Effect of CB2 Stimulation on Gene Expression in Pediatric B-Acute Lymphoblastic Leukemia: New Possible Targets
Abstract
:1. Introduction
2. Results
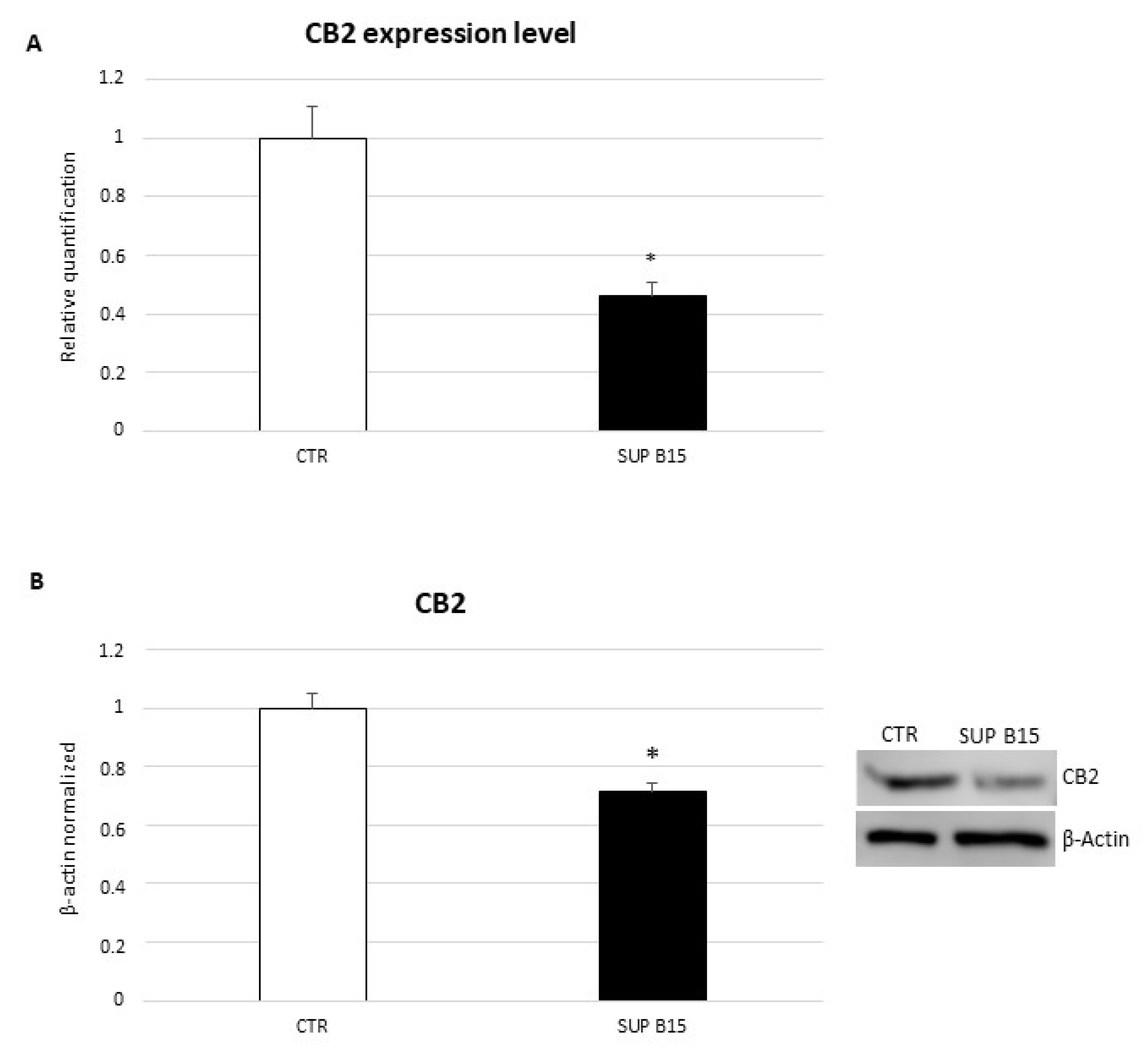
2.1. CB2 Receptor Expression in SUP-B15 Cell Line
2.2. Effect of CB2 Stimulation on Gene Expression: RNA-Sequencing Analysis
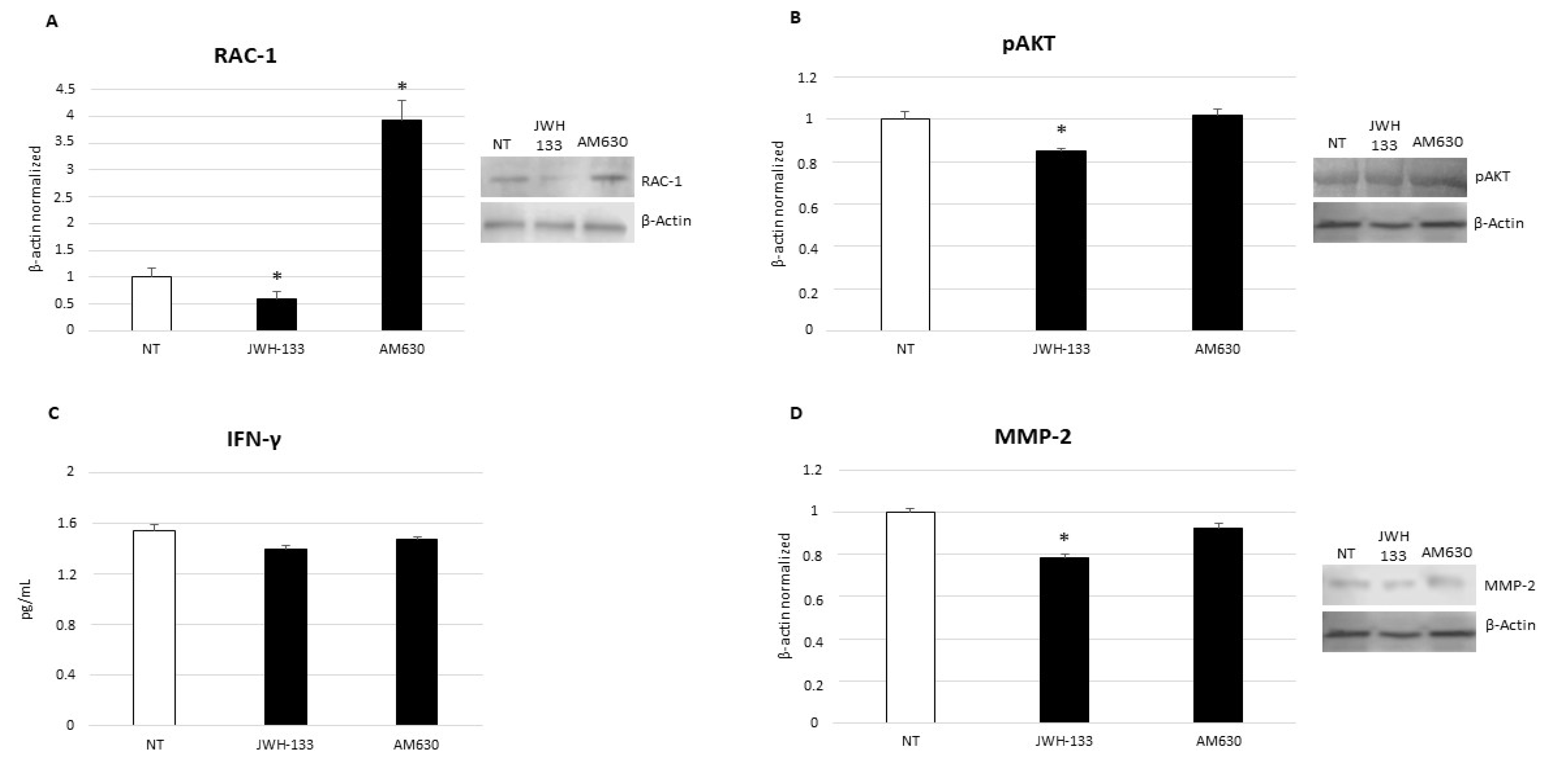
2.3. Effects of CB2 Receptor Stimulation on CD9-RAC1 Signaling Pathway
2.4. Effects of CB2 Receptor Stimulation on Leukemia Cell Proliferation
2.5. Effects of CB2 Receptor Stimulation on Leukemia Cell Survival
2.6. Effects of CB2 Receptor Stimulation on Invasion Capacity of Leukemia Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Drugs and Treatments
4.3. RNA Isolation and Real Time PCR
4.4. RNA-Sequencing Library Preparation and Sequencing
4.5. RNA-Sequencing Data Analysis
4.6. Protein Extraction and Western Blotting
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Count and Viability Assay
4.9. Annexin V & Dead Cell Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, H.; Yu, H.; Jin, R.M.; Wu, X.Y.; Chen, H.B. Genetic and Epigenetic Targeting Therapy for Pediatric Acute Lymphoblastic Leukemia. Cells 2021, 10, 3349. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Wang, B.Y.; Zhang, W.N.; Huang, J.Y.; Li, B.S.; Zhang, M.; Jiang, L.; Li, J.F.; Wang, M.J.; Dai, Y.J.; et al. Genomic Profiling of Adult and Pediatric B-cell Acute Lymphoblastic Leukemia. Ebiomedicine 2016, 8, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Muntada, M.; Trincado, J.L.; Blanco, J.; Bueno, C.; Rodriguez-Cortez, V.C.; Bataller, A.; Lopez-Millan, B.; Schwab, C.; Ortega, M.; Velasco, P.; et al. Clonal heterogeneity and rates of specific chromosome gains are risk predictors in childhood high-hyperdiploid B-cell acute lymphoblastic leukemia. Mol. Oncol. 2022. [Google Scholar] [CrossRef]
- Inaba, H.; Mullighan, C.G. Pediatric acute lymphoblastic leukemia. Haematologica 2020, 105, 2524–2539. [Google Scholar] [CrossRef]
- Teachey, D.T.; Hunger, S.P.; Loh, M.L. Optimizing therapy in the modern age: Differences in length of maintenance therapy in acute lymphoblastic leukemia. Blood 2021, 137, 168–177. [Google Scholar] [CrossRef]
- Rossi, F.; Di Paola, A.; Pota, E.; Argenziano, M.; Di Pinto, D.; Marrapodi, M.M.; Di Leva, C.; Di Martino, M.; Tortora, C. Biological Aspects of Inflamm-Aging in Childhood Cancer Survivors. Cancers 2021, 13, 4933. [Google Scholar] [CrossRef]
