Virus-Like Particles or Chimeric Virions. In Structure and Physics of Viruses: An Integrated Textbook; Subcellular Biochemistry; Mateu, M.G., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 631–665. ISBN 978-94-007-6552-8. [Google Scholar]
- Shirbaghaee, Z.; Bolhassani, A. Different Applications of Virus-like Particles in Biology and Medicine: Vaccination and Delivery Systems. Biopolymers 2016, 105, 113–132. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Müller, K.M. In Vitro Assembly of Virus-Like Particles and Their Applications. Life 2021, 11, 334. [Google Scholar] [CrossRef] [PubMed]
- Grzelczak, M.; Vermant, J.; Furst, E.M.; Liz-Marzán, L.M. Directed Self-Assembly of Nanoparticles. ACS Nano 2010, 4, 3591–3605. [Google Scholar] [CrossRef] [PubMed]
- Pushko, P.; Pumpens, P.; Grens, E. Development of Virus-Like Particle Technology from Small Highly Symmetric to Large Complex Virus-Like Particle Structures. INT 2013, 56, 141–165. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Marasini, B.; Chen, X.; Ding, L.; Wang, J.-J.; Xiao, P.; Villinger, F.; Spearman, P. A Bivalent, Spherical Virus-Like Particle Vaccine Enhances Breadth of Immune Responses against Pathogenic Ebola Viruses in Rhesus Macaques. J. Virol. 2020, 94, e01884-19. [Google Scholar] [CrossRef] [PubMed]
- Lua, L.H.L.; Connors, N.K.; Sainsbury, F.; Chuan, Y.P.; Wibowo, N.; Middelberg, A.P.J. Bioengineering Virus-like Particles as Vaccines. Biotechnol. Bioeng. 2014, 111, 425–440. [Google Scholar] [CrossRef]
- Derdak, S.V.; Kueng, H.J.; Leb, V.M.; Neunkirchner, A.; Schmetterer, K.G.; Bielek, E.; Majdic, O.; Knapp, W.; Seed, B.; Pickl, W.F. Direct Stimulation of T Lymphocytes by Immunosomes: Virus-like Particles Decorated with T Cell Receptor/CD3 Ligands plus Costimulatory Molecules. Proc. Natl. Acad. Sci. USA 2006, 103, 13144–13149. [Google Scholar] [CrossRef] [Green Version]
- Hills, R.A.; Howarth, M. Virus-like Particles against Infectious Disease and Cancer: Guidance for the Nano-Architect. Curr. Opin. Biotechnol. 2022, 73, 346–354. [Google Scholar] [CrossRef]
- Cayetano-Cruz, M.; Coffeen, C.F.; Valadez-García, J.; Montiel, C.; Bustos-Jaimes, I. Decoration of Virus-like Particles with an Enzymatic Activity of Biomedical Interest. Virus Res. 2018, 255, 1–9. [Google Scholar] [CrossRef]
- Smith, M.T.; Hawes, A.K.; Bundy, B.C. Reengineering Viruses and Virus-like Particles through Chemical Functionalization Strategies. Curr. Opin. Biotechnol. 2013, 24, 620–626. [Google Scholar] [CrossRef]
- Kawano, M.; Matsui, M.; Handa, H. Chapter 15—Technologies That Generate and Modify Virus-like Particles for Medical Diagnostic and Therapy Purposes. In Design and Development of New Nanocarriers; Grumezescu, A.M., Ed.; William Andrew Publishing: Norwich, NY, USA, 2018; pp. 555–594. ISBN 978-0-12-813627-0. [Google Scholar]
- Akdis, M.; Akdis, C.A. Therapeutic Manipulation of Immune Tolerance in Allergic Disease. Nat. Rev. Drug Discov. 2009, 8, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Frietze, K.M.; Peabody, D.S.; Chackerian, B. Engineering Virus-like Particles as Vaccine Platforms. Curr. Opin. Virol. 2016, 18, 44–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Rourke, J.P.; Peabody, D.S.; Chackerian, B. Affinity Selection of Epitope-Based Vaccines Using a Bacteriophage Virus-like Particle Platform. Curr. Opin. Virol. 2015, 11, 76–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.D.; Fiedler, J.D.; Finn, M.G. Assembly of Hybrid Bacteriophage Qβ Virus-like Particles. Biochemistry 2009, 48, 11155–11157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, J.C.; Acar, H.; Mellas, M.J.; Tirrell, M.V. 11—Peptides in Immunoengineering. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Koutsopoulos, S., Ed.; Woodhead Publishing: Sawston, UK, 2018; pp. 287–326. ISBN 978-0-08-100736-5. [Google Scholar]
- Heise, M.T. Viral Pathogenesis. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 978-0-12-801238-3. [Google Scholar]
- Kato, J.; Svensson, C.I. Chapter Nine—Role of Extracellular Damage-Associated Molecular Pattern Molecules (DAMPs) as Mediators of Persistent Pain. In Progress in Molecular Biology and Translational Science; Molecular and Cell Biology of Pain; Price, T.J., Dussor, G., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 131, pp. 251–279. [Google Scholar]
- Karandikar, S.; Mirani, A.; Waybhase, V.; Patravale, V.B.; Patankar, S. Chapter 10—Nanovaccines for Oral Delivery-Formulation Strategies and Challenges. In Nanostructures for Oral Medicine; Micro and Nano Technologies; Andronescu, E., Grumezescu, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 263–293. ISBN 978-0-323-47720-8. [Google Scholar]
