Transcriptional and Post-Translational Regulation of Junctional Adhesion Molecule-B (JAM-B) in Leukocytes under Inflammatory Stimuli
Abstract
:1. Introduction
2. Results
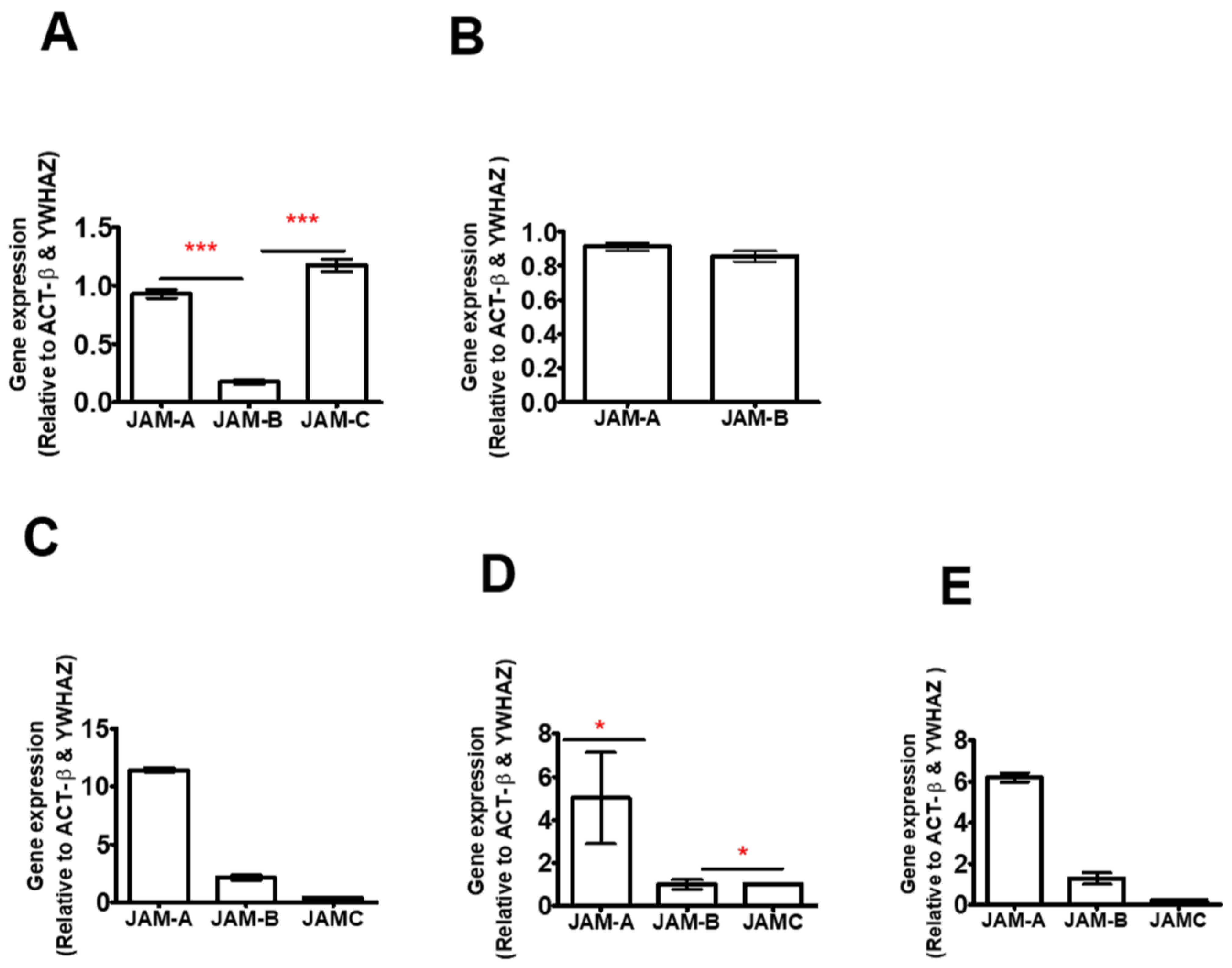
2.1. JAM-B Is Expressed in THP-1 Cells, Human PBMCs and Macrophages at mRNA Level
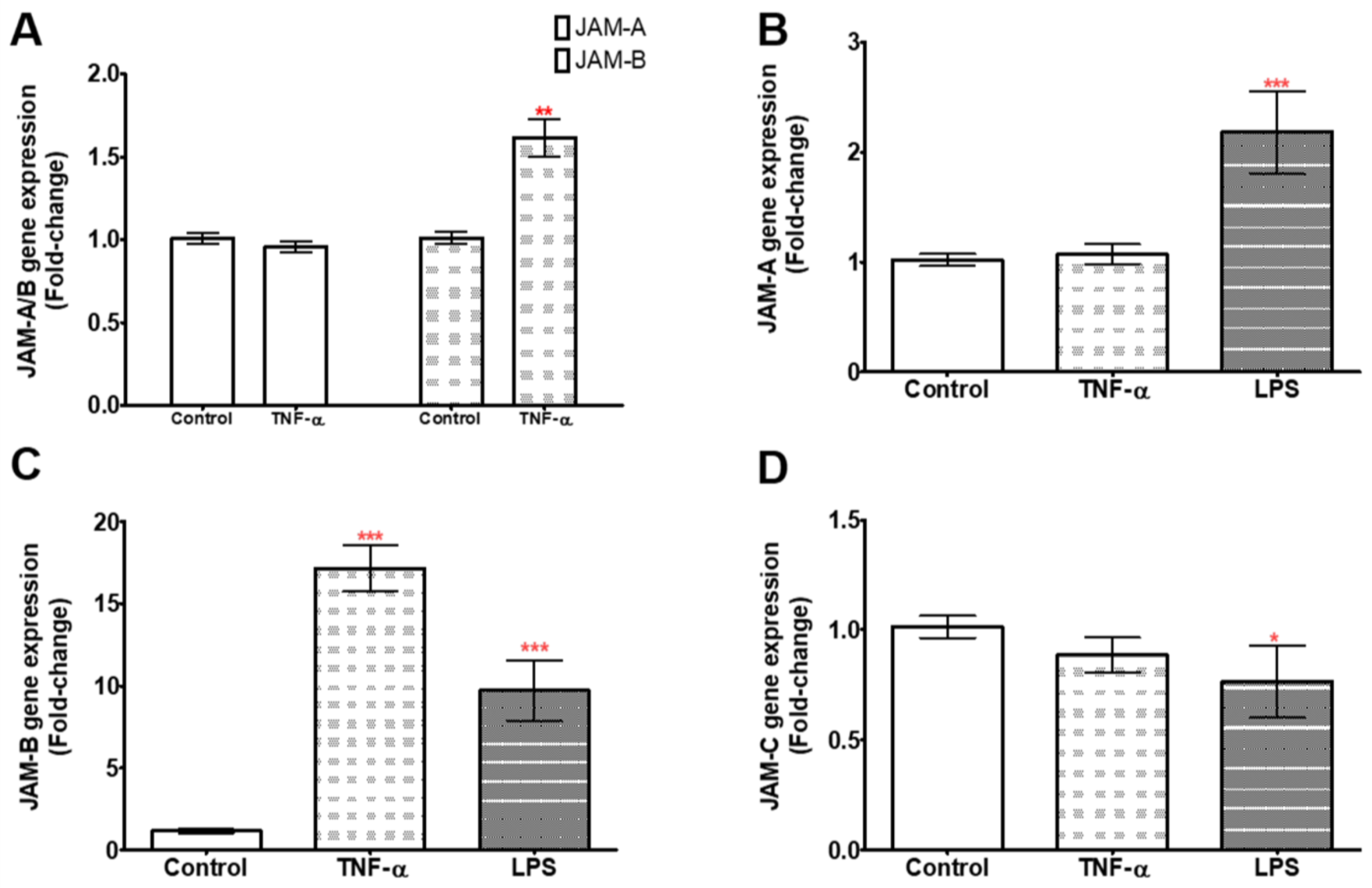
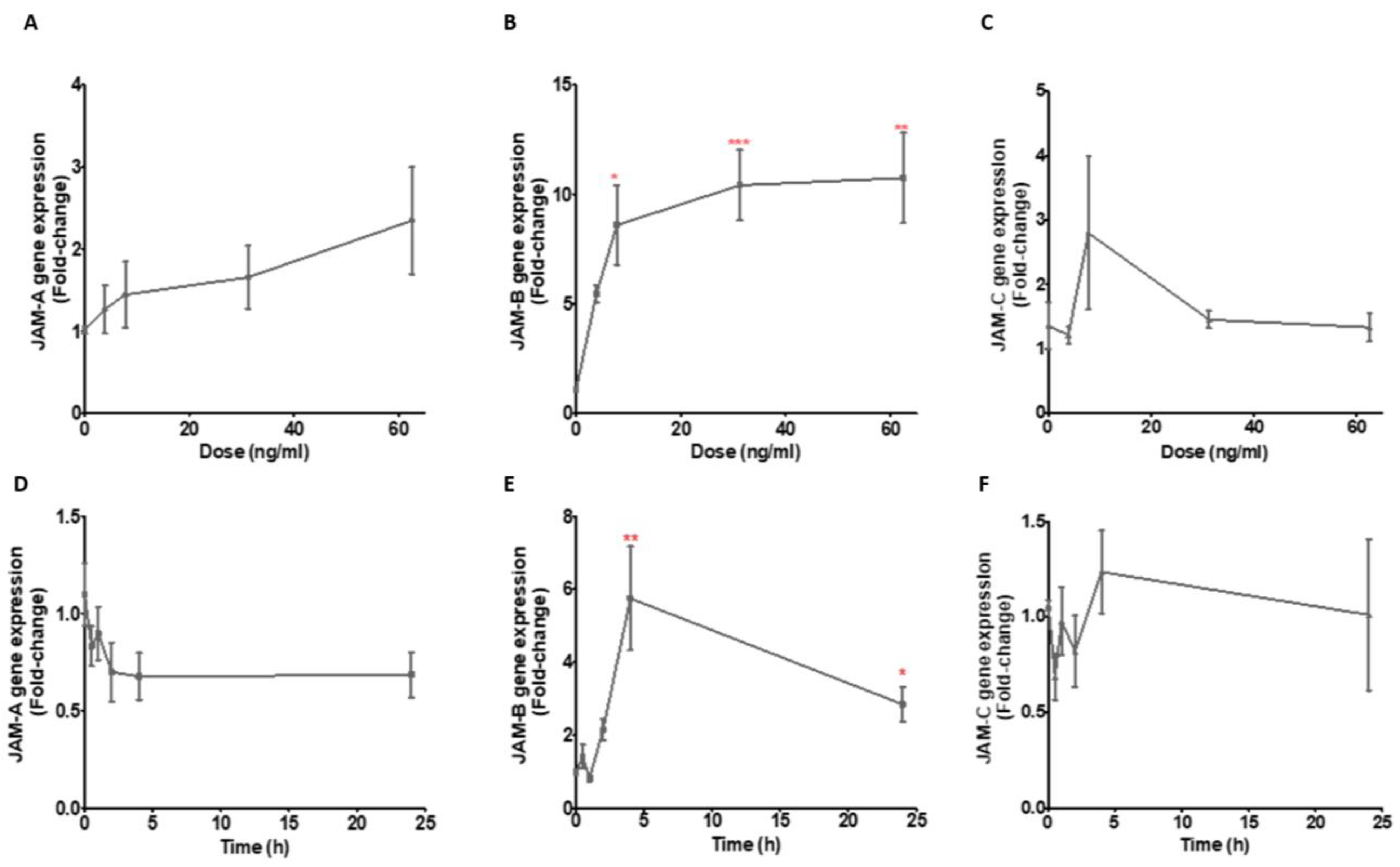
2.2. TNF-α and LPS Up-Regulate JAM-B Gene Expression in THP-1 Monocytes and PMA Differentiated THP-1 Macrophages
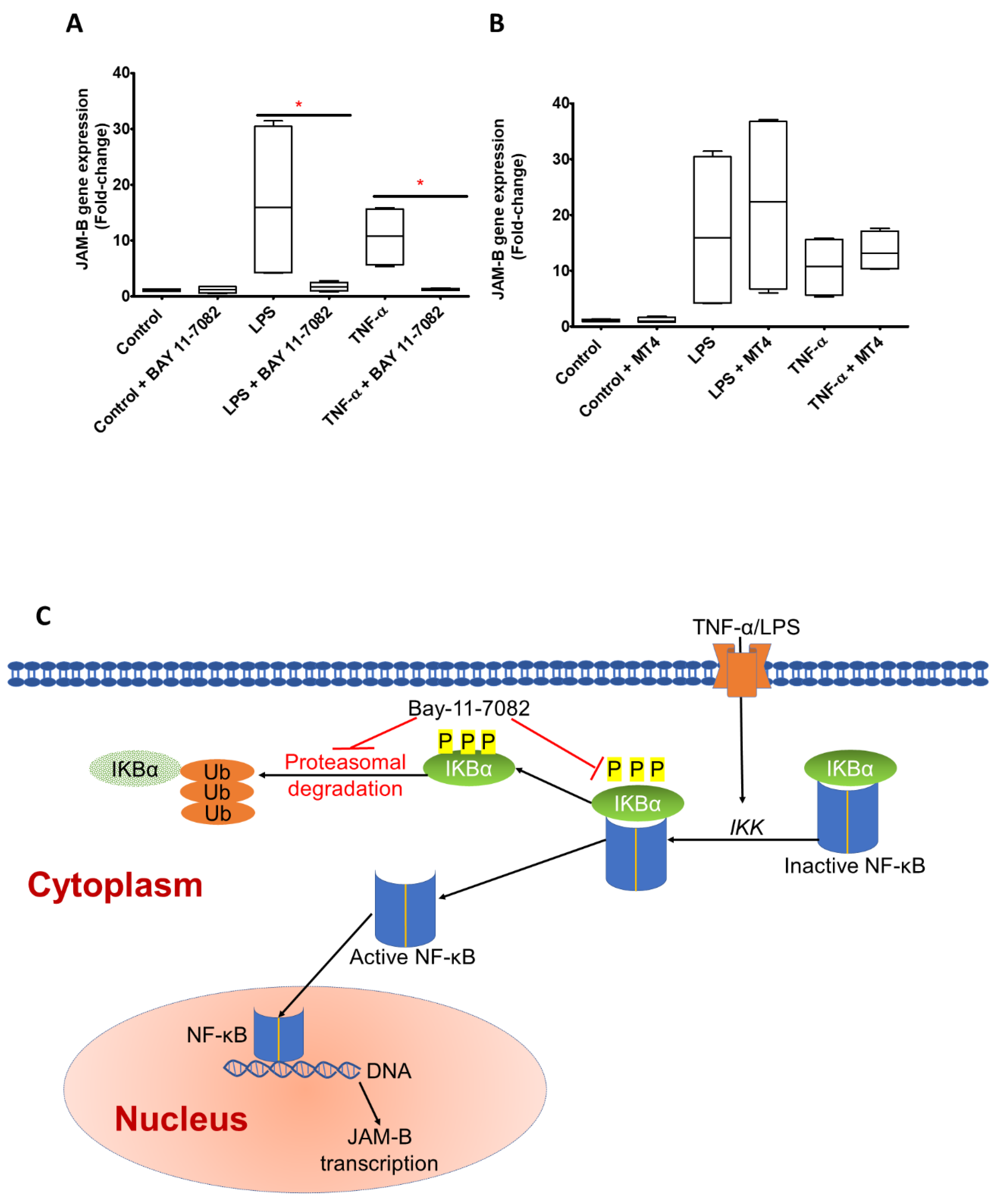
2.3. JAM-B Gene Expression Is Regulated by Nuclear Factor-κB (NF-κB)
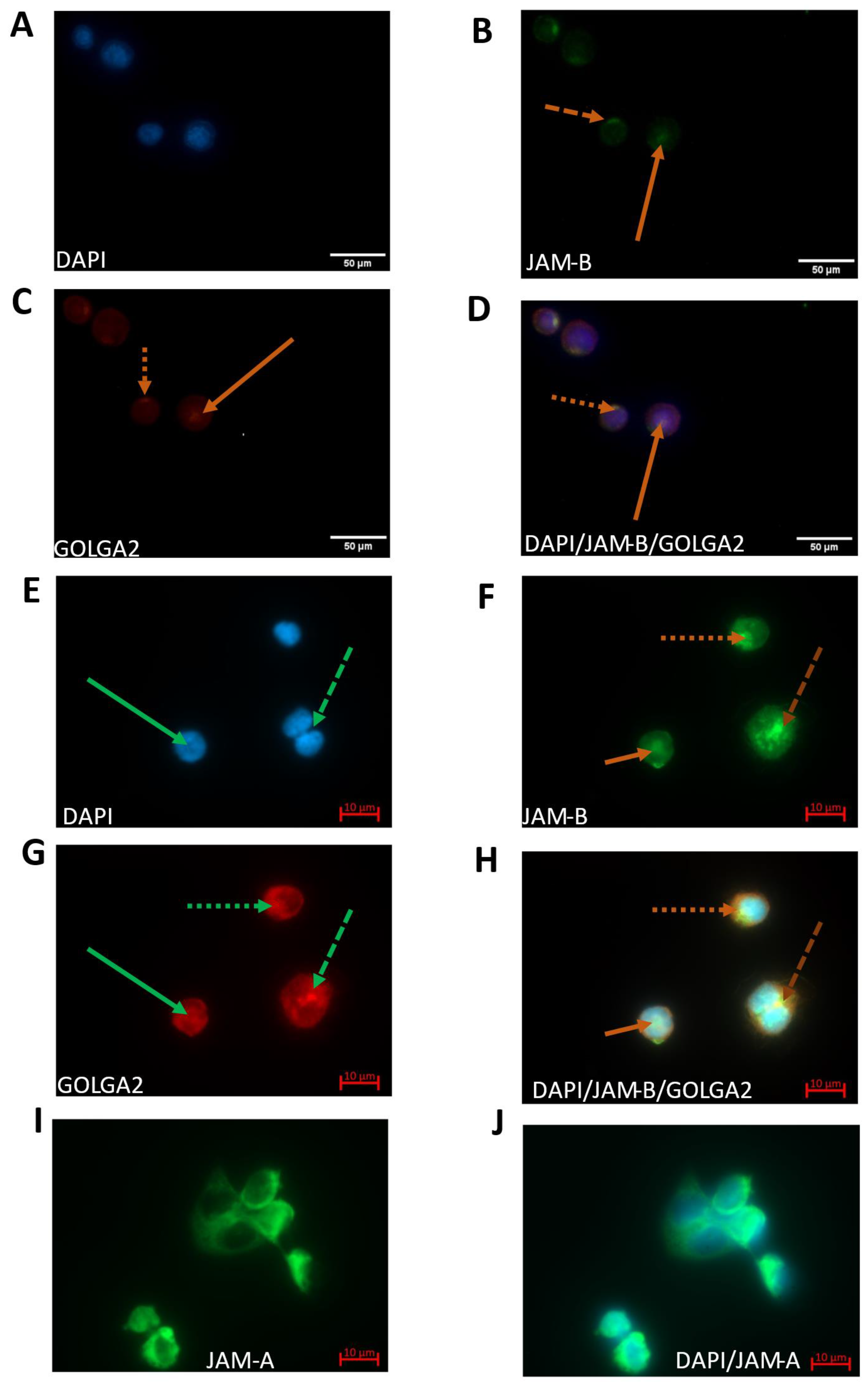
2.4. JAM-B Has Polarised Localisation at the Cell Surface and Is Localised to the Cis-Golgi Network as Well as the Cytoplasm and the Nucleus
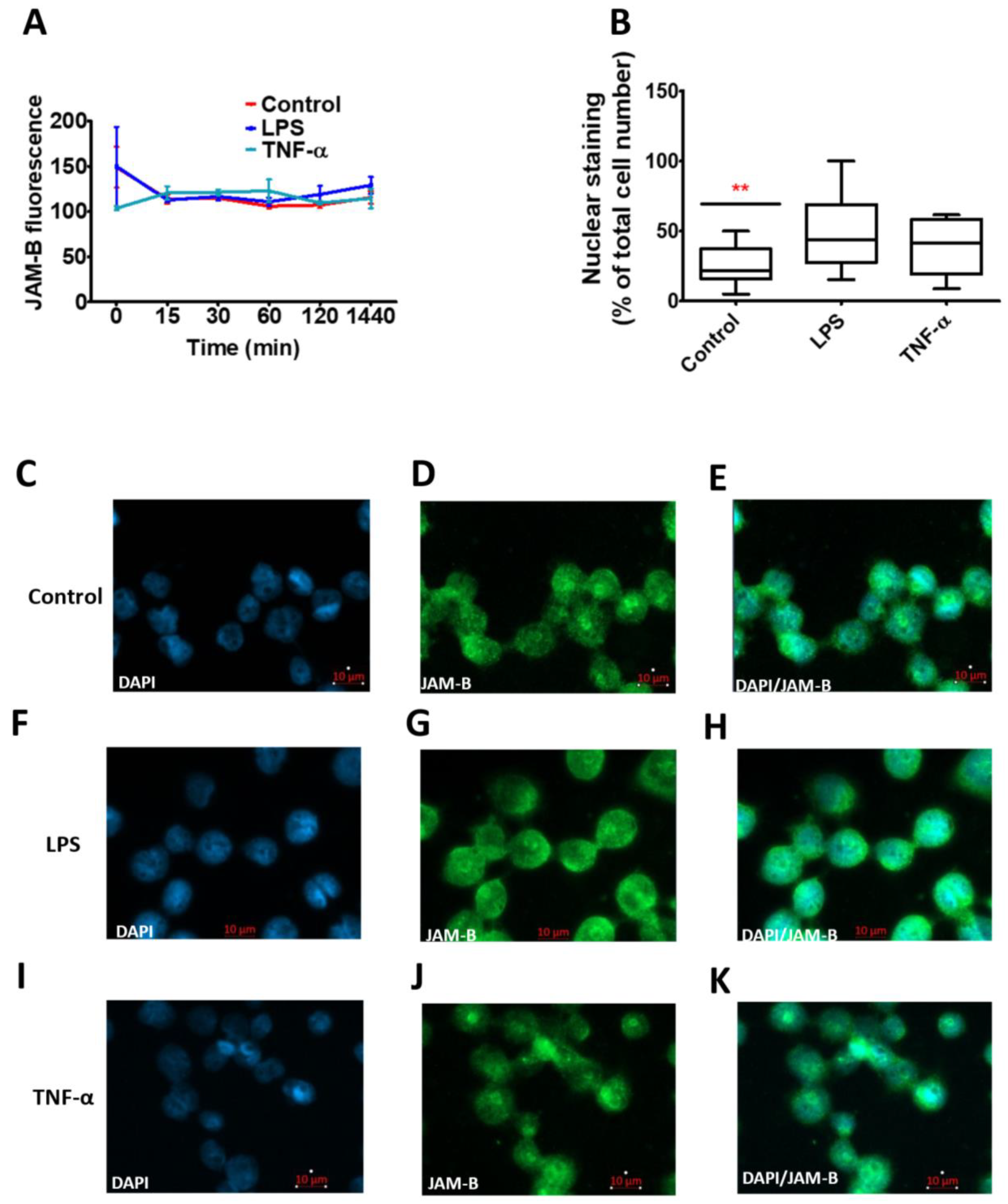
2.5. Total JAM-B Protein Fluorescence Is Not Affected by Inflammatory Stimuli, but JAM-B Nuclear Localisation Is Enhanced by Inflammatory Stimuli and Inhibition of Poly-Ubiquitination
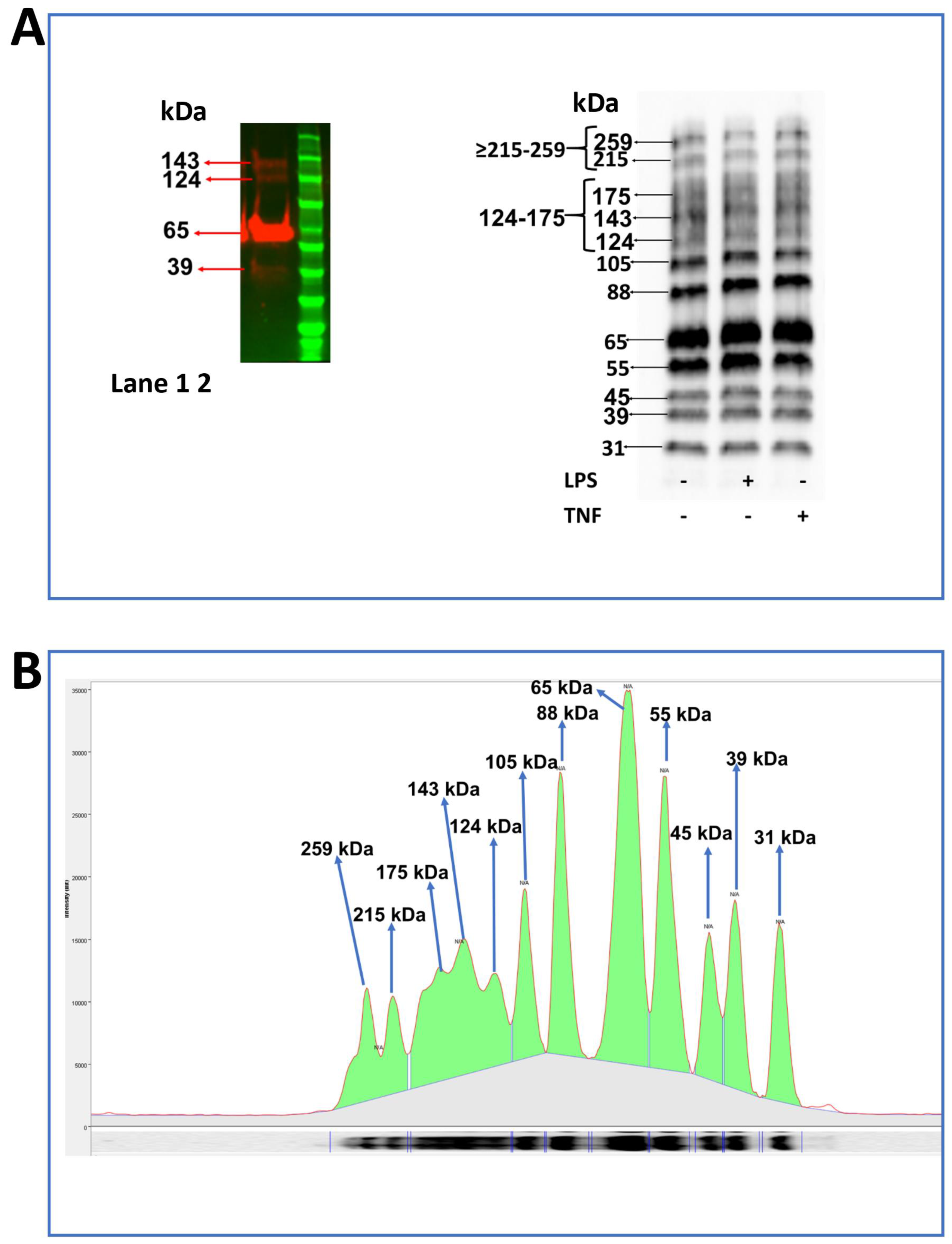
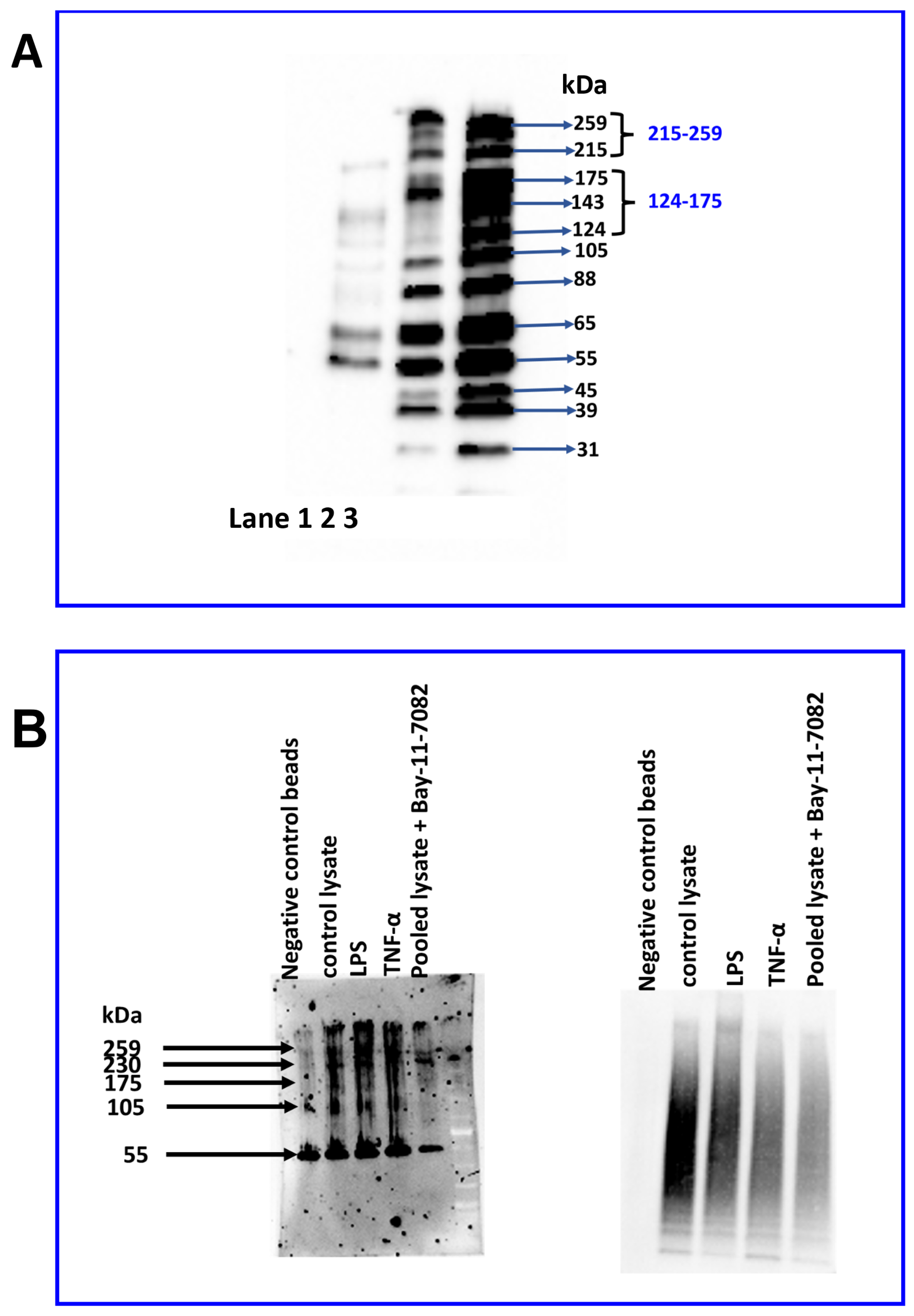
2.6. Identification of JAM-B Protein in THP-1 Monocytes Using Immunoblotting
2.7. Distinct Expression Profile of JAM-B Protein Species in Sub-Cellular THP-1 Cell Compartments and JAM-B Post-Translational Ubiquitination
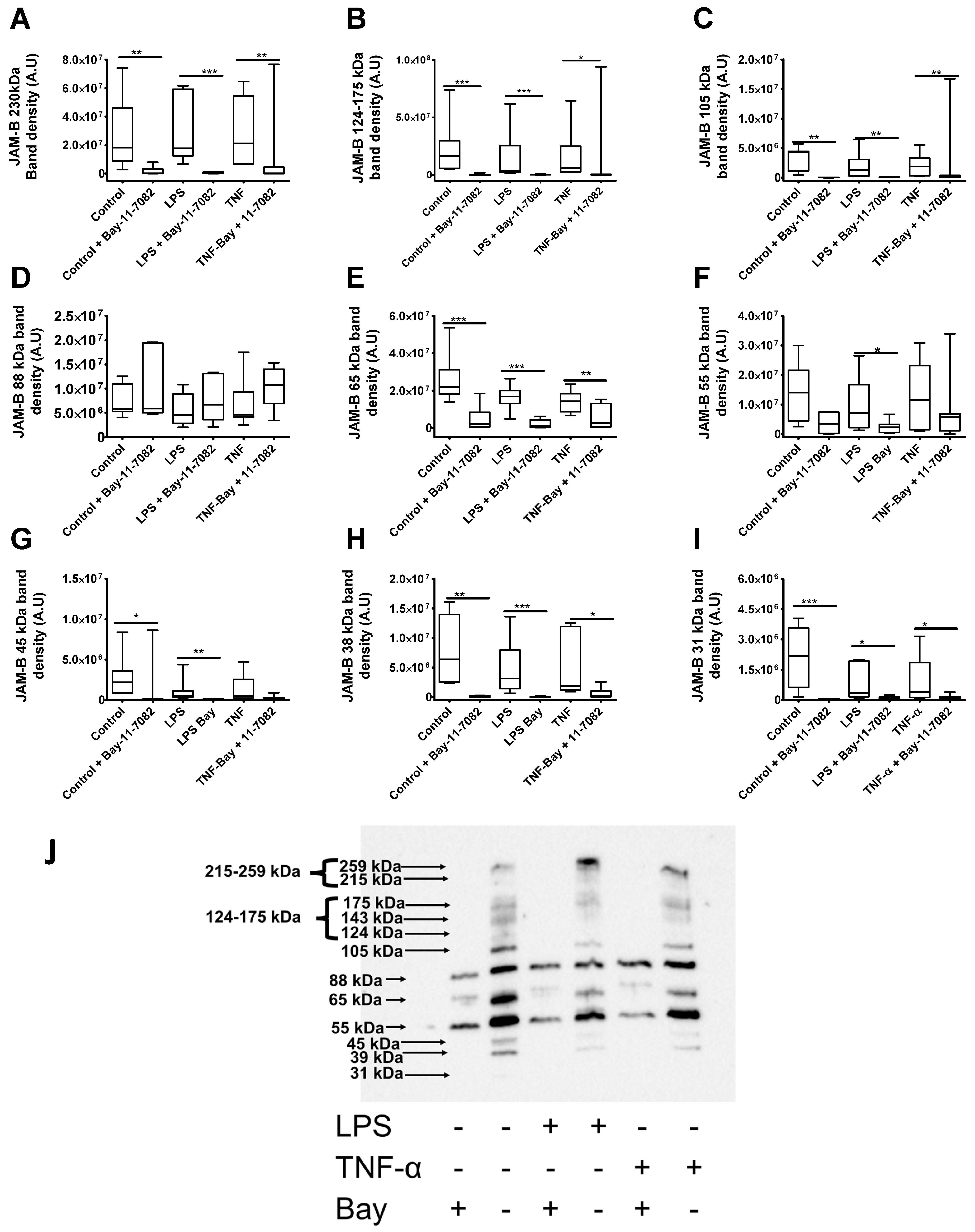
2.8. LPS Treatment Reduces the Expression of the 88 and 31 kDa JAM-B Protein Species
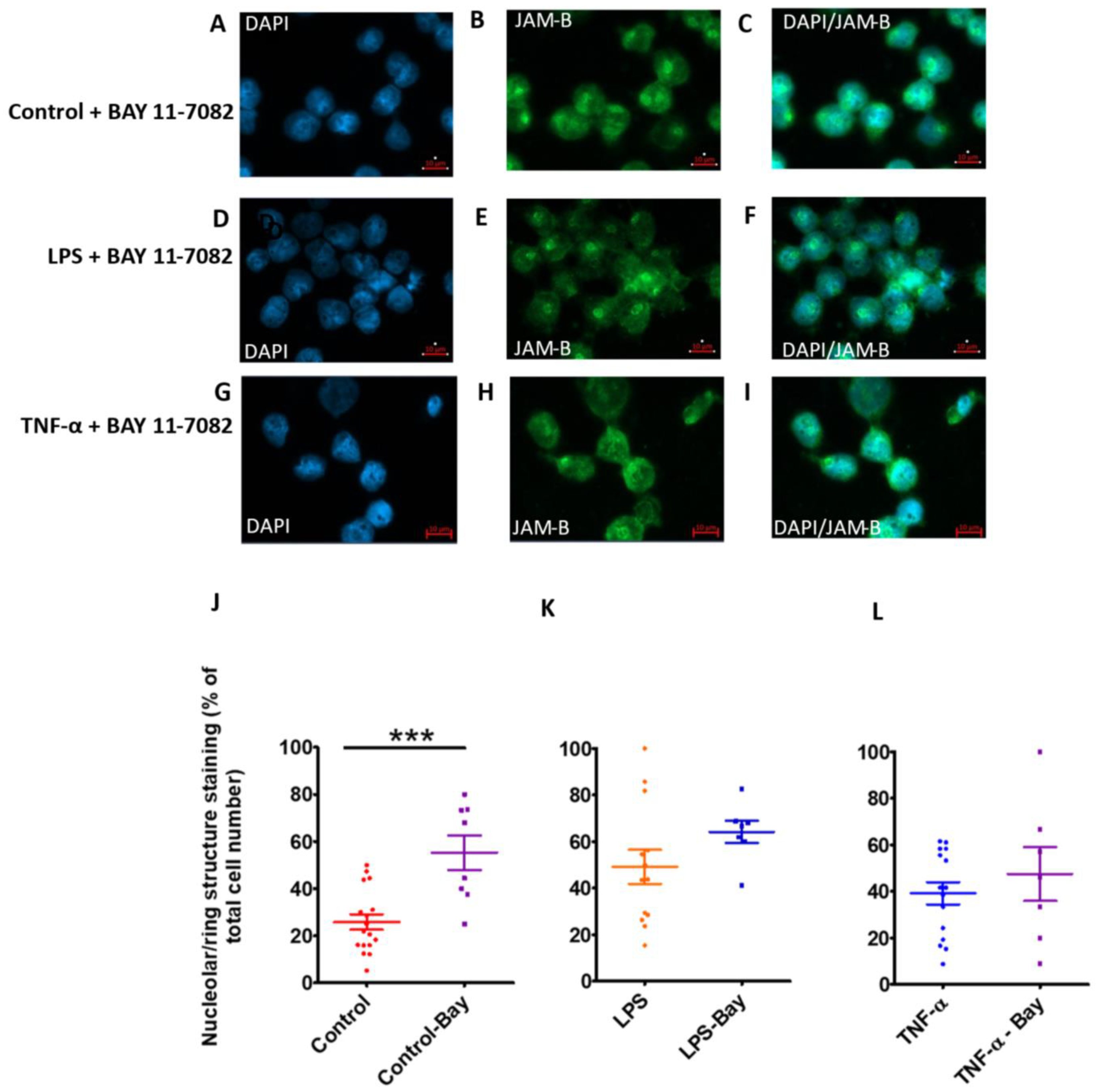