- Gupta, V.; Dash, S.; Aggarwal, P.; Singh, S.K. Alterations in Bone Turnover during Chemotherapy in Children with Acute Lymphoblastic Leukemia. South Asian J. Cancer 2021, 10, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Ibrahimova, A.; Pommert, L.; Breese, E.H. Acute Leukemia in Infants. Curr. Oncol. Rep. 2021, 23, 27. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.C.; Cleary, M.L. Novel methods and approaches to acute lymphoblastic leukemia drug discovery. Expert Opin. Drug Dis. 2014, 9, 1435–1446. [Google Scholar] [CrossRef]
- Cooper, S.L.; Brown, P.A. Treatment of Pediatric Acute Lymphoblastic Leukemia. Pediatr. Clin. N. Am. 2015, 62, 61–73. [Google Scholar] [CrossRef] [Green Version]
- Teachey, D.T.; Pui, C.H. Comparative features and outcomes between paediatric T-cell and B-cell acute lymphoblastic leukaemia. Lancet Oncol. 2019, 20, E142–E154. [Google Scholar] [CrossRef]
- Dreyer, Z.E.; Hilden, J.M.; Jones, T.L.; Devidas, M.; Winick, N.J.; Willman, C.L.; Harvey, R.C.; Chen, I.M.; Behm, F.G.; Pullen, J.; et al. Intensified Chemotherapy Without Sct in Infant All: Results From COG P9407 (Cohort 3). Pediatr. Blood Cancer 2015, 62, 419–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabissi, M.; Morelli, M.B.; Offidani, M.; Amantini, C.; Gentili, S.; Soriani, A.; Cardinali, C.; Leoni, P.; Santoni, G. Cannabinoids synergize with carfilzomib, reducing multiple myeloma cells viability and migration. Oncotarget 2016, 7, 77543–77557. [Google Scholar] [CrossRef] [Green Version]
- Kampa-Schittenhelm, K.M.; Salitzky, O.; Akmut, F.; Illing, B.; Kanz, L.; Salih, H.R.; Schittenhelm, M.M. Dronabinol has preferential antileukemic activity in acute lymphoblastic and myeloid leukemia with lymphoid differentiation patterns. BMC Cancer 2016, 16, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Punzo, F.; Tortora, C.; Di Pinto, D.; Manzo, I.; Bellini, G.; Casale, F.; Rossi, F. Anti-proliferative, pro-apoptotic and anti-invasive effect of EC/EV system in human Osteosarcoma. Oncotarget 2017, 8, 54459–54471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Punzo, F.; Tortora, C.; Di Pinto, D.; Pota, E.; Argenziano, M.; Di Paola, A.; Casale, F.; Rossi, F. Bortezomib and endocannabinoid/endovanilloid system: A synergism in osteosarcoma. Pharmacol. Res. 2018, 137, 25–33. [Google Scholar] [CrossRef]
- Parsons, L.H.; Hurd, Y.L. Endocannabinoid signalling in reward and addiction. Nat. Rev. Neurosci. 2015, 16, 579–594. [Google Scholar] [CrossRef] [Green Version]