- Crisci, E.; Bárcena, J.; Montoya, M. Virus-like Particles: The New Frontier of Vaccines for Animal Viral Infections. Vet. Immunol. Immunopathol. 2012, 148, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Garg, H.; Mehmetoglu-Gurbuz, T.; Joshi, A. Virus Like Particles (VLP) as Multivalent Vaccine Candidate against Chikungunya, Japanese Encephalitis, Yellow Fever and Zika Virus. Sci. Rep. 2020, 10, 4017. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-W.; Saubi, N.; Joseph-Munné, J. Design Concepts of Virus-Like Particle-Based HIV-1 Vaccines. Front. Immunol. 2020, 11, 573157. [Google Scholar] [CrossRef] [PubMed]
- Pijlman, G.P. Enveloped Virus-like Particles as Vaccines against Pathogenic Arboviruses. Biotechnol. J. 2015, 10, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Mukhopadhyay, S. Generating Enveloped Virus-like Particles with in Vitro Assembled Cores. Virology 2011, 413, 153–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, T.; Yui, M.; Deo, V.K.; Park, E.Y. Development of Rous Sarcoma Virus-like Particles Displaying HCC49 ScFv for Specific Targeted Drug Delivery to Human Colon Carcinoma Cells. Pharm. Res. 2015, 32, 3699–3707. [Google Scholar] [CrossRef] [PubMed]
- Ong, H.K.; Tan, W.S.; Ho, K.L. Virus like Particles as a Platform for Cancer Vaccine Development. PeerJ 2017, 5, e4053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Parham, N.J.; Zhang, F.; Aasa-Chapman, M.; Gould, E.A.; Zhang, H. Vaccination with Coxsackievirus B3 Virus-like Particles Elicits Humoral Immune Response and Protects Mice against Myocarditis. Vaccine 2012, 30, 2301–2308. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, N.; Robinson, M.K.; Bernard, C.; Kawaguchi, Y.; Koujin, Y.; Koen, A.; Madhi, S.; Polasek, T.M.; McNeal, M.; Dargis, M.; et al. Safety and Immunogenicity of a Plant-Derived Rotavirus-like Particle Vaccine in Adults, Toddlers and Infants. Vaccine 2021, 39, 5513–5523. [Google Scholar] [CrossRef]
- Rutkowska, D.A.; Mokoena, N.B.; Tsekoa, T.L.; Dibakwane, V.S.; O’Kennedy, M.M. Plant-Produced Chimeric Virus-like Particles—A New Generation Vaccine against African Horse Sickness. BMC Vet. Res. 2019, 15, 432. [Google Scholar] [CrossRef]
- Grgacic, E.V.L.; Anderson, D.A. Virus-like Particles: Passport to Immune Recognition. Methods 2006, 40, 60–65. [Google Scholar] [CrossRef]
- Deo, V.K.; Kato, T.; Park, E.Y. Chimeric Virus-Like Particles Made Using GAG and M1 Capsid Proteins Providing Dual Drug Delivery and Vaccination Platform. Mol. Pharm. 2015, 12, 839–845. [Google Scholar] [CrossRef]
- Pattinson, D.J.; Apte, S.H.; Wibowo, N.; Rivera-Hernandez, T.; Groves, P.L.; Middelberg, A.P.J.; Doolan, D.L. Chimeric Virus-Like Particles and Capsomeres Induce Similar CD8+ T Cell Responses but Differ in Capacity to Induce CD4+ T Cell Responses and Antibody Responses. Front. Immunol. 2020, 11, 564627. [Google Scholar] [CrossRef]
- Fontana, D.; Garay, E.; Cervera, L.; Kratje, R.; Prieto, C.; Gòdia, F. Chimeric VLPs Based on HIV-1 Gag and a Fusion Rabies Glycoprotein Induce Specific Antibodies against Rabies and Foot-and-Mouth Disease Virus. Vaccines 2021, 9, 251. [Google Scholar] [CrossRef]
- Clark, D.P.; Pazdernik, N.J. Chapter 3—Recombinant DNA Technology. In Biotechnology, 2nd ed.; Clark, D.P., Pazdernik, N.J., Eds.; Academic Cell: Boston, MA, USA, 2016; pp. 63–95. ISBN 978-0-12-385015-7. [Google Scholar]
- Yaniz-Galende, E.; Hajjar, R.J. 16—Stem Cell and Gene Therapy for Cardiac Regeneration. In Cardiac Regeneration and Repair; Li, R.-K., Weisel, R.D., Eds.; Woodhead Publishing: Sawston, UK, 2014; pp. 347–379. ISBN 978-0-85709-658-6. [Google Scholar]
- Nayak, S.; Herzog, R.W. Progress and Prospects: Immune Responses to Viral Vectors. Gene Ther. 2010, 17, 295–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brondyk, W.H. Chapter 11 Selecting an Appropriate Method for Expressing a Recombinant Protein. In Methods in Enzymology, 2nd ed.; Guide to Protein Purification; Burgess, R.R., Deutscher, M.P., Eds.; Academic Press: Cambridge, MA, USA, 2009; Volume 463, pp. 131–147. [Google Scholar]
- Burnett, M.J.B.; Burnett, A.C. Therapeutic Recombinant Protein Production in Plants: Challenges and Opportunities. Plants People Planet 2020, 2, 121–132. [Google Scholar] [CrossRef]
- Pham, P.V. Chapter 19—Medical Biotechnology: Techniques and Applications. In Omics Technologies and Bio-Engineering; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 449–469. ISBN 978-0-12-804659-3. [Google Scholar]
