Deubiquitinating Enzyme USP7 Is Required for Self-Renewal and Multipotency of Human Bone Marrow-Derived Mesenchymal Stromal Cells
Abstract
:1. Introduction
2. Results
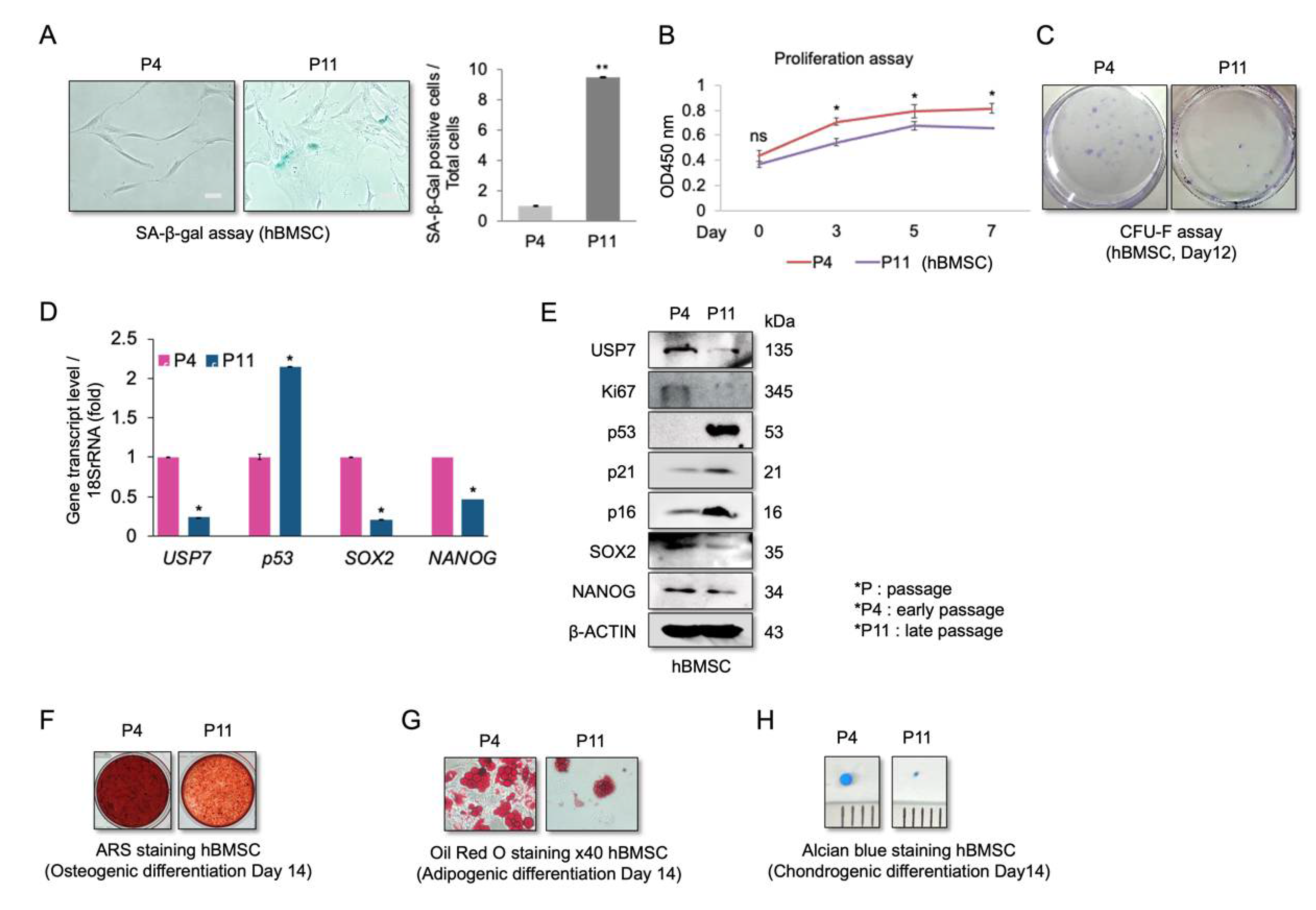
2.1. Reduced USP7 Levels Are Associated with Senescence, Decreased Self-Renewal, and Compromised Differentiation Potential of hBMSCs
2.2. USP7-Deficient hBMSCs Have a Compromised Self-Renewal Capacity and Multipotency
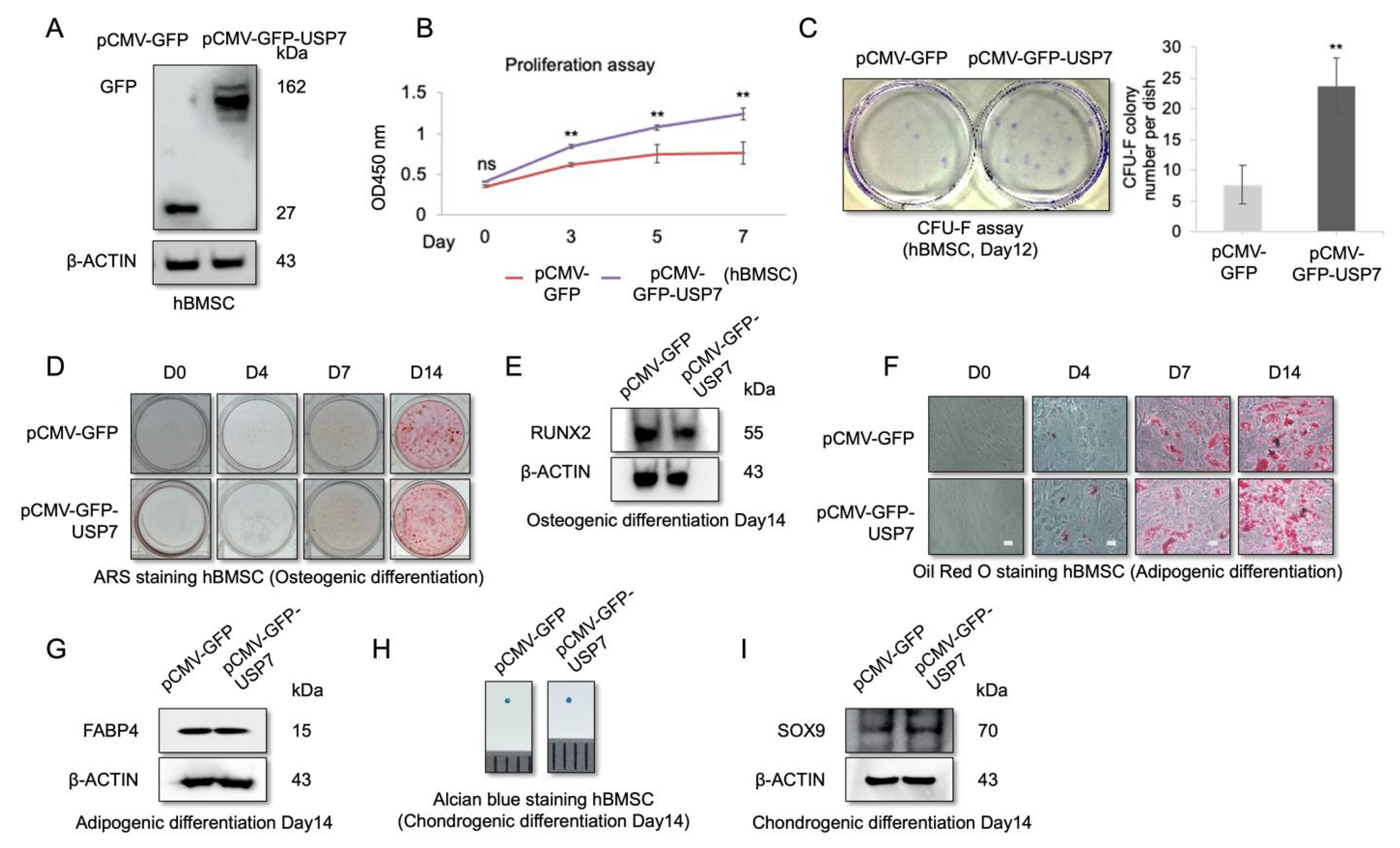
2.3. USP7 Overexpression Enhances Proliferation and Self-Renewal Capability of hBMSCs, but Did Not Affect Multipotency
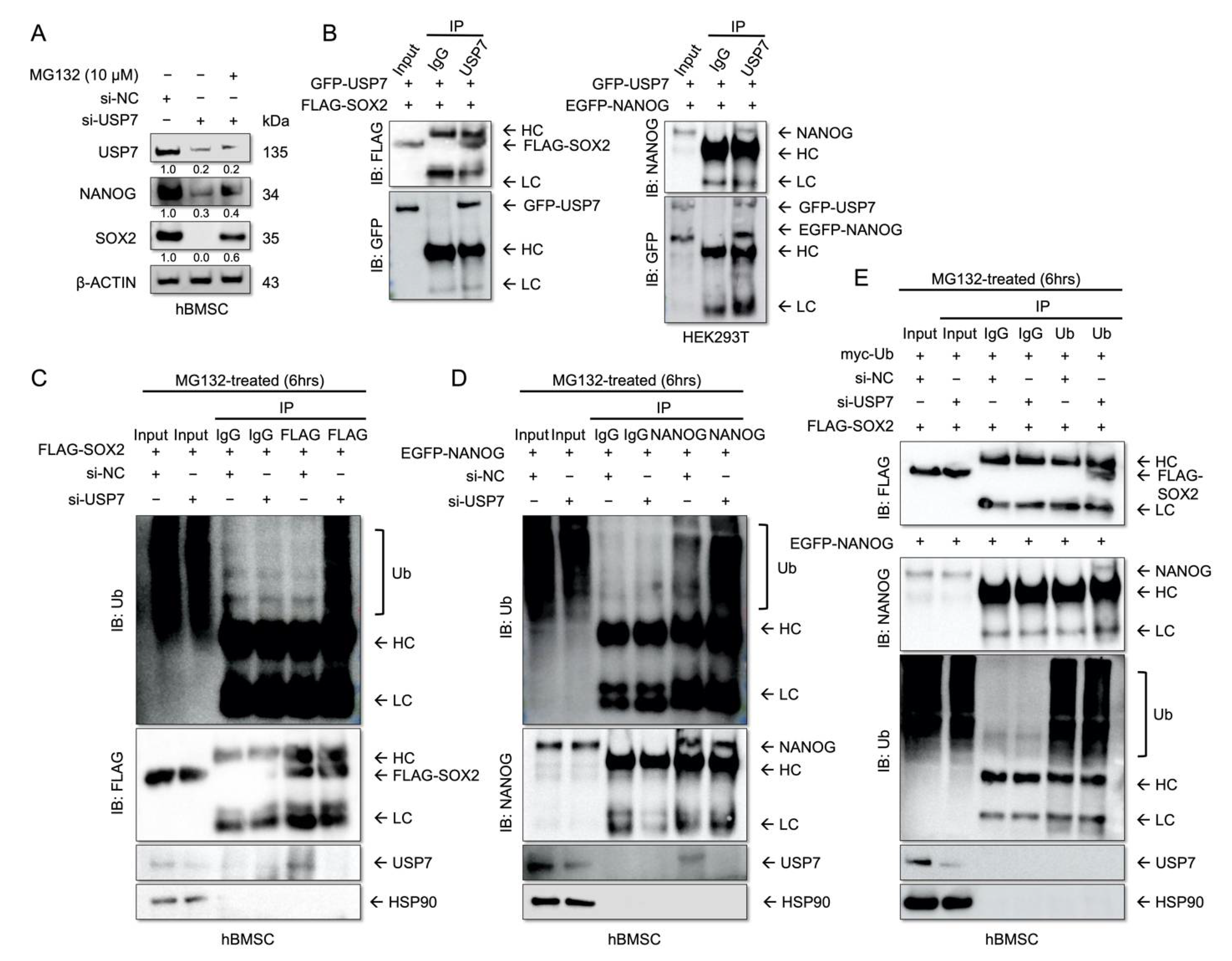
2.4. USP7 Interacts with and Stabilizes SOX2 and NANOG
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Senescence-Associated-β-Gal Assay (SA-β-Gal Assay)
4.3. Cell Proliferation Assay
4.4. Colony-Forming Unit Fibroblast (CFU-F) Assay
4.5. Osteogenic Differentiation
4.6. Adipogenic Differentiation
4.7. Chondrogenic Differentiation
4.8. Western Blotting
4.9. Quantitative Real-Time-Polymerase Chain Reaction (qRT-PCR)
4.10. Plasmids and siRNAs
4.11. Immunoprecipitation Analysis
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Ding, Y.; Liu, Z.; Liang, X. Senescence in mesenchymal stem cells: Functional alterations, molecular mechanisms, and rejuvenation strategies. Front. Cell Dev. Biol. 2020, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Deb, A. How stem cells turn into bone and fat. N. Engl. J. Med. 2019, 380, 2268–2270. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, E.; Mazziotta, C.; Iaquinta, M.R.; Lanzillotti, C.; Fortini, F.; D’Agostino, A.; Trevisiol, L.; Nocini, R.; Barbanti-Brodano, G.; Mescola, A.; et al. Enhanced osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by a hybrid hydroxylapatite/collagen scaffold. Front. Cell Dev. Biol. 2020, 8, 610570. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, I.; Larson, B.L.; Vuoristo, J.T.; Cui, J.G.; Prockop, D.J. Adipogenic differentiation of human adult stem cells from bone marrow stroma (mscs). J. Bone Miner. Res. 2004, 19, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Mahboudi, H.; Kazemi, B.; Soleimani, M.; Hanaee-Ahvaz, H.; Ghanbarian, H.; Bandehpour, M.; Enderami, S.E.; Kehtari, M.; Barati, G. Enhanced chondrogenesis of human bone marrow mesenchymal stem cell (bmsc) on nanofiber-based polyethersulfone (pes) scaffold. Gene 2018, 643, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.; Zou, S.; Wang, J.; Han, Y.; Zhang, L.; Lv, C.; Jiang, B.; Ren, Q.; Chen, L.; Yang, L.; et al. The rna-binding protein musashi2 governs osteoblast-adipocyte lineage commitment by suppressing pparγ signaling. Bone Res. 2022, 10, 31. [Google Scholar] [CrossRef]
- Ross, C.L.; Siriwardane, M.; Almeida-Porada, G.; Porada, C.D.; Brink, P.; Christ, G.J.; Harrison, B.S. The effect of low-frequency electromagnetic field on human bone marrow stem/progenitor cell differentiation. Stem Cell Res. 2015, 15, 96–108. [Google Scholar] [CrossRef] [Green Version]
- Ramos, T.L.; Sánchez-Abarca, L.I.; Muntión, S.; Preciado, S.; Puig, N.; López-Ruano, G.; Hernández-Hernández, Á.; Redondo, A.; Ortega, R.; Rodríguez, C.; et al. Msc surface markers (cd44, cd73, and cd90) can identify human msc-derived extracellular vesicles by conventional flow cytometry. Cell Commun. Signal. 2016, 14, 2. [Google Scholar] [CrossRef] [Green Version]
- Mushahary, D.; Spittler, A.; Kasper, C.; Weber, V.; Charwat, V. Isolation, cultivation, and characterization of human mesenchymal stem cells. Cytometry A 2018, 93, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Pittenger, M.F.; Discher, D.E.; Péault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Alves, H.; Mentink, A.; Le, B.; van Blitterswijk, C.A.; de Boer, J. Effect of antioxidant supplementation on the total yield, oxidative stress levels, and multipotency of bone marrow-derived human mesenchymal stromal cells. Tissue Eng. Part A 2013, 19, 928–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denu, R.A.; Hematti, P. Effects of oxidative stress on mesenchymal stem cell biology. Oxid. Med. Cell Longev. 