2.9. Bay-11-7082 Increases the Abundance of the 88 kDa JAM-B Species and Reduces the Abundance of Other Higher and Lower Mass JAM-B Species
3. Materials and Methods
3.1. Cell Culture and Treatments
3.2. Isolation of Human PBMCs and Primary Leukocytes
3.3. RNA Extraction
3.4. Quantitative PCR
3.5. Immunostaining
3.6. Protein Extraction and Western Blotting
3.7. Ubiquitin Enrichment and Immunoprecipitation
3.8. Nuclear Export and Localisation and Motif Prediction
3.9. Statistics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sugano, Y.; Takeuchi, M.; Hirata, A.; Matsushita, H.; Kitamura, T.; Tanaka, M.; Miyajima, A. Junctional adhesion molecule-A, JAM-A, is a novel cell-surface marker for long-term repopulating hematopoietic stem cells. Blood 2008, 111, 1167–1172. [Google Scholar] [CrossRef]
- Li, X.; Stankovic, M.; Lee, B.P.-L.; Aurrand-Lions, M.; Hahn, C.N.; Lu, Y.; Imhof, B.A.; Vadas, M.A.; Gamble, J.R. JAM-C induces endothelial cell permeability through its association and regulation of β3 integrins. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 1200–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vonlaufen, A.; Aurrand-Lions, M.; Pastor, C.; Lamagna, C.; Hadengue, A.; Imhof, B.; Frossard, J.-L. The role of junctional adhesion molecule C (JAM-C) in acute pancreatitis. J. Pathol. 2006, 209, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Barton, E.S.; Forrest, J.C.; Connolly, J.L.; Chappell, J.D.; Liu, Y.; Schnell, F.J.; Nusrat, A.; Parkos, C.A.; Dermody, T.S. Junction adhesion molecule is a receptor for reovirus. Cell 2001, 104, 441–451. [Google Scholar] [CrossRef] [Green Version]