- Kant, R.; Rayaprolu, V.; McDonald, K.; Bothner, B. Curating Viscoelastic Properties of Icosahedral Viruses, Virus-Based Nanomaterials, and Protein Cages. J. Biol. Phys. 2018, 44, 211–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Twarock, R.; Luque, A. Structural Puzzles in Virology Solved with an Overarching Icosahedral Design Principle. Nat. Commun. 2019, 10, 4414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, N.P.; Demo, G.; Agnello, E.; Kelch, B.A. Principles for Enhancing Virus Capsid Capacity and Stability from a Thermophilic Virus Capsid Structure. Nat. Commun. 2019, 10, 4471. [Google Scholar] [CrossRef] [Green Version]
- Dokland, T. Scaffolding Proteins and Their Role in Viral Assembly. CMLS Cell. Mol. Life Sci. 1999, 56, 580–603. [Google Scholar] [CrossRef]
- Strable, E.; Finn, M.G. Chemical Modification of Viruses and Virus-Like Particles. In Viruses and Nanotechnology; Current Topics in Microbiology and Immunology; Manchester, M., Steinmetz, N.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–21. ISBN 978-3-540-69379-6. [Google Scholar]
- Zdanowicz, M.; Chroboczek, J. Virus-like Particles as Drug Delivery Vectors. Acta Biochim. Pol. 2016, 63, 469–473. [Google Scholar] [CrossRef]
- Buzón, P.; Maity, S.; Roos, W.H. Physical Virology: From Virus Self-Assembly to Particle Mechanics. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1613. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Sánchez, L.; Tapia-Moreno, A.; Juarez-Moreno, K.; Patterson, D.P.; Cadena-Nava, R.D.; Douglas, T.; Vazquez-Duhalt, R. Design of a VLP-Nanovehicle for CYP450 Enzymatic Activity Delivery. J. Nanobiotechnol. 2015, 13, 66. [Google Scholar] [CrossRef] [Green Version]
- Villagrana-Escareño, M.V.; Reynaga-Hernández, E.; Galicia-Cruz, O.G.; Durán-Meza, A.L.; De la Cruz-González, V.; Hernández-Carballo, C.Y.; Ruíz-García, J. VLPs Derived from the CCMV Plant Virus Can Directly Transfect and Deliver Heterologous Genes for Translation into Mammalian Cells. BioMed Res. Int. 2019, 2019, e4630891. [Google Scholar] [CrossRef]
- Masarapu, H.; Patel, B.K.; Chariou, P.L.; Hu, H.; Gulati, N.M.; Carpenter, B.L.; Ghiladi, R.A.; Shukla, S.; Steinmetz, N.F. Physalis Mottle Virus-Like Particles as Nanocarriers for Imaging Reagents and Drugs. Biomacromolecules 2017, 18, 4141–4153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panda, S.K.; Kapur, N.; Paliwal, D.; Durgapal, H. Recombinant Hepatitis E Virus like Particles Can Function as RNA Nanocarriers. J. Nanobiotechnol. 2015, 13, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamin, C.; Brohlin, O.; Shahrivarkevishahi, A.; Gassensmith, J.J. Chapter 11—Virus like Particles: Fundamental Concepts, Biological Interactions, and Clinical Applications. In Nanoparticles for Biomedical Applications; Micro and Nano Technologies; Chung, E.J., Leon, L., Rinaldi, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 153–174. ISBN 978-0-12-816662-8. [Google Scholar]
- Sainsbury, F. Virus-like Nanoparticles: Emerging Tools for Targeted Cancer Diagnostics and Therapeutics. Ther. Deliv. 2017, 8, 1019–1021. [Google Scholar] [CrossRef] [Green Version]
- Destito, G.; Schneemann, A.; Manchester, M. Biomedical Nanotechnology Using Virus-Based Nanoparticles. In Viruses and Nanotechnology; Current Topics in Microbiology and Immunology; Manchester, M., Steinmetz, N.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 95–122. ISBN 978-3-540-69379-6. [Google Scholar]
- Mansour, H.M.; Sohn, M.; Al-Ghananeem, A.; DeLuca, P.P. Materials for Pharmaceutical Dosage Forms: Molecular Pharmaceutics and Controlled Release Drug Delivery Aspects. Int. J. Mol. Sci. 2010, 11, 3298–3322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, W.; Zhang, D.; Wu, Y.; Lv, X.; Hu, B.; Zhou, X.; Ye, S.; Bi, S.; Ren, L.; Zhang, X. Modularized Peptides Modified HBc Virus-like Particles for Encapsulation and Tumor-Targeted Delivery of Doxorubicin. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Ashley, C.E.; Carnes, E.C.; Phillips, G.K.; Durfee, P.N.; Buley, M.D.; Lino, C.A.; Padilla, D.P.; Phillips, B.; Carter, M.B.; Willman, C.L.; et al. Cell-Specific Delivery of Diverse Cargos by Bacteriophage MS2 Virus-like Particles. ACS Nano 2011, 5, 5729–5745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, X.; Cai, X.; Yang, Y. Genetic Engineering Strategies for Construction of Multivalent Chimeric VLPs Vaccines. Expert Rev. Vaccines 2020, 19, 235–246. [Google Scholar] [CrossRef] [Green Version]
- Fiedler, J.D.; Higginson, C.; Hovlid, M.L.; Kislukhin, A.A.; Castillejos, A.; Manzenrieder, F.; Campbell, M.G.; Voss, N.R.; Potter, C.S.; Carragher, B.; et al. Engineered Mutations Change the Structure and Stability of a Virus-Like Particle. Biomacromolecules 2012, 13, 2339–2348. [Google Scholar] [CrossRef] [Green Version]