- Rossi, F.; Bellini, G.; Tortora, C.; Bernardo, M.E.; Luongo, L.; Conforti, A.; Starc, N.; Manzo, I.; Nobili, B.; Locatelli, F.; et al. CB2 and TRPV1 receptors oppositely modulate in vitro human osteoblast activity. Pharmacol. Res. 2015, 99, 194–201. [Google Scholar] [CrossRef]
- Bellini, G.; Torella, M.; Manzo, I.; Tortora, C.; Luongo, L.; Punzo, F.; Colacurci, N.; Nobili, B.; Maione, S.; Rossi, F. PKC beta II-mediated cross-talk of TRPV1/CB2 modulates the glucocorticoid-induced osteoclast overactivity. Pharmacol. Res. 2017, 115, 267–274. [Google Scholar] [CrossRef]
- Argenziano, M.; Tortora, C.; Bellini, G.; Di Paola, A.; Punzo, F.; Rossi, F. The Endocannabinoid System in Pediatric Inflammatory and Immune Diseases. Int. J. Mol. Sci. 2019, 20, 5875, Correction in Int. J. Mol. Sci. 2020, 21, 2757. [Google Scholar] [CrossRef] [Green Version]
- Velasco, G.; Sanchez, C.; Guzman, M. Anticancer mechanisms of cannabinoids. Curr. Oncol. 2016, 23, S23–S32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, S.; Liu, Y.; Su, M.; Ling, X.; Liu, F.; Ge, Y.; Bai, M. Combined CB2 receptor agonist and photodynamic therapy synergistically inhibit tumor growth in triple negative breast cancer. Photodiagnosis Photodyn. Ther. 2018, 24, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, K.; Wang, X.; Severa, D.; Eriksson, M.; Kimby, E.; Merup, M.; Christensson, B.; Flygare, J.; Sander, B. Expression of cannabinoid receptors type 1 and type 2 in non-Hodgkin lymphoma: Growth inhibition by receptor activation. Int. J. Cancer 2008, 123, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Soto-Mercado, V.; Mendivil-Perez, M.; Jimenez-Del-Rio, M.; Fox, J.E.; Velez-Pardo, C. Cannabinoid CP55940 selectively induces apoptosis in Jurkat cells and in ex vivo T-cell acute lymphoblastic leukemia through H2O2 signaling mechanism. Leuk. Res. 2020, 95, 106389. [Google Scholar] [CrossRef] [PubMed]
- Khunluck, T.; Lertsuwan, K.; Chutoe, C.; Sooksawanwit, S.; Inson, I.; Teerapornpuntakit, J.; Tohtong, R.; Charoenphandhu, N. Activation of cannabinoid receptors in breast cancer cells improves osteoblast viability in cancer-bone interaction model while reducing breast cancer cell survival and migration. Sci. Rep. 2022, 12, 7398. [Google Scholar] [CrossRef]