2016, 2016, 2989076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saez, I.; Koyuncu, S.; Gutierrez-Garcia, R.; Dieterich, C.; Vilchez, D. Insights into the ubiquitin-proteasome system of human embryonic stem cells. Sci. Rep. 2018, 8, 4092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006, 17, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhou, Z.; Shah, A.A.; Zou, H.; Tao, J.; Chen, Q.; Wan, Y. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 2016, 6, 62. [Google Scholar] [CrossRef] [Green Version]
- Baek, D.; Park, K.H.; Lee, K.M.; Jung, S.; Joung, S.; Kim, J.; Lee, J.W. Ubiquitin-specific protease 53 promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Cell Death Dis. 2021, 12, 238. [Google Scholar] [CrossRef]
- Suresh, B.; Lee, J.; Kim, H.; Ramakrishna, S. Regulation of pluripotency and differentiation by deubiquitinating enzymes. Cell Death Differ. 2016, 23, 1257–1264. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, A.P.; Suresh, B.; Kim, H.H.; Kim, K.S.; Ramakrishna, S. Concise review: Fate determination of stem cells by deubiquitinating enzymes. Stem Cells 2017, 35, 9–16. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Chakraborty, D.; Basu, M.; Ghosh, M.K. Emerging insights into hausp (usp7) in physiology, cancer and other diseases. Signal. Transduct. Target. Ther. 2018, 3, 17. [Google Scholar] [CrossRef]
- Pozhidaeva, A.; Bezsonova, I. Usp7: Structure, substrate specificity, and inhibition. DNA Repair 2019, 76, 30–39. [Google Scholar] [CrossRef]
- Gao, Y.; Koppen, A.; Rakhshandehroo, M.; Tasdelen, I.; van de Graaf, S.F.; van Loosdregt, J.; van Beekum, O.; Hamers, N.; van Leenen, D.; Berkers, C.R.; et al. Early adipogenesis is regulated through usp7-mediated deubiquitination of the histone acetyltransferase tip60. Nat. Commun. 2013, 4, 2656. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Lv, L.; Li, W.; Zhang, X.; Jiang, Y.; Ge, W.; Zhou, Y. Protein deubiquitinase usp7 is required for osteogenic differentiation of human adipose-derived stem cells. Stem Cell Res. Ther. 2017, 8, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X.; Xu, X.; Yang, C.; Luo, Y.; Wu, Y.; Wang, J. Usp7 regulates the proliferation and differentiation of atdc5 cells through the sox9-pthrp-pth1r axis. Bone 2021, 143, 115714. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhai, K.; Wu, Q.; Fang, X.; Huang, Q.; Tao, W.; Lathia, J.D.; Yu, J.S.; Rich, J.N.; Bao, S. Hausp stabilizes sox2 through deubiquitination to maintain self-renewal and tumorigenic potential of glioma stem cells. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wei, X.; Guo, J.; Li, Q.; Jia, Q.; Jing, Q.; Li, Y.; Zhou, B.; Chen, J.; Gao, S.; Zhang, X.; et al. Bach1 regulates self-renewal and impedes mesendodermal differentiation of human embryonic stem cells. Sci. Adv. 2019, 5, eaau7887. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Zhao, H.; Yu, C.; Kang, Y.; Yang, X. Targeting ubiquitin-specific protease 7 (usp7) in cancer: A new insight to overcome drug resistance. Front. Pharmacol. 2021, 12, 648491. [Google Scholar] [CrossRef]
- Dai, X.; Lu, L.; Deng, S.; Meng, J.; Wan, C.; Huang, J.; Sun, Y.; Hu, Y.; Wu, B.; Wu, G.; et al. Usp7 targeting modulates anti-tumor immune response by reprogramming tumor-associated macrophages in lung cancer. Theranostics 2020, 10, 9332–9347. [Google Scholar] [CrossRef]
- Wang, Z.; Kang, W.; You, Y.; Pang, J.; Ren, H.; Suo, Z.; Liu, H.; Zheng, Y. Usp7: Novel drug target in cancer therapy. Front. Pharmacol. 2019, 10, 427. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, J.; Chen, C.; Yuan, H.; Wen, X.; Sun, H. Usp7: Target validation and drug discovery for cancer therapy. Med. Chem. 2018, 14, 3–18. [Google Scholar] [CrossRef]
- Turnbull, A.P.; Ioannidis, S.; Krajewski, W.W.; Pinto-Fernandez, A.; Heride, C.; Martin, A.C.L.; Tonkin, L.M.; Townsend, E.C.; Buker, S.M.; Lancia, D.R.; et al. Molecular basis of usp7 inhibition by selective small-molecule inhibitors. Nature 2017, 550, 481–486. [Google Scholar] [CrossRef]