- Sainsbury, F.; Saxena, P.; Aljabali, A.A.A.; Saunders, K.; Evans, D.J.; Lomonossoff, G.P. Genetic Engineering and Characterization of Cowpea Mosaic Virus Empty Virus-Like Particles. In Virus Hybrids as Nanomaterials: Methods and Protocols; Methods in Molecular Biology; Lin, B., Ratna, B., Eds.; Humana Press: Totowa, NJ, USA, 2014; pp. 139–153. ISBN 978-1-62703-751-8. [Google Scholar]
- Ramazi, S.; Zahiri, J. Post-Translational Modifications in Proteins: Resources, Tools and Prediction Methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef]
- Farley, A.R.; Link, A.J. Chapter 40 Identification and Quantification of Protein Posttranslational Modifications. In Methods in Enzymology, 2nd ed.; Guide to Protein Purification; Burgess, R.R., Deutscher, M.P., Eds.; Academic Press: Cambridge, MA, USA, 2009; Volume 463, pp. 725–763. [Google Scholar]
- Müller, M.M. Post-Translational Modifications of Protein Backbones: Unique Functions, Mechanisms, and Challenges. Biochemistry 2018, 57, 177–185. [Google Scholar] [CrossRef]
- Margolin, E.; Crispin, M.; Meyers, A.; Chapman, R.; Rybicki, E.P. A Roadmap for the Molecular Farming of Viral Glycoprotein Vaccines: Engineering Glycosylation and Glycosylation-Directed Folding. Front. Plant Sci. 2020, 11, 609207. [Google Scholar] [CrossRef] [PubMed]
- Maphis, N.M.; Peabody, J.; Crossey, E.; Jiang, S.; Jamaleddin Ahmad, F.A.; Alvarez, M.; Mansoor, S.K.; Yaney, A.; Yang, Y.; Sillerud, L.O.; et al. Qβ Virus-like Particle-Based Vaccine Induces Robust Immunity and Protects against Tauopathy. NPJ Vaccines 2019, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.J.; Landry, N.; Trépanier, S.; Mercier, G.; Dargis, M.; Couture, M.; D’Aoust, M.-A.; Vézina, L.-P. Human Antibody Response to N-Glycans Present on Plant-Made Influenza Virus-like Particle (VLP) Vaccines. Vaccine 2014, 32, 6098–6106. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wu, X.; Li, L.; Liu, Z.; Wang, Z. Use of Baculovirus Expression System for Generation of Virus-like Particles: Successes and Challenges. Protein Expr. Purif. 2013, 90, 104–116. [Google Scholar] [CrossRef]
- Kramer, K.; Al-Barwani, F.; Baird, M.A.; Young, V.L.; Larsen, D.S.; Ward, V.K.; Young, S.L. Functionalisation of Virus-Like Particles Enhances Antitumour Immune Responses. J. Immunol. Res. 2019, 2019, 5364632. [Google Scholar] [CrossRef]
- Lerouge, P.; Bardor, M.; Pagny, S.; Gomord, V.; Faye, L. N-Glycosylation of Recombinant Pharmaceutical Glycoproteins Produced in Transgenic Plants Towards an Humanisation of Plant N-Glycans. Curr. Pharm. Biotechnol. 2000, 1, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Mourdikoudis, S.; Pallares, R.M.; Thanh, N.T.K. Characterization Techniques for Nanoparticles: Comparison and Complementarity upon Studying Nanoparticle Properties. Nanoscale 2018, 10, 12871–12934. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.-W.; Cambre, M.; Lee, H.-J. The Toxicity of Nanoparticles Depends on Multiple Molecular and Physicochemical Mechanisms. Int. J. Mol. Sci. 2017, 18, 2702. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, B.J.; Bonar, M.M.; Costas-Cancelas, I.N.; Hunt, K.; Makarkov, A.I.; Chierzi, S.; Krawczyk, C.M.; Landry, N.; Ward, B.J.; Rouiller, I. Morphological Characterization of a Plant-Made Virus-like Particle Vaccine Bearing Influenza Virus Hemagglutinins by Electron Microscopy. Vaccine 2018, 36, 2147–2154. [Google Scholar] [CrossRef]
- Jeoung, H.-Y.; Lee, W.-H.; Jeong, W.; Shin, B.-H.; Choi, H.-W.; Lee, H.S.; An, D.-J. Immunogenicity and Safety of Virus-like Particle of the Porcine Encephalomyocarditis Virus in Pig. Virol. J. 2011, 8, 170. [Google Scholar] [CrossRef] [Green Version]
- Naupu, P.N.; van Zyl, A.R.; Rybicki, E.P.; Hitzeroth, I.I. Immunogenicity of Plant-Produced Human Papillomavirus (HPV) Virus-Like Particles (VLPs). Vaccines 2020, 8, 740. [Google Scholar] [CrossRef]
- Ren, F.; Swevers, L.; Lu, Q.; Zhao, Y.; Yan, J.; Li, H.; Sun, J. Effect of Mutations in Capsid Shell Protein on the Assembly of BmCPV Virus-like Particles. J. Gen. Virol. 2021, 102, 1542. [Google Scholar] [CrossRef] [PubMed]