- Lee, H.S.; Tamia, G.; Song, H.J.; Amarakoon, D.; Wei, C.I.; Lee, S.H. Cannabidiol exerts anti-proliferative activity via a cannabinoid receptor 2-dependent mechanism in human colorectal cancer cells. Int. Immunopharmacol. 2022, 108, 108865. [Google Scholar] [CrossRef]
- Moreno, E.; Cavic, M.; Krivokuca, A.; Casado, V.; Canela, E. The Endocannabinoid System as a Target in Cancer Diseases: Are We There Yet? Front. Pharmacol. 2019, 10, 339. [Google Scholar] [CrossRef] [Green Version]
- Punzo, F.; Manzo, I.; Tortora, C.; Pota, E.; Angelo, V.; Bellini, G.; Di Paola, A.; Verace, F.; Casale, F.; Rossi, F. Effects of CB2 and TRPV1 receptors’ stimulation in pediatric acute T-lymphoblastic leukemia. Oncotarget 2018, 9, 21244–21258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Xing, H.Y.; Gong, Y.P.; Yang, X.; Guo, Y.; Shan, Q.Q.; Zhou, R.Q. Multidrug resistant gene MDR1 contributes to development of imatinib-resistance in Ph (+) acute lymphoblastic leukemia cell line SUP-BS15RI. J. Sichuan Univ. Med. Sci. Ed. 2012, 43, 657–660, 665. [Google Scholar]
- Arnaud, M.P.; Vallee, A.; Robert, G.; Bonneau, J.; Leroy, C.; Varin-Blank, N.; Rio, A.G.; Troadec, M.B.; Galibert, M.D.; Gandemer, V. CD9, a key actor in the dissemination of lymphoblastic leukemia, modulating CXCR4-mediated migration via RAC1 signaling. Blood 2015, 126, 1802–1812. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.Z.; Chen, Y.P.; Gong, X.C.; Guo, Q.J.; Lin, C.Y.; Luo, Q.F.; Tu, Z.W.; Pan, J.J.; Li, J.G. SEC61G overexpression and DNA amplification correlates with prognosis and immune cell infiltration in head and neck squamous cell carcinoma. Cancer Med. 2021, 10, 7847–7862. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, D.M.; Hwang, E.S.; Shahsafaei, A.; Glimcher, L.H. T-bet, a T-cell-associated transcription factor, is expressed in a subset of B-cell lymphoproliferative disorders. Am. J. Clin. Pathol. 2004, 122, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Harashima, A.; Matsuo, Y.; Drexler, H.G.; Okochi, A.; Motoda, R.; Tanimoto, M.; Orita, K. Transcription factor expression in B-cell precursor-leukemia cell lines: Preferential expression of T-bet. Leuk. Res. 2005, 29, 841–848. [Google Scholar] [CrossRef] [Green Version]
- Wirsching, H.G.; Krishnan, S.; Florea, A.M.; Frei, K.; Krayenbuhl, N.; Hasenbach, K.; Reifenberger, G.; Weller, M.; Tabatabai, G. Thymosin beta 4 gene silencing decreases stemness and invasiveness in glioblastoma. Brain 2014, 137, 433–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.Y.; Hou, F.; Zhang, Z.P.; Nuo, M.T.; Liang, H.; Cang, M.; Wang, Z.G.; Wang, X.; Xu, T.; Yan, L.Y.; et al. Role of thymosin beta 4 in hair growth. Mol. Genet. Genom. 2016, 291, 1639–1646. [Google Scholar] [CrossRef]