- Williams, L.; Martin-Padura, I.; Dejana, E.; Hogg, N.; Simmons, D. Identification and characterisation of human Junctional Adhesion Molecule (JAM). Mol. Immunol. 1999, 36, 1175–1188. [Google Scholar] [CrossRef]
- Kummer, D.; Ebnet, K. Junctional adhesion molecules (JAMs): The JAM-integrin connection. Cells 2018, 7, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Maschio, A.; De Luigi, A.; Martin-Padura, I.; Brockhaus, M.; Bartfai, T.; Fruscella, P.; Adorini, L.; Martino, G.; Furlan, R.; de Simoni, M.G.; et al. Leukocyte recruitment in the cerebrospinal fluid of mice with experimental meningitis is inhibited by an antibody to junctional adhesion molecule (JAM). J. Exp. Med. 1999, 190, 1351–1356. [Google Scholar] [CrossRef] [PubMed]
- Koval, M. Chapter 1—Junctional interplay in lung epithelial barrier function. In Lung Epithelial Biology in the Pathogenesis of Pulmonary Disease; Sidhaye, V.K., Koval, M., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 1–20. [Google Scholar]
- Chopyk, D.M.; Kumar, P.; Raeman, R.; Liu, Y.; Smith, T.; Anania, F.A. Dysregulation of junctional adhesion molecule—A contributes to ethanol-induced barrier disruption in intestinal epithelial cell monolayers. Physiol. Rep. 2017, 5, e13541. [Google Scholar] [CrossRef] [Green Version]