- Plescia, C.B.; David, E.A.; Patra, D.; Sengupta, R.; Amiar, S.; Su, Y.; Stahelin, R.V. SARS-CoV-2 Viral Budding and Entry Can Be Modeled Using BSL-2 Level Virus-like Particles. J. Biol. Chem. 2021, 296, 100103. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-M.J.; Chien, C.-Y.; Liu, M.-T.; Fang, Z.-S.; Chang, S.-Y.; Juang, R.-H.; Chang, S.-C.; Chen, H.-W. Multi-Antigen Avian Influenza a (H7N9) Virus-like Particles: Particulate Characterizations and Immunogenicity Evaluation in Murine and Avian Models. BMC Biotechnol. 2017, 17, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic Light Scattering: A Practical Guide and Applications in Biomedical Sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Shnoudeh, A.J.; Hamad, I.; Abdo, R.W.; Qadumii, L.; Jaber, A.Y.; Surchi, H.S.; Alkelany, S.Z. Chapter 15—Synthesis, Characterization, and Applications of Metal Nanoparticles. In Biomaterials and Bionanotechnology; Tekade, R.K., Ed.; Advances in Pharmaceutical Product Development and Research; Academic Press: Amsterdam, The Netherlands, 2019; pp. 527–612. ISBN 978-0-12-814427-5. [Google Scholar]
- Hawe, A.; Weinbuch, D.; Zölls, S.; Reichel, A.; Carpenter, J.F. Chapter 10—Submicrometer, Micrometer and Visible Particle Analysis in Biopharmaceutical Research and Development. In Biophysical Characterization of Proteins in Developing Biopharmaceuticals, 2nd ed.; Houde, D.J., Berkowitz, S.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 285–310. ISBN 978-0-444-64173-1. [Google Scholar]
- Lorber, B.; Fischer, F.; Bailly, M.; Roy, H.; Kern, D. Protein Analysis by Dynamic Light Scattering: Methods and Techniques for Students. Biochem. Mol. Biol. Educ. 2012, 40, 372–382. [Google Scholar] [CrossRef]
- Al-Ghobashy, M.A.; Mostafa, M.M.; Abed, H.S.; Fathalla, F.A.; Salem, M.Y. Correlation between Dynamic Light Scattering and Size Exclusion High Performance Liquid Chromatography for Monitoring the Effect of PH on Stability of Biopharmaceuticals. J. Chromatogr. B 2017, 1060, 1–9. [Google Scholar] [CrossRef]
- Ausar, S.F.; Foubert, T.R.; Hudson, M.H.; Vedvick, T.S.; Middaugh, C.R. Conformational Stability and Disassembly of Norwalk Virus-like Particles: Effect of ph and temperature. J. Biol. Chem. 2006, 281, 19478–19488. [Google Scholar] [CrossRef] [Green Version]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate Secondary Structure Prediction and Fold Recognition for Circular Dichroism Spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, S. DLS and Zeta Potential—What They Are and What They Are Not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.C.; Crist, R.M.; Clogston, J.D.; McNeil, S.E. Zeta Potential: A Case Study of Cationic, Anionic, and Neutral Liposomes. Anal. Bioanal. Chem. 2017, 409, 5779–5787. [Google Scholar] [CrossRef] [PubMed]
- Marsalek, R.; Kotyrba, M.; Volna, E.; Jarusek, R. Neural Network Modelling for Prediction of Zeta Potential. Mathematics 2021, 9, 3089. [Google Scholar] [CrossRef]
- Doan, C.D.; Ghosh, S. Formation and Stability of Pea Proteins Nanoparticles Using Ethanol-Induced Desolvation. Nanomaterials 2019, 9, 949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skoglund, S.; Hedberg, J.; Yunda, E.; Godymchuk, A.; Blomberg, E.; Wallinder, I.O. Difficulties and Flaws in Performing Accurate Determinations of Zeta Potentials of Metal Nanoparticles in Complex Solutions—Four Case Studies. PLoS ONE 2017, 12, e0181735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derman, S.; Akdeste, Z.M. Particle Size and Zeta Potential Investigation of Synthetic Peptide-Protein Conjugates/Sentetik Peptid-Protein Konjugatlarının Parçacık Boyutu ve Zeta Potensiyel Analizi. Turk. J. Biochem. 2015, 40, 282–289. [Google Scholar] [CrossRef]
- Samandoulgou, I.; Fliss, I.; Jean, J. Zeta Potential and Aggregation of Virus-Like Particle of Human Norovirus and Feline Calicivirus Under Different Physicochemical Conditions. Food Environ. Virol. 2015, 7, 249–260. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using Circular Dichroism Spectra to Estimate Protein Secondary Structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.K.; Siddiqi, M.K.; Alam, P.; Khan, R.H. Protein Misfolding and Aggregation: Mechanism, Factors and Detection. Process Biochem. 2016, 51, 1183–1192. [Google Scholar] [CrossRef]
- Rogers, D.M.; Jasim, S.B.; Dyer, N.T.; Auvray, F.; Réfrégiers, M.; Hirst, J.D. Electronic Circular Dichroism Spectroscopy of Proteins. Chem 2019, 5, 2751–2774. [Google Scholar] [CrossRef]
- Samandoulgou, I.; Hammami, R.; Morales Rayas, R.; Fliss, I.; Jean, J. Stability of Secondary and Tertiary Structures of Virus-Like Particles Representing Noroviruses: Effects of PH, Ionic Strength, and Temperature and Implications for Adhesion to Surfaces. Appl. Environ. Microbiol. 2015, 81, 7680–7686. [Google Scholar] [CrossRef] [Green Version]
- Ninyio, N.N.; Ho, K.L.; Yong, C.Y.; Chee, H.Y.; Hamid, M.; Ong, H.K.; Mariatulqabtiah, A.R.; Tan, W.S. Chimeric Virus-Like Particles of Prawn Nodavirus Displaying Hepatitis B Virus Immunodominant Region: Biophysical Properties and Cytokine Response. Int. J. Mol. Sci. 2021, 22, 1922. [Google Scholar] [CrossRef] [PubMed]