- Inaba, H.; Pui, C.H. Advances in the Diagnosis and Treatment of Pediatric Acute Lymphoblastic Leukemia. J. Clin. Med. 2021, 10, 1926. [Google Scholar] [CrossRef]
- Tanasi, I.; Ba, I.; Sirvent, N.; Braun, T.; Cuccuini, W.; Ballerini, P.; Duployez, N.; Tanguy-Schmidt, A.; Tamburini, J.; Maury, S.; et al. Efficacy of tyrosine kinase inhibitors in Ph-like acute lymphoblastic leukemia harboring ABL-class rearrangements. Blood 2019, 134, 1351–1355. [Google Scholar] [CrossRef]
- Kapoor, I.; Bodo, J.; Hill, B.T.; Hsi, E.D.; Almasan, A. Targeting BCL-2 in B-cell malignancies and overcoming therapeutic resistance. Cell Death Dis. 2020, 11, 941. [Google Scholar] [CrossRef]
- Wayne, A.S.; Shin-Kashiyama, E.; Sposto, R.; Gaynon, P. Therapeutic Advances in Childhood Leukemia and Lymphoma (TACL): Overview and introduction to the proceedings of the 2016 TACL investigator meeting. Pediatric Hematol. Oncol. 2017, 34, 349–354. [Google Scholar] [CrossRef]
- Inaba, H.; Pui, C.H. Immunotherapy in pediatric acute lymphoblastic leukemia. Cancer Metastasis Rev. 2019, 38, 595–610. [Google Scholar] [CrossRef]
- Ramer, R.; Wittig, F.; Hinz, B. The Endocannabinoid System as a Pharmacological Target for New Cancer Therapies. Cancers 2021, 13, 5701. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.N.; Chen, Z.S.; Braas, D.; Lee, J.W.; Xiao, G.; Geng, H.M.; Cosgun, K.N.; Hurtz, C.; Shojaee, S.; Cazzaniga, V.; et al. Metabolic gatekeeper function of B-lymphoid transcription factors (vol. 542, page 479, 2017). Nature 2018, 558, E5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacher, R.; Chu, L.F.; Argus, C.; Bolin, J.M.; Knight, P.; Thomson, J.A.; Stewart, R.; Kendziorski, C. Enhancing biological signals and detection rates in single-cell RNA-seq experiments with cDNA library equalization. Nucleic Acids Res. 2022, 50, e12. [Google Scholar] [CrossRef] [PubMed]
- Sicking, M.; Zivna, M.; Bhadra, P.; Baresova, V.; Tirincsi, A.; Hadzibeganovic, D.; Hodanova, K.; Vyletal, P.; Sovova, J.; Jedlickova, I.; et al. Phenylbutyrate rescues the transport defect of the Sec61 alpha mutations V67G and T185A for renin. Life Sci. Alliance 2022, 5, e202101150. [Google Scholar] [CrossRef]