- Yoon, D.S.; Kim, Y.H.; Jung, H.S.; Paik, S.; Lee, J.W. Importance of sox2 in maintenance of cell proliferation and multipotency of mesenchymal stem cells in low-density culture. Cell Prolif. 2011, 44, 428–440. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, T.; Woiwode, I.; Guerreiro, R.F.; Vogt, R.; Lammers, M.; Hofmann, K. An evolutionary approach to systematic discovery of novel deubiquitinases, applied to legionella. Life Sci. Alliance 2020, 3. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [Green Version]
- Matic, I.; Antunovic, M.; Brkic, S.; Josipovic, P.; Mihalic, K.C.; Karlak, I.; Ivkovic, A.; Marijanovic, I. Expression of oct-4 and sox-2 in bone marrow-derived human mesenchymal stem cells during osteogenic differentiation. Open Access Maced. J. Med. Sci. 2016, 4, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funato, N.; Ohtani, K.; Ohyama, K.; Kuroda, T.; Nakamura, M. Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors. Mol. Cell Biol. 2001, 21, 7416–7428. [Google Scholar] [CrossRef] [Green Version]
- Squillaro, T.; Peluso, G.; Galderisi, U. Clinical trials with mesenchymal stem cells: An update. Cell Transplant. 2016, 25, 829–848. [Google Scholar] [CrossRef] [Green Version]
- Galderisi, U.; Calarco, A.; Melone, M.A.B.; Peluso, G. Is it possible to improve the success rate of cellular therapy based on mesenchymal stem cells? J. Stem Cells Res. Rev. Rep. 2014, 1, 1017. [Google Scholar]
- Lee, S.; Yoon, D.S.; Paik, S.; Lee, K.M.; Jang, Y.; Lee, J.W. Microrna-495 inhibits chondrogenic differentiation in human mesenchymal stem cells by targeting sox9. Stem Cells Dev. 2014, 23, 1798–1808. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.H.; Yoon, D.S.; Kim, H.O.; Lee, J.W. Characterization of different subpopulations from bone marrow-derived mesenchymal stromal cells by alkaline phosphatase expression. Stem Cells Dev. 2012, 21, 2958–2968. [Google Scholar] [CrossRef] [Green Version]