- Antosiewicz, J.M.; Shugar, D. UV–Vis Spectroscopy of Tyrosine Side-Groups in Studies of Protein Structure. Part 2: Selected Applications. Biophys. Rev. 2016, 8, 163–177. [Google Scholar] [CrossRef] [Green Version]
- Das Graças Nascimento Amorim, A.; Sánchez-Paniagua, M.; de Oliveira, T.M.; Mafud, A.C.; da Silva, D.A.; de Souza de Almeida Leite, J.R.; López-Ruiz, B. Synthesis, Characterization and Use of Enzyme Cashew Gum Nanoparticles for Biosensing Applications. J. Mater. Chem. B 2021, 9, 6825–6835. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Uchida, M.; Waghwani, H.K.; Douglas, T. Synthetic Virus-like Particles for Glutathione Biosynthesis. ACS Synth. Biol. 2020, 9, 3298–3310. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.; Young, V.L.; Donaldson, B.C.; Woodall, M.J.; Shields, N.J.; Walker, G.F.; Ward, V.K.; Young, S.L. Delivering Two Tumour Antigens Survivin and Mucin-1 on Virus-Like Particles Enhances Anti-Tumour Immune Responses. Vaccines 2021, 9, 463. [Google Scholar] [CrossRef] [PubMed]
- Farré, M.; Jha, A.N. Chapter 9—Metabolomics Effects of Nanomaterials: An Ecotoxicological Perspective. In Environmental Metabolomics; Álvarez-Muñoz, D., Farré, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 259–281. ISBN 978-0-12-818196-6. [Google Scholar]
- Frueh, D.P.; Goodrich, A.C.; Mishra, S.H.; Nichols, S.R. NMR Methods for Structural Studies of Large Monomeric and Multimeric Proteins. Curr. Opin. Struct. Biol. 2013, 23, 734–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purslow, J.A.; Khatiwada, B.; Bayro, M.J.; Venditti, V. NMR Methods for Structural Characterization of Protein-Protein Complexes. Front. Mol. Biosci. 2020, 7, 9. [Google Scholar] [CrossRef]
- Marbella, L.E.; Millstone, J.E. NMR Techniques for Noble Metal Nanoparticles. Chem. Mater. 2015, 27, 2721–2739. [Google Scholar] [CrossRef]
- Leung, R.L.C.; Robinson, M.D.M.; Ajabali, A.A.A.; Karunanithy, G.; Lyons, B.; Raj, R.; Raoufmoghaddam, S.; Mohammed, S.; Claridge, T.D.W.; Baldwin, A.J.; et al. Monitoring the Disassembly of Virus-like Particles by 19F-NMR. J. Am. Chem. Soc. 2017, 139, 5277–5280. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Huang, P.-H.; Zhang, H.; Rich, J.; Bachman, H.; Ye, J.; Zhang, W.; Chen, C.; Xie, Z.; Tian, Z.; et al. Fabrication of Tunable, High-Molecular-Weight Polymeric Nanoparticles via Ultrafast Acoustofluidic Micromixing. Lab Chip 2021, 21, 2453–2463. [Google Scholar] [CrossRef]
- Smith, D.M. Protein Separation and Characterization Procedures. In Food Analysis; Food Science Tex Seriest; Nielsen, S.S., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 431–453. ISBN 978-3-319-45776-5. [Google Scholar]
- Chakavarti, B.; Chakavarti, D. Electrophoretic Separation of Proteins. J. Vis. Exp. 2008, e758. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Ge, L.; Yuan, K.; Li, C.; Zhen, X.; Cai, W.; Cheng, R.; Jiang, X. Targeting and Microenvironment-Improving of Phenylboronic Acid-Decorated Soy Protein Nanoparticles with Different Sizes to Tumor. Theranostics 2019, 9, 7417–7430. [Google Scholar] [CrossRef] [PubMed]
- Gollapudi, D.; Wycuff, D.L.; Schwartz, R.M.; Cooper, J.W.; Cheng, K. Development of High-Throughput and High Sensitivity Capillary Gel Electrophoresis Platform Method for Western, Eastern, and Venezuelan Equine Encephalitis (WEVEE) Virus like Particles (VLPs) Purity Determination and Characterization. Electrophoresis 2017, 38, 2610–2621. [Google Scholar] [CrossRef] [PubMed]
- Martino, M.L.; Crooke, S.N.; Manchester, M.; Finn, M.G. Single-Point Mutations in Qβ Virus-like Particles Change Binding to Cells. Biomacromolecules 2021, 22, 3332–3341. [Google Scholar] [CrossRef]
- Büyükköroğlu, G.; Dora, D.D.; Özdemir, F.; Hızel, C. Chapter 15—Techniques for Protein Analysis. In Omics Technologies and Bio-Engineering; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 317–351. ISBN 978-0-12-804659-3. [Google Scholar]
- Williams, D.A. Chapter 3—Fundamentals of Mass Spectrometry. In Pharmacochemistry Library; Browne, T.R., Ed.; Stable Isotopes in Pharmaceutical Research; Elsevier: Amsterdam, The Netherlands, 1997; Volume 26, pp. 19–45. [Google Scholar]
- Glish, G.L.; Vachet, R.W. The Basics of Mass Spectrometry in the Twenty-First Century. Nat. Rev. Drug Discov. 2003, 2, 140–150. [Google Scholar] [CrossRef]
- Bustos, A.R.M.; Encinar, J.R.; Sanz-Medel, A. Mass Spectrometry for the Characterisation of Nanoparticles. Anal. Bioanal. Chem. 2013, 405, 5637–5643. [Google Scholar] [CrossRef] [PubMed]