- Arrate, M.P.; Rodriguez, J.M.; Tran, T.M.; Brock, T.A.; Cunningham, S.A. Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J. Biol. Chem. 2001, 276, 45826–45832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazzoni, G.; Martinez-Estrada, O.M.; Mueller, F.; Nelboeck, P.; Schmid, G.; Bartfai, T.; Dejana, E.; Brockhaus, M. Hemophilic interaction of junctional adhesion molecule. J. Biol. Chem. 2000, 275, 30970–30976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozaki, H.; Ishiia, K.; Arai, H.; Horiuchia, H.; Kawamotoa, T.; Suzukib, H.; Kitaa, T. Junctional Adhesion Molecule (JAM) is phosphorylated by protein kinase C upon platelet activation. Biochem. Biophys. Res. Commun. 2000, 276, 873–878. [Google Scholar] [CrossRef]
- Martìn-Padura, I.; Lostaglio, S.; Schneemann, M.; Williams, L.; Romano, M.; Fruscella, P.; Panzeri, M.C.; Stoppacciaro, A.; Ruco, L.; Villa, A.; et al. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 1998, 142, 117–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, S.A.; Arrate, M.P.; Rodriguez, J.M.; Bjercke, R.J.; Vanderslice, P.; Morris, A.P.; Brock, T.A. A novel protein with homology to the junctional adhesion molecule—Characterization of leukocyte interactions. J. Biol. Chem. 2000, 275, 34750–34756. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, H.; Ishii, K.; Kawamoto, T.; Ashida, N.; Arai, H.; Kita, T. Phosphorylation of junctional adhesion molecule in activated platelets. Circulation 1999, 100, 404. [Google Scholar]