- Zoller, M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 2009, 9, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.F.; Cai, S.Y.; Li, R.; Cao, G.B. Cannabinoid receptor 2 agonist attenuates blood-brain barrier damage in a rat model of intracerebral hemorrhage by activating the Rac1 pathway. Int. J. Mol. Med. 2018, 42, 2914–2922. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.M.; Zhou, L.; Killela, P.; Rasheed, A.B.; Di, C.H.; Poe, W.E.; McLendon, R.E.; Bigner, D.D.; Nicchitta, C.; Yan, H. Glioblastoma Proto-oncogene SEC61 gamma Is Required for Tumor Cell Survival and Response to Endoplasmic Reticulum Stress. Cancer Res. 2009, 69, 9105–9111. [Google Scholar] [CrossRef] [Green Version]
- Malfitano, A.M.; Laezza, C.; Pisanti, S.; Manera, C.; Bifulco, M. Immuno-Modulatory Properties of a Quinolin-2-(1H)-on-3-Carboxamide Derivative: Relevance in Multiple Sclerosis. Recent Pat. CNS Drug Discov. 2016, 10, 113–121. [Google Scholar] [CrossRef]
- Tiberi, M.; Evron, T.; Saracini, S.; Boffa, L.; Mercuri, N.B.; Chintalacharuvu, S.R.; Atamas, S.P.; Chiurchiu, V. Potent T cell-mediated anti-inflammatory role of the selective CB2 agonist lenabasum in multiple sclerosis. Neuropathol. Appl. Neurobiol. 2022, 48, e12768. [Google Scholar] [CrossRef]
- Todisco, E.; Gaipa, G.; Biagi, E.; Bonamino, M.; Gramigna, R.; Introna, M.; Biondi, A. CD40 ligand-stimulated B cell precursor leukemic cells elicit interferon-gamma production by autologous bone marrow T cells in childhood acute lymphoblastic leukemia. Leukemia 2002, 16, 2046–2054. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.J.; Kim, J.E.; Kim, M.J.; Kwon, H.J.; Suh, S.W.; Song, H.K.; Kang, T.C. The protective effects of interleukin-18 and interferon-gamma on neuronal damages in the rat hippocampus following status epilepticus. Neuroscience 2010, 170, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.; Khalil, M.; Abdellateif, M.S.; Ebeid, E.; Madney, Y.; Kandeel, E.Z. Role of matrix metalloproteinase MMP-2, MMP-9 and tissue inhibitor of metalloproteinase (TIMP-1) in the clinical progression of pediatric acute lymphoblastic leukemia. Hematology 2021, 26, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Arriaga, P.; Pascual, T.; Garcia-Alvarez, A.; Fernandez-Somoano, A.; Lopez-Cima, M.F.; Tardon, A. Genetic polymorphisms in MMP 2, 9 and 3 genes modify lung cancer risk and survival. BMC Cancer 2012, 12, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gholizadeh, F.; Ghahremani, M.H.; Aliebrahimi, S.; Shadboorestan, A.; Ostad, S.N. Assessment of Cannabinoids Agonist and Antagonist in Invasion Potential of K562 Cancer Cells. Iran. Biomed. J. 2019, 23, 153–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