- Comby-Zerbino, C.; Dagany, X.; Chirot, F.; Dugourd, P.; Antoine, R. The Emergence of Mass Spectrometry for Characterizing Nanomaterials. Atomically Precise Nanoclusters and Beyond. Mater. Adv. 2021, 2, 4896–4913. [Google Scholar] [CrossRef]
- Weiss, V.U.; Pogan, R.; Zoratto, S.; Bond, K.M.; Boulanger, P.; Jarrold, M.F.; Lyktey, N.; Pahl, D.; Puffler, N.; Schelhaas, M.; et al. Virus-like Particle Size and Molecular Weight/Mass Determination Applying Gas-Phase Electrophoresis (Native NES GEMMA). Anal. Bioanal. Chem. 2019, 411, 5951–5962. [Google Scholar] [CrossRef] [Green Version]
- Miller, L.M.; Bond, K.M.; Draper, B.E.; Jarrold, M.F. Characterization of Classical Vaccines by Charge Detection Mass Spectrometry. Anal. Chem. 2021, 93, 11965–11972. [Google Scholar] [CrossRef]
- Fu, K.; March, K.; Alexaki, A.; Fabozzi, G.; Moysi, E.; Petrovas, C. Immunogenicity of Protein Therapeutics: A Lymph Node Perspective. Front. Immunol. 2020, 11, 791. [Google Scholar] [CrossRef]
- Hong, S.; Choi, D.W.; Kim, H.N.; Park, C.G.; Lee, W.; Park, H.H. Protein-Based Nanoparticles as Drug Delivery Systems. Pharmaceutics 2020, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.O.; Augusto, G.; Bachmann, M.F. The 3Ds in Virus-like Particle Based-Vaccines: “Design, Delivery and Dynamics”. Immunol. Rev. 2020, 296, 155–168. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, L.; Zhang, C.; Lu, Y. Exploration on the Expression and Assembly of Virus-like Particles. Biotechnol. Notes 2021, 2, 51–58. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, H.-J. Yeast as an Expression System for Producing Virus-like Particles: What Factors Do We Need to Consider? Lett. Appl. Microbiol. 2017, 64, 111–123. [Google Scholar] [CrossRef]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Akwiditya, M.A.; Yong, C.Y.; Yusof, M.T.; Mariatulqabtiah, A.R.; Ho, K.L.; Tan, W.S. Hepatitis B Virus-Like Particle: Targeted Delivery of Plasmid Expressing Short Hairpin RNA for Silencing the Bcl-2 Gene in Cervical Cancer Cells. Int. J. Mol. Sci. 2021, 22, 2320. [Google Scholar] [CrossRef] [PubMed]
- Puente-Massaguer, E.; Grau-Garcia, P.; Strobl, F.; Grabherr, R.; Striedner, G.; Lecina, M.; Gòdia, F. Stable Sf9 Cell Pools as a System for Rapid HIV-1 Virus-like Particle Production. J. Chem. Technol. Biotechnol. 2021, 96, 3388–3397. [Google Scholar] [CrossRef]
- He, J.; Lai, H.; Esqueda, A.; Chen, Q. Plant-Produced Antigen Displaying Virus-Like Particles Evokes Potent Antibody Responses against West Nile Virus in Mice. Vaccines 2021, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Lai, H. Plant-Derived Virus-like Particles as Vaccines. Hum. Vaccines Immunother. 2013, 9, 26–49. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Wang, H.; Deng, F. Advances and Challenges in Enveloped Virus-like Particle (VLP)-Based Vaccines. J. Immunol. Sci. 2018, 2, 36–41. [Google Scholar]
- Fernandes, F.; Teixeira, A.P.; Carinhas, N.; Carrondo, M.J.; Alves, P.M. Insect Cells as a Production Platform of Complex Virus-like Particles. Expert Rev. Vaccines 2013, 12, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.H.; Cai, H.; Steinmetz, N.F. Viral Nanoparticles for Drug Delivery, Imaging, Immunotherapy, and Theranostic Applications. Adv. Drug Deliv. Rev. 2020, 156, 214–235. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Nolte, R.J.M.; Cornelissen, J.J.L.M. Virus-Based Nanocarriers for Drug Delivery. Adv. Drug Deliv. Rev. 2012, 64, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Koyani, R.; Pérez-Robles, J.; Cadena-Nava, R.D.; Vazquez-Duhalt, R. Biomaterial-Based Nanoreactors, an Alternative for Enzyme Delivery. Nanotechnol. Rev. 2017, 6, 405–419. [Google Scholar] [CrossRef]
- Tapia-Moreno, A.; Juarez-Moreno, K.; Gonzalez-Davis, O.; Cadena-Nava, R.D.; Vazquez-Duhalt, R. Biocatalytic Virus Capsid as Nanovehicle for Enzymatic Activation of Tamoxifen in Tumor Cells. Biotechnol. J. 2017, 12, 1600706. [Google Scholar] [CrossRef]
- Chauhan, K.; Sengar, P.; Juarez-Moreno, K.; Hirata, G.A.; Vazquez-Duhalt, R. Camouflaged, Activatable and Therapeutic Tandem Bionanoreactors for Breast Cancer Theranosis. J. Colloid Interface Sci. 2020, 580, 365–376. [Google Scholar] [CrossRef]
- Li, W.; Jing, Z.; Wang, S.; Li, Q.; Xing, Y.; Shi, H.; Li, S.; Hong, Z. P22 Virus-like Particles as an Effective Antigen Delivery Nanoplatform for Cancer Immunotherapy. Biomaterials 2021, 271, 120726. [Google Scholar] [CrossRef]
- Hu, H.; Masarapu, H.; Gu, Y.; Zhang, Y.; Yu, X.; Steinmetz, N.F. Physalis Mottle Virus-like Nanoparticles for Targeted Cancer Imaging. ACS Appl. Mater. Interfaces 2019, 11, 18213–18223. [Google Scholar] [CrossRef]