- Alessio, N.; Aprile, D.; Cappabianca, S.; Peluso, G.; Di Bernardo, G.; Galderisi, U. Different stages of quiescence, senescence, and cell stress identified by molecular algorithm based on the expression of ki67, rps6, and beta-galactosidase activity. Int. J. Mol. Sci. 2021, 22, 3102. [Google Scholar] [CrossRef]
- Zia, S.; Cavallo, C.; Vigliotta, I.; Parisi, V.; Grigolo, B.; Buda, R.; Marrazzo, P.; Alviano, F.; Bonsi, L.; Zattoni, A.; et al. Effective label-free sorting of multipotent mesenchymal stem cells from clinical bone marrow samples. Bioengineering 2022, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.S.; Lee, K.M.; Choi, Y.; Ko, E.A.; Lee, N.H.; Cho, S.; Park, K.H.; Lee, J.H.; Kim, H.W.; Lee, J.W. Tlr4 downregulation by the rna-binding protein pum1 alleviates cellular aging and osteoarthritis. Cell Death Differ. 2022, 29, 1364–1378. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.J.; Park, K.H.; Lee, K.-M.; Chun, Y.-M.; Lee, J.W. Deubiquitinating Enzyme USP7 Is Required for Self-Renewal and Multipotency of Human Bone Marrow-Derived Mesenchymal Stromal Cells. Int. J. Mol. Sci. 2022, 23, 8674. https://doi.org/10.3390/ijms23158674
Kim YJ, Park KH, Lee K-M, Chun Y-M, Lee JW. Deubiquitinating Enzyme USP7 Is Required for Self-Renewal and Multipotency of Human Bone Marrow-Derived Mesenchymal Stromal Cells. International Journal of Molecular Sciences. 2022; 23(15):8674. https://doi.org/10.3390/ijms23158674
Chicago/Turabian StyleKim, You Ji, Kwang Hwan Park, Kyoung-Mi Lee, Yong-Min Chun, and Jin Woo Lee. 2022. "Deubiquitinating Enzyme USP7 Is Required for Self-Renewal and Multipotency of Human Bone Marrow-Derived Mesenchymal Stromal Cells" International Journal of Molecular Sciences 23, no. 15: 8674. https://doi.org/10.3390/ijms23158674
APA StyleKim, Y. J., Park, K. H., Lee, K. -M., Chun, Y. -M., & Lee, J. W. (2022). Deubiquitinating Enzyme USP7 Is Required for Self-Renewal and Multipotency of Human Bone Marrow-Derived Mesenchymal Stromal Cells. International Journal of Molecular Sciences, 23(15), 8674. https://doi.org/10.3390/ijms23158674