- Gupta, S.K.; Pillarisetti, K.; Ohlstein, E.H. Platelet agonist F11 receptor is a member of the immunoglobulin superfamily and identical with junctional adhesion molecule (JAM): Regulation of expression in human endothelial cells and macrophages. IUBMB Life 2000, 50, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Ebnet, K.; Suzuki, A.; Ohno, S.; Vestweber, D. Junctional adhesion molecules (JAMs): More molecules with dual functions? J. Cell Sci. 2004, 117, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Luissint, A.-C.; Nusrat, A.; Parkos, C.A. JAM-related proteins in mucosal homeostasis and inflammation. Semin. Immunopathol. 2014, 36, 211–226. [Google Scholar] [CrossRef] [Green Version]
- Stelzer, I.A.; Mori, M.; DeMayo, F.; Lydon, J.; Arck, P.C.; Solano, M.E. Differential mouse-strain specific expression of Junctional Adhesion Molecule (JAM)-B in placental structures. Cell Adhes. Migr. 2016, 10, 2–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.N.; He, R.; Chen, S.; Zhang, L.; Cao, G.; Yang, W.; Li, J. The JAM-B/c-src/MMP9 pathway is associated with progression and regulates the invasion of pancreatic cancer. J. Cancer 2020, 11, 3246–3255. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Yu, H.; Martin, T.A.; Teng, X.; Jiang, W.G. The role of JAM-B in cancer and cancer metastasis (Review). Oncol. Rep. 2016, 36, 3–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauko, A.; Mu, Z.; Gutmann, D.H.; Naik, U.P.; Lathia, J.D. Junctional adhesion molecules in cancer: A paradigm for the diverse functions of cell–cell interactions in tumor progression. Cancer Res. 2020, 80, 4878–4885. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, S.A.; Rodriguez, J.M.; Arrate, M.P.; Tran, T.M.; Brock, T.A. JAM2 interacts with α4β1—Facilitation by JAM3. J. Biol. Chem. 2002, 277, 27589–27592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, R.J.; Hardt, K.; Hatting, M.; Bistrian, R.; Diehl, S.; Radeke, H.H.; Podda, M.; Schön, M.P.; Kaufmann, R.; Henschler, R.; et al. Junctional adhesion molecule (JAM)-B supports lymphocyte rolling and adhesion through interaction with α4β1 integrin. Immunology 2009, 128, 196–205. [Google Scholar] [CrossRef]
- Yang, N.; Sin, D.D.; Dorscheid, D.R. Various factors affect lipopolysaccharide sensitization in cell cultures. BioTechniques 2020, 69, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.W.; Schoenleber, R.; Jesmok, G.; Best, J.; Moore, S.A.; Collins, T.; Gerritsen, M.E. Novel inhibitors of cytokine-induced IκBα phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J. Biol. Chem. 1997, 272, 21096–21103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Liu, Z.; Xia, Y.; Xu, J.; Lv, G.; Wang, L.; Ma, T.; Jiang, L.; Mou, Y.; Jiang, X.; et al. HOXB7 overexpression promotes cell proliferation and correlates with poor prognosis in gastric cancer patients by inducing expression of both AKT and MARKs. Oncotarget 2017, 8, 1247–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juliana, C.; Fernandes-Alnemri, T.; Wu, J.; Datta, P.; Solorzano, L.; Yu, J.-W.; Meng, R.; Quong, A.A.; Latz, E.; Scott, C.P.; et al. Anti-inflammatory compounds parthenolide and bay 11-7082 are direct inhibitors of the inflammasome. J. Biol. Chem. 2010, 285, 9792–9802. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.J. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 2005, 7, 758–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, C.J. The structure of the mitotic spindle and nucleolus during mitosis in the amebo-flagellate naegleria. PLoS ONE 2012, 7, e34763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abadía-Molina, F.; Calvente, V.M.; Baird, S.D.; Shamim, F.; Martin, F.; MacKenzie, A. Neuronal apoptosis inhibitory protein (NAIP) localizes to the cytokinetic machinery during cell division. Sci. Rep. 2017, 7, 39981. [Google Scholar] [CrossRef] [Green Version]
- Lordier, L.; Chang, Y.; Jalil, A.; Aurade, F.; Garçon, L.; Lécluse, Y.; Larbret, F.; Kawashima, T.; Kitamura, T.; Larghero, J.; et al. Aurora B is dispensable for megakaryocyte polyploidization, but contributes to the endomitotic process. Blood 2010, 116, 2345–2355. [Google Scholar] [CrossRef]
- Lonard, D.M.; O’Malley, B.W. Chapter 4. Emerging roles of the ubiquitin proteasome system in nuclear hormone receptor signaling. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2009; Volume 87, pp. 117–135. [Google Scholar] [CrossRef]
- Von Mikecz, A. The nuclear ubiquitin-proteasome system. J. Cell Sci. 2006, 119, 1977–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R & D Systems. Recombinant Human JAM-B/VE-JAM Fc Chimera. 02/6/2018 Product Datasheet. 2018. Available online: https://resources.rndsystems.com/pdfs/datasheets/1074-vj.pdf?v=20210505&_ga=2.217420374.1550705995.1620203287-255883908.1570118152 (accessed on 5 May 2021).