- Sapsford, K.E.; Algar, W.R.; Berti, L.; Gemmill, K.B.; Casey, B.J.; Oh, E.; Stewart, M.H.; Medintz, I.L. Functionalizing Nanoparticles with Biological Molecules: Developing Chemistries That Facilitate Nanotechnology. Chem. Rev. 2013, 113, 1904–2074. [Google Scholar] [CrossRef]
- Mateu, M.G. Virus Engineering: Functionalization and Stabilization. Protein Eng. Des. Sel. 2011, 24, 53–63. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Tagliamonte, M.; Buonaguro, F.M.; Tornesello, M.L.; Buonaguro, L. Virus-like Particles as Preventive and Therapeutic Cancer Vaccines. Vaccines 2022, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Brune, K.D.; Howarth, M. New Routes and Opportunities for Modular Construction of Particulate Vaccines: Stick, Click, and Glue. Front. Immunol. 2018, 9, 1432. [Google Scholar] [CrossRef] [PubMed]
- Thiruppathi, R.; Mishra, S.; Ganapathy, M.; Padmanabhan, P.; Gulyás, B. Nanoparticle Functionalization and Its Potentials for Molecular Imaging. Adv. Sci. 2017, 4, 1600279. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M. The Need for Robust Characterization of Nanomaterials for Nanomedicine Applications. Nat. Commun. 2021, 12, 5246. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Jennings, G.T. Vaccine Delivery: A Matter of Size, Geometry, Kinetics and Molecular Patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- Lee, B.; Yoon, S.; Lee, J.W.; Kim, Y.; Chang, J.; Yun, J.; Ro, J.C.; Lee, J.-S.; Lee, J.H. Statistical Characterization of the Morphologies of Nanoparticles through Machine Learning Based Electron Microscopy Image Analysis. ACS Nano 2020, 14, 17125–17133. [Google Scholar] [CrossRef]
- Mahmood, S.; Mandal, U.K.; Chatterjee, B.; Taher, M. Advanced Characterizations of Nanoparticles for Drug Delivery: Investigating Their Properties through the Techniques Used in Their Evaluations. Nanotechnol. Rev. 2017, 6, 355–372. [Google Scholar] [CrossRef]
- Quentin, D.; Raunser, S. Electron Cryomicroscopy as a Powerful Tool in Biomedical Research. J. Mol. Med. 2018, 96, 483–493. [Google Scholar] [CrossRef] [Green Version]
- Benjin, X.; Ling, L. Developments, Applications, and Prospects of Cryo-electron Microscopy. Protein Sci. 2020, 29, 872–882. [Google Scholar] [CrossRef]
- Vera-Velasco, N.M.; García-Murria, M.J.; Sánchez del Pino, M.M.; Mingarro, I.; Martinez-Gil, L. Proteomic Composition of Nipah Virus-like Particles. J. Proteom. 2018, 172, 190–200. [Google Scholar] [CrossRef]
- Vergara Barragán, E.; Bach, H.; Meza-Reyes, S.; Montiel-Smith, S.; Sánchez-Arreola, E.; Hernández, L.R. Bioactivities of Flavonoids from Lopezia racemosa. BioMed Res. Int. 2019, 2019, 3286489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tracanna, M.I.; Fortuna, A.M.; Contreras Cárdenas, A.V.; Marr, A.K.; McMaster, W.R.; Gómez-Velasco, A.; Sánchez-Arreola, E.; Hernández, L.R.; Bach, H. Anti-Leishmanial, Anti-Inflammatory and Antimicrobial Activities of Phenolic Derivatives from Tibouchina paratropica. Phytother. Res. 2015, 29, 393–397. [Google Scholar] [CrossRef] [PubMed]





| Name | Expression System | Shape | Features and Biomedical Applications | Reference |
|---|---|---|---|---|
| tHBcAg VLPs | Icosahedral |
| ||
| E. coli strain W3110IQ | [162] | |||
| SARS-CoV-2 VLPs | Corona-like |
| ||
| HEK-293T and Vero E6 cells | [161] | |||
| HIV-1 Gag-eGFP VLPs | Not reported |
| ||
| Sf9 cells |  | [163] | ||
| HBcAg-wDIII VLPs | Spherical |
| ||
| Nicotiana benthamiana |  | [164] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mejía-Méndez, J.L.; Vazquez-Duhalt, R.; Hernández, L.R.; Sánchez-Arreola, E.; Bach, H. Virus-like Particles: Fundamentals and Biomedical Applications. Int. J. Mol. Sci. 2022, 23, 8579. https://doi.org/10.3390/ijms23158579
Mejía-Méndez JL, Vazquez-Duhalt R, Hernández LR, Sánchez-Arreola E, Bach H. Virus-like Particles: Fundamentals and Biomedical Applications. International Journal of Molecular Sciences. 2022; 23(15):8579. https://doi.org/10.3390/ijms23158579
Chicago/Turabian StyleMejía-Méndez, Jorge L., Rafael Vazquez-Duhalt, Luis R. Hernández, Eugenio Sánchez-Arreola, and Horacio Bach. 2022. "Virus-like Particles: Fundamentals and Biomedical Applications" International Journal of Molecular Sciences 23, no. 15: 8579. https://doi.org/10.3390/ijms23158579
APA StyleMejía-Méndez, J. L., Vazquez-Duhalt, R., Hernández, L. R., Sánchez-Arreola, E., & Bach, H. (2022). Virus-like Particles: Fundamentals and Biomedical Applications. International Journal of Molecular Sciences, 23(15), 8579. https://doi.org/10.3390/ijms23158579