| Differentially Expressed Genes | LogFC JWH-133 vs. NT | FDR JWH-133 vs. NT |
|---|---|---|
| CD9 | −2.436 | 0 |
| SEC61G | −1.636 | 0.001 |
| TBX21 | −1.193 | 0.025 |
| TMSB4X | −1.134 | 0.031 |
| Total Number of Cells × 106 | Total Number of Viable Cells × 106 | % Cell Viability | |
|---|---|---|---|
| SUP-B15 NT | 1.28 | 1.12 | 88.10 |
| SUP-B15 JWH-133 | 1.27 | 1.07 | 84.30 |
| SUP-B15 AM630 | 1.27 | 1.10 | 86.60 |
| % Live | % Early Apoptosis | % Late Apoptosis | % Dead | % TOT | |
|---|---|---|---|---|---|
| SUP-B15 NT | 86.80 | 5.70 | 0 | 7.50 | 5.70 |
| SUP-B15 JWH-133 | 85.85 | 7.60 | 0 | 6.55 | 7.60 |
| SUP-B15 AM630 | 87.50 | 4.90 | 0 | 7.58 | 4.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Punzo, F.; Argenziano, M.; Tortora, C.; Di Paola, A.; Mutarelli, M.; Pota, E.; Di Martino, M.; Di Pinto, D.; Marrapodi, M.M.; Roberti, D.; et al. Effect of CB2 Stimulation on Gene Expression in Pediatric B-Acute Lymphoblastic Leukemia: New Possible Targets. Int. J. Mol. Sci. 2022, 23, 8651. https://doi.org/10.3390/ijms23158651
Punzo F, Argenziano M, Tortora C, Di Paola A, Mutarelli M, Pota E, Di Martino M, Di Pinto D, Marrapodi MM, Roberti D, et al. Effect of CB2 Stimulation on Gene Expression in Pediatric B-Acute Lymphoblastic Leukemia: New Possible Targets. International Journal of Molecular Sciences. 2022; 23(15):8651. https://doi.org/10.3390/ijms23158651
Chicago/Turabian StylePunzo, Francesca, Maura Argenziano, Chiara Tortora, Alessandra Di Paola, Margherita Mutarelli, Elvira Pota, Martina Di Martino, Daniela Di Pinto, Maria Maddalena Marrapodi, Domenico Roberti, and et al. 2022. "Effect of CB2 Stimulation on Gene Expression in Pediatric B-Acute Lymphoblastic Leukemia: New Possible Targets" International Journal of Molecular Sciences 23, no. 15: 8651. https://doi.org/10.3390/ijms23158651
APA StylePunzo, F., Argenziano, M., Tortora, C., Di Paola, A., Mutarelli, M., Pota, E., Di Martino, M., Di Pinto, D., Marrapodi, M. M., Roberti, D., & Rossi, F. (2022). Effect of CB2 Stimulation on Gene Expression in Pediatric B-Acute Lymphoblastic Leukemia: New Possible Targets. International Journal of Molecular Sciences, 23(15), 8651. https://doi.org/10.3390/ijms23158651