- ProSci. JAM2 Antibody, Cat. No.: 15-397. Antibody Datasheet. 2021. Available online: https://www.prosci-inc.com/ProductLeaflet/file/getpdf/name/JAM2_Antibody.pdf?fileId=110121 (accessed on 5 May 2021).
- Strickson, S.; Campbell, D.G.; Emmerich, C.H.; Knebel, A.; Plater, L.; Ritorto, M.S.; Shpiro, N.; Cohen, P. The anti-inflammatory drug BAY 11-7082 suppresses the MyD88-dependent signalling network by targeting the ubiquitin system. Biochem. J. 2013, 451, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Day, P.; Burrows, L.; Richards, D.; Fountain, S.J. Inhibitors of DAG metabolism suppress CCR2 signalling in human monocytes. Br. J. Pharmacol. 2019, 176, 2736–2749. [Google Scholar] [CrossRef]
- Layhadi, J.A.; Fountain, S.J. ATP-evoked intracellular Ca2+ responses in M-CSF differentiated human monocyte-derived macrophage are mediated by P2X4 and P2Y11 receptor activation. Int. J. Mol. Sci. 2019, 20, 5113. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Miranda, J.; Martín-Tapia, D.; Valdespino-Vázquez, Y.; Alarcón, L.; Espejel-Nuñez, A.; Guzmán-Huerta, M.; Muñoz-Medina, J.E.; Shibayama, M.; Chávez-Munguía, B.; Estrada-Gutiérrez, G.; et al. Syncytiotrophoblast of placentae from women with zika virus infection has altered tight junction protein expression and increased paracellular permeability. Cells 2019, 8, 1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redmond, S.A.; Mei, F.; Eshed-Eisenbach, Y.; Osso, L.A.; Leshkowitz, D.; Shen, Y.-A.A.; Kay, J.N.; Aurrand-Lions, M.; Lyons, D.A.; Peles, E.; et al. Somatodendritic expression of JAM2 inhibits oligodendrocyte myelination. Neuron 2016, 91, 824–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Cour, T.; Kiemer, L.; Mølgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.; Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Čalyševa, J.; et al. ELM—The eukaryotic linear motif resource in 2020. Nucleic Acids Res. 2020, 48, D296–D306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Flores, J.M.; Silva-Ayala, D.; Espinoza, M.A.; López, S.; Arias, C.F. The tight junction protein JAM-A functions as coreceptor for rotavirus entry into MA104 cells. Virology 2015, 475, 172–178. [Google Scholar] [CrossRef] [Green Version]
- Ueki, T.; Iwasawa, K.; Ishikawa, H.; Sawa, Y. Expression of junctional adhesion molecules on the human lymphatic endothelium. Microvasc. Res. 2008, 75, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Bryan, N.B.; Dorfleutner, A.; Rojanasakul, Y.; Stehlik, C. Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J. Immunol. 2009, 182, 3173–3182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filardy, A.; He, J.; Bennink, J.R.; Yewdell, J.W.; Kelsall, B.L. Posttranscriptional control of NLRP3 inflammasome activation in colonic macrophages. Mucosal Immunol. 2016, 9, 850–858. [Google Scholar] [CrossRef] [Green Version]
- Fehr, A.R.; Yu, D. Control the host cell cycle: Viral regulation of the anaphase-promoting complex. J. Virol. 2013, 87, 8818–8825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtake, F.; Saeki, Y.; Ishido, S.; Kanno, J.; Tanaka, K. The K48–K63 branched ubiquitin chain regulates NF-κB signaling. Mol. Cell 2016, 64, 251–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene | Forward Primer | Reverse Primer | Product Size (bp) |
|---|---|---|---|
| JAM-A | 5′GGGTGACCTTCTTGCCAACT′3 | 5′GATGGAGGCACAAGCACGAT′3 | 142 |
| JAM-B | 5′AGGCCTATGGGTTTTCTGCC′3 | 5′CAAAGGAGACACTCCGACCC′3 | 144 |
| JAM-B | 5′AATTCAGGGAGACTTGGCGG′3 | 5′TTTCGAGCAACGACCTCACA′3 | 114 |
| YWHAZ | 5′GCAATTACTGAGAGACAACTTGACA′3 | 5′TGGAAGGCCGGTTAATTTT′3 | 96 |
| ACT-β | 5′GCACCCAGCACAATGAAGA′3 | 5′CGATCCACACGGAGTACTTG′3 | 64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Day-Walsh, P.E.; Keeble, B.; Pirabagar, G.; Fountain, S.J.; Kroon, P.A. Transcriptional and Post-Translational Regulation of Junctional Adhesion Molecule-B (JAM-B) in Leukocytes under Inflammatory Stimuli. Int. J. Mol. Sci. 2022, 23, 8646. https://doi.org/10.3390/ijms23158646
Day-Walsh PE, Keeble B, Pirabagar G, Fountain SJ, Kroon PA. Transcriptional and Post-Translational Regulation of Junctional Adhesion Molecule-B (JAM-B) in Leukocytes under Inflammatory Stimuli. International Journal of Molecular Sciences. 2022; 23(15):8646. https://doi.org/10.3390/ijms23158646
Chicago/Turabian StyleDay-Walsh, Priscilla E., Bryony Keeble, Gothai Pirabagar, Samuel J. Fountain, and Paul A. Kroon. 2022. "Transcriptional and Post-Translational Regulation of Junctional Adhesion Molecule-B (JAM-B) in Leukocytes under Inflammatory Stimuli" International Journal of Molecular Sciences 23, no. 15: 8646. https://doi.org/10.3390/ijms23158646






